Abstract
The Drosophila Gene Disruption Project (GDP) has created a public collection of mutant strains containing single transposon insertions associated with different genes. These strains often disrupt gene function directly, allow production of new alleles, and have many other applications for analyzing gene function. Here we describe the addition of ∼7600 new strains, which were selected from >140,000 additional P or piggyBac element integrations and 12,500 newly generated insertions of the Minos transposon. These additions nearly double the size of the collection and increase the number of tagged genes to at least 9440, approximately two-thirds of all annotated protein-coding genes. We also compare the site specificity of the three major transposons used in the project. All three elements insert only rarely within many Polycomb-regulated regions, a property that may contribute to the origin of “transposon-free regions” (TFRs) in metazoan genomes. Within other genomic regions, Minos transposes essentially at random, whereas P or piggyBac elements display distinctive hotspots and coldspots. P elements, as previously shown, have a strong preference for promoters. In contrast, piggyBac site selectivity suggests that it has evolved to reduce deleterious and increase adaptive changes in host gene expression. The propensity of Minos to integrate broadly makes possible a hybrid finishing strategy for the project that will bring >95% of Drosophila genes under experimental control within their native genomic contexts.
DROSOPHILA has served as an important model organism for >100 years, in large part, because of the wealth of mutants available and the ease with which they can be manipulated experimentally. Mutagenesis using single insertions of an engineered transposon offers many advantages for analyzing gene regulation and function (Cooley et al. 1988; Bellen et al. 1989; Bier et al. 1989). The insertions frequently interfere directly with gene function and can also be remobilized to generate additional useful mutations in the genomic region where they reside through the processes of local jumping or imprecise excision. By incorporating useful internal sequences, transposons can be used to report or manipulate gene expression, sense chromatin structure, or function as sites for site-specific recombination.
The Drosophila Gene Disruption Project (GDP) was established in 1991 to bring the advantages of this method to the research community by generating transposon mutations in most Drosophila genes. During phase 1 of the project, we characterized insertions causing recessive phenotypes (Spradling et al. 1999). The availability of an annotated genome sequence (Adams et al. 2000; Misra et al. 2002) enabled phase 2, where insertions were associated with predicted genes solely on the basis of their genomic location. By 2004, ∼40% of known Drosophila genes had one or more associated GDP insertion alleles (Bellen et al. 2004). Several large collections of insertion lines were independently generated as well, further increasing the potential gene coverage (Thibault et al. 2004; Kim et al. 2010).
Several different approaches may help further increase the number of disrupted genes. Transposable elements differ in their target site specificity (Bellen et al. 2004; Thibault et al. 2004); hence, generating insertions using new transposons might provide greater efficiency than continued mutagenesis using P and piggyBac elements. The Minos transposon, a mariner family member, is a particularly attractive candidate (Metaxakis et al. 2005). The site-specific recombinase from phage ϕC31 provides the ability to efficiently integrate even large DNAs into genomic attP target sites (Groth et al. 2004; Bateman et al. 2006; Venken et al. 2006; 2009). Including a ϕC31 attP site in the elements used for mutagenesis would offer many advantages for genomic manipulation, including increased mutagenicity (Groth et al. 2004; Bateman et al. 2006; Venken et al. 2006). Previous studies of the capabilities of integrated attP-containing transposons illustrate their exceptional utility (Venken and Bellen 2007; Venken et al. 2009).
Transposon site specificity represents a critically important factor in determining the optimum strategy for completing the GDP project. The size and quality of the data collected by the GDP provide a special opportunity to characterize the insertional preference of specific transposons in detail. It is well established that some transposons hit certain sites, “hotspots,” much more frequently than expected by chance, while other regions, “coldspots,” are avoided. P elements frequently insert near promoters, an advantage for mutagenesis and misexpression screening, but also preferentially target hotspots (Spradling et al. 1995; Liao et al. 2000; Bellen et al. 2004). In addition, a significant fraction of Drosophila genes, including many clustered and tissue-specific genes, appear almost refractory to disruption by P insertion (Bellen et al. 2004). piggyBac elements also target hotspots, but show less regional and promoter bias (Bellen et al. 2004; Thibault et al. 2004). However, piggyBac elements do not excise imprecisely to create local deletions, a significant disadvantage compared to P elements.
Here we summarize the status of the GDP collection at the completion of phase 2. We have added P-element, piggyBac, and Minos insertions to the publicly available GDP collection to provide genetic access to at least 9440 genes. In addition to expanding this resource of mutants for researchers, our studies also provide new insights into transposon site selectivity and document an influence of chromatin structure. We show that, because of very low site specificity, it should be feasible to tile the Drosophila genome with Minos insertions that would facilitate the site-directed mutagenesis of almost all Drosophila genes and functional elements by homologous recombination.
Materials and Methods
The EY collection
The construction of the P-element–based EY transposon (P{EPgy2}, Table 1), the generation of 10,310 insertion lines, and the mapping of their insertion sites have been described (Bellen et al. 2004). Using the same methods, we generated an additional 11,830 EY lines (strain names EY10505–EY16964 and EY18301–EY23670) and mapped 9585 insertions to unique sites on the reference Drosophila genomic sequence (release 5, http://www.fruitfly.org). This brought the total number of unselected EY transpositions generated and uniquely localized in the genome to 18,214. The new EY insertions that we selected for the GDP collection were balanced and their insertion sites were verified by resequencing of flanking DNA as described (Bellen et al. 2004).
Table 1.
Mutator transposons
| Line name | Marker | Transposon | Reference | Map |
| EY | white, yellow | P{EPgy2} | Bellen et al. 2004 | 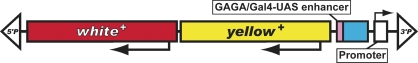 |
| HP | white | P{EPg} | Staudt et al. 2005 | 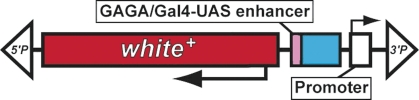 |
| DP, GG | yellow | P{Mae-UAS.6.11} | Beinert et al. 2004; Staudt et al. 2005 | 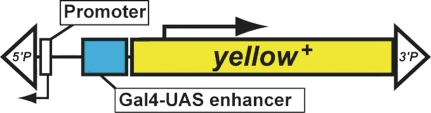 |
| d | white | P{XP} | Thibault et al. 2004 |  |
| c | white | PBac{PB} | Thibault et al. 2004 |  |
| e | white | PBac{RB} | Thibault et al. 2004 |  |
| f | white | PBac{WH} | Thibault et al. 2004 |  |
| G | white | P{EP} | Røørth 1996; Kim et al. 2010; GenExel Library at KAIST (http://genexel.kaist.ac.kr/mapview3/index.html) | 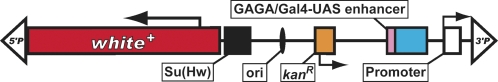 |
| G0, SH | white | P{lacW} | Peter et al. 2002; Oh et al. 2003 | 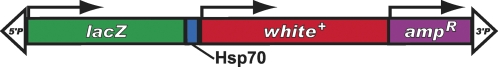 |
| MB | EGFP | Mi{ET1} | Metaxakis et al. 2005 | 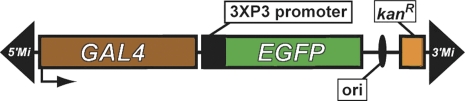 |
The schematic diagrams are not drawn to scale and are meant only to indicate the components present in each transposon. Thin lines separating some components have been added to prevent labels from overlapping and are not intended to indicate spacers between components. Please refer to the original publications and curated FlyBase reports for details.
The Exelixis collection
The generation and properties of 26,540 P- and piggyBac insertion lines that were mapped by Exelixis to unique sites in the Drosophila reference genomic sequence (release 2) have been described (Thibault et al. 2004). These lines probably do not represent a completely random collection of insertions, because some lines disrupting major hotspots appear to have been culled by Exelixis. However, we found many cases where at least two lines bearing identical piggyBac insertion sites had been retained, suggesting that such culling was limited or incomplete. Most of these stocks, as well as insertion site data, were generously made available to the GDP so that the most useful lines could be distributed publicly. Approximately 400 base pairs (bp) of the genomic reference sequence surrounding the insertion site(s) in these lines along with a coordinate or range of coordinates denoting the insertion site were reported (Thibault et al. 2004). We selected ∼2100 lines from the Exelixis collection for distribution by the Bloomington Drosophila Stock Center (BDSC), on the basis of the insertion site coordinates reported by Exelixis. Exelixis subsequently provided us with 52,183 flanking sequence reads derived from 22,144 strains with associated phred quality scores (Ewing and Green 1998). In April 2005, 24,678 of these flanking sequence reads were submitted to GenBank by the GDP. We subsequently realigned the flanking sequences to the Drosophila reference genomic sequence (release 5) on the basis of more stringent criteria using our standard pipeline and mapped 16,073 insertions to unique sites. While there was usually close agreement, insertion site coordinates deduced by the GDP and Exelixis sometimes varied by several hundred base pairs, and 535 strains lacked any sequence reads. Some strains had multiple sequence reads from one or both flanks and these sometimes mapped to different sites. After changes due to the reanalyzed sequence flanks, updated annotation, strain losses, and line substitutions, 1859 Exelixis lines are currently part of the GDP collection at the BDSC, while 357 Exelixis GDP lines are maintained at Harvard Medical School (https://drosophila.med.harvard.edu/) (Table 2).
Table 2.
Summary of GDP lines
| Collection | BDSC Lines | In genes | Intergenic | New genes |
| Spradling et al. 1999 | 934 | 898 | 36 | 936 |
| Bellen et al. 2004 | 6062 | 5118 | 944 | 3910 |
| New EYs | 1193 | 1059 | 134 | 641 |
| Exelixis | 1859 | 1800 | 59 | 1983a |
| Staudt et al. 2005b | 284 | 276 | 8 | 109 |
| MB | 2658 | 2147 | 511 | 1155 |
| GenExel | 1136 | 1120 | 16 | 616 |
| Other | 530 | 514 | 16 | 90 |
| Total | 14,656 | 12,932 | 1724 | 9440 |
The numbers of strains from the indicated sources selected for the GDP collection and currently available at the BDSC are shown. The numbers of strains containing insertions in genes (see Methods) or within intergenic regions are also given. The New Genes column gives the number of genes hit by insertions in that collection that are not hit by insertions from the collections above it in the table. The values reflect the current status of the GDP collection; the values for the Spradling et al. 1999 and Bellen et al. 2004 collections are lower than those originally reported, due to loss or replacement with strains hitting the same gene from later collections (see Methods).
Includes 357 genes hit by lines that were sent to the Harvard Stock Center, rather than BDSC.
The Staudt et al. 2005 collection is also referred to as the Max Plank/EMBL/DeveloGen collection in Methods.
The MB collection
To generate new insertions of a Minos element, we used the Mi{ET1} element described in Metaxakis et al. (2005) (Table 1). It contains the Minos 255-bp inverted repeats and a minimal hsp70 promoter upstream of the GAL4 gene and may function as an enhancer detector/trap (hence “ET”) if inserted in the appropriate location. The GFP gene, driven in the eye and brain of adults and larvae by the 3xP3 promoter (Horn et al. 2000), is the marker used for selection. The stocks were generated and balanced in the w1118 isogenic background described in Ryder et al. (2004). The Minos Mi{ET1} mutator [FlyBase identification (ID) FBtp0021506; referred to as MiET1 by Metaxakis et al. 2005], which we refer to as the MB element, was inserted on a TM3, Sb Ser balancer chromosome. The starting site of the mutator was mapped by flanking sequence (GenBank accession ET202027) to a site corresponding to coordinate 3L:12580323 of the Drosophila melanogaster reference genomic sequence. The MB mutator was mobilized using a transgenic source of transposase under the control of a heat-shock promoter (P{hsILMiT}, FlyBase ID FBtp0021508, referred to as PhsILMiT by Metaxakis et al. (2005) inserted on a second chromosome balancer (P{hsILMiT}2.4; FlyBase ID FBti0073645).
We generated 12,426 strains containing new insertions of the MB transposon (nearly always single insertions) and mapped 10,781 insertions from 10,630 strains to a unique site in the genome. Lines that were selected for the GDP collection were balanced and their insertion sites verified by resequencing before delivery to the BDSC. Sequences flanking MB insertions were determined by inverse PCR and DNA sequencing, as described in Bellen et al. 2004 with the following modifications. Genomic DNA was digested with HpaII; 5′ flanks were amplified with the primers MI.5.F (CAAAAGCAACTAATGTAACGG) and MI.5.R (TTGCTCTTCTTGAGATTAAGGTA) at an annealing temperature of 50°; 3′ flanks were amplified with MI.3.F (ATGATAGTAAATCACATTACG) and MI.3.R (CAATAATTTAATTAATTTCCC) at an annealing temperature of 50°; and 5′ and 3′ flanks were sequenced with MI.seq (TTTCGTCGTGAAGAGAAT). A detailed protocol is available on the GDP Website (http://flypush.imgen.bcm.tmc.edu/pscreen/). Insertion-bearing chromosomes were balanced using P{RS3}l(1)CB-6411-31, w1118/FM7h (X chromosome), w1118/Dp(1;Y)y+; nocSco/SM6a (2nd chromosome), and w1118/Dp(1;Y)y+; TM2/TM6C, Sb1 (3rd chromosome), which are all in the “iso31” isogenic background (Ryder et al. 2004) and were obtained from the BDSC. A Meme analysis failed to uncover any significant target sequence preference beyond the requirement for “TA.”
The GenExel (Aprogen) collection
GenExel, now Aprogen, generated a very large collection of lines bearing insertions of the P-element construct P{EP} (Røørth 1996; FlyBase ID FBtp0001317) at the Korean Advanced Institute of Science and Technology (KAIST); (Table 1, “G”; see http://www.oxfordjournals.org/nar/database/summary/677; Kim et al. 2010). Initially, ∼27,000 lines were selected from a starting set of ∼100,000 transpositions by requiring a minimum spacing of 200 bp between insertions to prune out lines with insertions in transposon hotspots. Most insertions were not balanced. Sequence coordinates for 24,789 insertions were provided to the GDP. GenExel subsequently sent us 1685 strains that we had identified as candidates. After balancing the insertions and sequencing their flanks, 1136 lines were added to the GDP collection at the BDSC.
The Max Planck/EMBL/DeveloGen collection
We received lines from a collection of P-element insertions generated by researchers at Max Planck Güöttingen, the EMBL labs at Heidelberg and DeveloGen (Staudt et al. 2005). The lines comprising this collection are indicated by the prefixes HP or DP (Table 1). Insertion site information was provided, and lines hitting novel genes were identified for transfer directly from Max Plank to the BDSC.
Other collections
The Göttingen collections of insertions on the X chromosome (Peter et al. 2002; Beinert et al. 2004) were screened. The elements comprising this collection are designated by the prefix G0 or GG (Table 1). Candidates from the P{lacW} insertion collection on FRT-bearing chromosomes described by Oh et al. (2003) were resequenced and screened. The elements comprising this collection are designated by the prefix SH (Table 1).
Strain selection
Strains were selected for inclusion essentially as described previously (Bellen et al. 2004). The GDP employs a strategy of continuous library improvement, both by adding new lines and by replacing/upgrading existing lines with better ones. Briefly, each new candidate insertion from the screens described is compared with the Drosophila genome annotation, as well as with the insertion sites of all existing GDP collection strains within the gene region in question. On the basis of the best judgment of an expert annotator, lines can be retained for several reasons. Of highest priority are lines likely to disrupt any gene lacking a current GDP insertion. Because the annotated 5′ end of many gene models may be truncated relative to the true 5′ end, insertions located within 500 bp of the annotated 5′ end or anywhere within the transcribed region are selected. In addition, a second insertion in a gene is saved if it is located in a distinct promoter, disrupts another transcript isoform, or provides another unique genetic property. The continued presence of unannotated protein-coding and RNA genes, and genetic regulatory elements, especially in annotations prior to modENCODE (Roy et al. 2010), provides the final reason for selecting lines. Since P elements show a strong preference for promoters, P-element insertions located 2 kb or more from the nearest annotated promoter or existing insertion are also retained. Similarly, a small number of piggyBac or MB lines that had insertions within regions >10 kb distant from any existing insertion have also been kept for use in genetically manipulating the surrounding genome. Many insertions thought initially to be within intergenic regions have subsequently been mapped to genes as the annotation improved. Many such lines have been used to functionally characterize novel genes, promoters, piRNA clusters, and small RNA genes (for example, Brennecke et al. 2003, 2007; Godfrey et al. 2006).
The GDP recognizes that lines added to the collection on the basis of the above criteria are not equally valuable. Hence, lines whose value is less certain are subject to replacement. For example, lines mapping upstream from annotated transcription units are replaced when lines become available whose insertions are located within the unit. Strains containing two insertions on the same chromosome are retained if one is located within a novel gene. However, such lines are also replaced as soon as a single-copy insertion in the gene becomes available. Other reasons for line replacement are restraints on distribution. Some donated collections cannot be distributed to for-profit corporations. These lines are subject to replacement whenever an equivalent line without such conditions becomes available.
Data handling and access
Genomic sequences flanking the P-element and piggyBac transposon insertions were determined as described in Bellen et al. (2004); sequences flanking MB insertions were determined as described above. The analysis and alignment of all flanking sequences were as described in Bellen et al. (2004). The genome sequence coordinates given here are based on the release 5 reference genome sequence. We consider an insertion to hit a gene if the insertion site is within the annotated transcription unit of the gene or within 500 bp upstream of the 5′ end, on the basis of the FlyBase gene annotation release FB2009_10.
The GDP Website (http://flypush.imgen.bcm.tmc.edu/pscreen/) has a searchable database of strains that are part of the GDP collection at the BDSC, as well as those that have been selected to be added to the collection and are in the process of being balanced and rechecked. Data presented are the transposon construct, line name, genomic insertion site, inferred cytogenetic map location, associated gene, FlyBase annotation reference, and BDSC stock number.
Project data are sent to FlyBase (http://flybase.bio.indiana.edu/) and GenBank (http://www.ncbi.nlm.nih.gov/) before the lines are transferred to the BDSC for public distribution (http://flystocks.bio.indiana.edu/). Insertion data are displayed using the University of California Santa Cruz (UCSC) genome browser (Fujita et al. 2010). Custom tracks for this display are available from the GDP Website. Complete insertion information on EY, MB, and Exelixis piggyBac insertions that were analyzed in this study for site specificity is available on the GDP Website.
Results
New P-element and piggyBac insertion lines
Previous efforts generated a GDP collection consisting of 7140 lines bearing P-element or piggyBac insertions that provided access to 5362 genes (Bellen et al. 2004). One approach to further expanding the collection is simply to screen more lines containing unselected insertions of these elements. To this end, 11,830 new insertions of the EY element, a modified P transposon that can be used to misexpress endogenous genes adjacent to its insertion site (Table 1), were generated. In addition, two large collections of insertion strains were donated to the project. Exelixis provided site coordinates for 6194 P-element and 18,668 piggyBac insertion lines. The structure of the P{XP}, PBac{PB}, PBac{RB}, and PBac{WH} transposons used to construct these lines (Thibault et al. 2004) is shown in Table 1. GenExel (currently, Aprogen) generously made available sequence coordinates from ∼24,789 P{EP} element insertions (Table 1) that they selected from a starting collection of ∼100,000 lines. Several other groups of investigators provided coordinates for smaller but significant collections (see Methods).
The insertion sites in all the new lines, which include the full genetic diversity generated by >140,000 P- and piggyBac element transpositions, were screened against the Drosophila genome annotation to identify lines that would expand the genetic diversity of the GDP collection. Overall, 5002 P or piggyBac lines were added to the collection because their insertions were located in novel genes (3439), in putative regulatory regions, or because they were more likely than a currently existing allele to strongly disrupt gene function (Table 2, see Methods for further details).
Generation of Minos insertion lines
Our results illustrate how random forward mutagenesis becomes increasingly inefficient as saturation is approached. About 50,000 P and piggyBac lines were required to identify insertions associated with the first 5362 genes (Bellen et al. 2004). Subsequently, our screening of nearly three times as many insertions yielded only 0.66 times as many new genes, highlighting the fact that P- and piggyBac insertion sites were becoming saturated. Indeed, <2% of newly generated EY insertions near the end of the screen disrupted genes not previously represented in the collection.
To continue improving the GDP collection and to further investigate the options for finishing the project, a screen was carried out using Minos, a mariner family transposon unrelated to either P or piggyBac. A previous study (Metaxakis et al. 2005) suggested that Minos integrates into the D. melanogaster genome with little site specificity. However, this conclusion was based on a small sample of ∼100 insertions. To exploit the properties of this element and to measure its behavior more accurately, we carried out a large screen to generate new insertions using the Minos-based Mi{ET1} element (Metaxakis et al. 2005; see Table 1). We refer to these as MB lines. Of the 12,426 MB lines with independent transpositions that were generated and sequenced, we recovered flanking sequence that could be unambiguously localized to a unique site in the genome from 10,630 lines (86%).
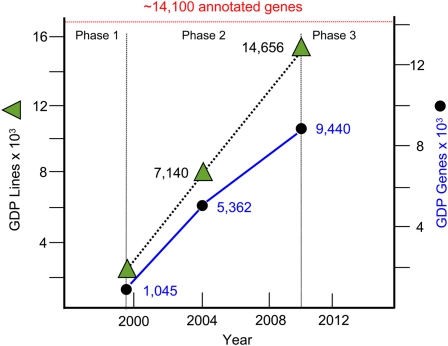
We added 2658 of the MB lines to the GDP collection (Table 2). Although lines were saved for a variety of reasons, 1155 of the MB lines hit genes new to the GDP collection, bringing the total number of disrupted genes to 9440, which is about two-thirds of currently annotated Drosophila protein-coding genes (Tweedie et al. 2009). Thus, since the last report (Bellen et al. 2004), the number of lines in the GDP collection has approximately doubled, and the number of disrupted genes has increased by 77% (Figure 1).
Figure 1 .
Growth of the GDP strain collection. The total number of GDP strains (green triangles) and the number of genes with one or more associated GDP lines (filled circles) are shown as a function of time beginning with the completion of the Drosophila genome sequence in 2000, which signaled the end of project phase 1. In 2010, the project completed phase 2 in which genes were targeted on the basis of the location of insertions from undirected forward screens.
Comparing the insertional specificities of P, piggyBac, and Minos elements
The high efficiency of the MB screen in generating useful new insertions provided further evidence that significant differences exist in the insertional specificities of P, piggyBac, and Minos elements. To further investigate whether to continue with forward Minos mutagenesis, we analyzed the site specificities of MB, EY, and piggyBac elements in detail. We used information from 18,214 EY insertions, 12,244 Exelixis piggyBac insertions that upon reanalysis by GDP were unambiguously mapped to unique sites, and 10,458 MB insertions. Both the EY and the MB screens incorporate data on all transpositions outside the chromosome bearing the starting insertion. In contrast, some redundant or nearly redundant piggyBac insertions may have been culled from the data sent by Exelixis (see Methods). However, removal of lines with similar insertion sites would only serve to increase the apparent randomness of piggyBac insertion. In addition, the methods we used in generating and analyzing these data minimize problems caused by insertions within repetitive sequences or within heterochromatic regions that suppress marker gene expression (see Discussion).
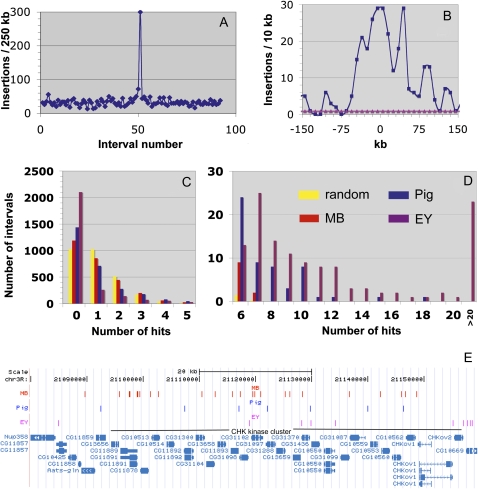
The MB screen showed one anomaly with the potential to skew our analysis. A general scan of the insertion distribution revealed the presence of a single large MB hotspot in chromosome 3L at 12.583 Mb, which corresponds to the site of the starting element located on a balancer chromosome homolog (Figure 2A). Such “homolog hotspots” have been observed previously in some, but not most P-element screens (Tower and Kurapati 1994; Bellen et al. 2004). Approximately 310 of the 10,458 insertions were located within 300 kb of the starting site in a peaked distribution (Figure 2B). A similar distribution of new insertions arising near the original insertion on the starting chromosome has previously been observed when transposons were experimentally remobilized, a phenomenon known as “local transposition.” However, homolog hotspots differ in that they result from hopping to nearby sites on the homolog, rather than the starting chromosome itself. No homolog hotspot was observed in the EY screen. Since this hotspot does not reflect the intrinsic site specificity of Minos elements, these 310 lines were not used in analyzing site specificity. However, these observations do provide evidence that Minos elements can undergo high-frequency local transposition.
Figure 2 .
Saturation behavior of P, piggyBac, and Minos insertions. (A) Plot of MB insertions per 250 kb vs. interval number along chromosome 3L reveals a large hotspot. (B) MB insertions within 10-kb intervals around the hotspot in A. The number per interval expected by chance is shown in pink. 0 corresponds to 3L:12580233, the site on the homolog of the mobilized element in the MB screen. (C and D) Distribution of MB (red), piggyBac (blue), or EY (purple) insertions within 10-kb genomic intervals on chromosome 3R, compared with random transposition (Poisson distribution, yellow). To facilitate comparison, the same numbers of insertions were analyzed in each case (2790; corresponding to 1 insertion per interval). The number of intervals with 0 insertions (C, “0”) is relevant to coldspot behavior; intervals hit more frequently than by random expectation (D) are indicative of piggyBac and P-element hotspots. (E) The Minos hotspot located within a cluster of genes encoding CHK kinases on chromosome 3R. The locations of MB (Minos), Pig (piggyBac), and EY (P) element insertions are shown by vertical bars above the gene map of the region.
The insertional specificities of P, piggyBac, and Minos elements differ
To visualize differences in transposition specificity, we divided the 117 Mb “core” genome (including all euchromatin and some telomeric and pericentric heterochromatin) into regular 10-kb intervals and determined how many times each interval was hit by MB, piggyBac, or EY insertions. To facilitate comparison, the same number of insertions was scored in each case (selected in numerical order by strain name), and this was set equal to the number of intervals (defined as λ = 1). Because the number of insertions on each arm varied, each arm was analyzed separately. The results for chromosome arm 3R, which are typical, are shown (Figure 2, C and D). From inspection of the fraction of intervals with no insertions (Figure 2C) and from the number of intervals with more insertions than expected by chance alone (Figure 2D), it is clear that the three transposons interact distinctively with the genome.
Minos (Figure 2, C and D, red) closely approximates a random distribution. Only 15% more genomic intervals lacked an insert than expected for perfectly random integration, and only a small number of weak candidate hotspots showed up as an excess of intervals with more insertions than expected. An interval could contain more insertions than average due to the presence of a single hotspot, several weaker hotspots, or many dispersed insertions. Candidate Minos hotspots were usually broader than a single gene. No relationship could be found between the genes located in different MB hotspots (supporting information, Table S1). The most striking one was located within a cluster of 25 genes encoding CHK-like kinases (Figure 2E). On either side of this cluster, the density of MB insertions returned to normal.
Both piggyBac (Figure 2C, blue) and P (Figure 2C, purple) elements showed much greater departures from random integration. Nonrandom piggyBac site specificity caused the number of unhit intervals to increase by ∼30%, whereas P insertions left more than twice as many intervals unhit than expected by chance. Both elements also showed a very large excess of hotspots, both in number and in hit frequency (Figure 2D). P-element hotspots have been analyzed previously (Bellen et al. 2004), but it is still unknown why they are targets for preferential insertion. Interestingly, the strongest piggyBac hotspot genes (Table S2) significantly differ from those preferentially targeted by P elements (Bellen et al. 2004). piggyBac target genes frequently encode transcription factors, chromatin factors, and genes involved in growth, nervous system development, and behavior.
Differences in transposition relative to genes
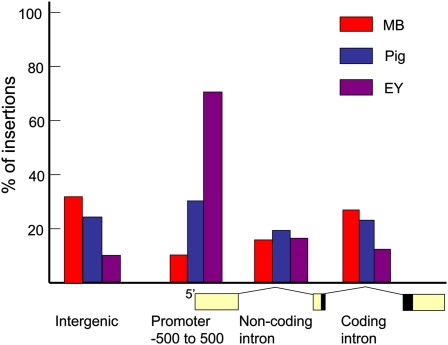
Comparing the location of insertions relative to annotated transcripts revealed additional aspects of how these elements target the genome (Figure 3). Intergenic insertions were defined as those lying outside the transcription unit and its promoter, which was assumed to extend 500 bp 5′ to the annotated transcription start. Minos transposed into such regions 36% of the time, more frequently than either P elements (12%) or piggyBac elements (24%). The low frequency of P-element insertion within intergenic regions may result from the strong proclivity of these elements to insert near promoters. About 73% of P-element insertions (83% of insertions in annotated genes) lie within 500 bp of an annotated 5′ transcription start site. In contrast, only 30% of piggyBac and 9% of MB insertions were in promoters by this definition. Each time the annotation is revised, new promoters are mapped to more of the orphan P-insertion sites.
Figure 3 .
Transposon insertion with respect to transcript structure. The percentage of MB, piggyBac (Pig), and EY insertions located in the indicated regions of annotated transcripts are shown. Numbers may not sum to 100% because an insertion may disrupt multiple transcripts in different positions. A region was scored positive if one or more annotated transcripts with the indicated character were hit by an insertion. To simplify calculation, only the first four annotated transcripts hit by the insertion were considered in determining these values. Because of the large N values, the 95% confidence intervals of these proportions were always less than ±1%. Consequently, the differences were significant except in the case of MB compared to EY insertion in noncoding introns.
One important potential use of transposon insertions is to generate new protein trap alleles (Morin et al. 2001; Buszczak et al. 2007; Quiññones-Coello et al. 2007). Protein fusions to GFP (protein traps) can be produced in vivo by the transposition of an element bearing splice donor and acceptor sites flanking a GFP-encoding exon. To generate a productive fusion, it is necessary that the transposon integrate into a coding intron of the appropriate splice frame and orientation. Despite the fact that 36% of MB elements insert outside of transcription units, MB elements produced the highest frequency of transposition into coding introns among the three elements tested (Figure 3). MB elements were much better than P elements (which were hampered by their promoter bias) but only slightly better than piggyBac.
Genome tiling for local mutagenesis
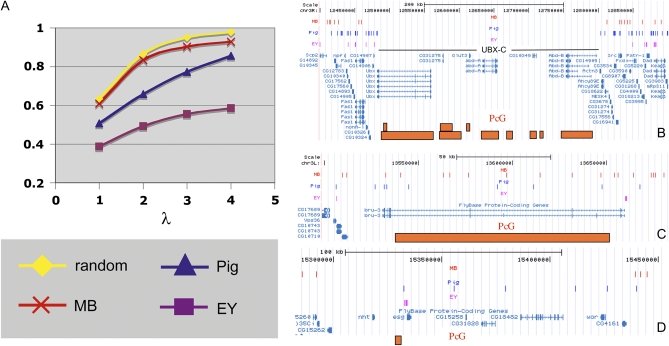
The ability of a transposon to integrate broadly and in effect to tile the genome is critically important for the insertions to be used to manipulate the surrounding region of the genome. To assess breadth of coverage, we plotted the fraction of 40-kb intervals hit at least once within chromosome 3R, as an example, as a function of lambda (λ), the ratio of number of insertions divided by the number of intervals (Figure 4A). At λ = 3, ∼95% of intervals will be hit by random insertion (yellow), and our experiments show that Minos elements (red) hit ∼90%. In contrast, the same number of P-element insertions (purple) hit only 55% of intervals and piggyBac insertions (blue) hit only 77%. How these curves approach saturation will be discussed below, but Figure 4A makes clear that the genome could be quite thoroughly tiled by generating a collection of Minos elements equivalent in size or only slightly larger than the current MB collection (λ = 10,458 × 40 kb/117,000 kb = 3.8).
Figure 4 .
Transposons nonrandomly avoid some genomic intervals, including regions with PcG-dependent repressive marks. (A) The saturation behavior of 40-kb genomic intervals for transposon insertion on chromosome 3R is plotted as λ (the ratio of number of insertions/number of intervals) increases. Poisson (random) expectation (yellow), MB (Minos) elements (red), piggyBac elements (blue), and EY (P) elements (purple). EY elements saturate well below 100%. In contrast, MB elements approach saturation only slightly more slowly than random, whereas piggyBacs appear intermediate. (B) MB, piggyBac, and EY elements insert with greatly reduced frequency in the Bithorax gene cluster. Regions of the Drosophila genome as displayed on the UCSC browser are shown. Insertion sites for these elements are shown in labeled tracks above the map as vertical lines of unit thickness (MB in red; piggyBac in blue; EY in purple; thicker lines denote multiple insertions). The orange boxes display the approximate position of PcG-target regions as mapped by Schwartz et al. (2010). (C) Similar display of the bru-3 gene region shows that not all Polycomb-regulated chromatin domains are transposon poor. (D) The esg gene cluster and its surrounding region illustrates that some PcG targets are largely refractory to MB insertion, but not to the other two elements.
Polycomb-regulated regions correspond to transposon coldspots
To determine whether the MB curve in Figure 4A will eventually reach 100% or whether there are intervals that cannot be hit by Minos insertions, we investigated all 30 examples where two or more adjacent 40-kb zones lacked any insertions at λ = 4 (excluding 10 basal chromosome regions whose high repetitive DNA content probably impeded mapping). The 30 double-negative regions were strikingly nonrandom and suggested a biological mechanism limiting Minos insertion (Table S3). The two largest transposon-free zones occurred on chromosome 3R and corresponded precisely with the BX-C (Figure 4B) and ANT-C homeotic gene complexes. These complexes are known to be regulated at the level of chromatin structure by Polycomb and Trithorax group genes (Ringrose and Paro 2007). The failure to recover insertions in these domains is not serendipitous, as 17 other MB-free sites also correspond to Polycomb group (PcG)-regulated gene clusters, including domains that house ct, ems, trh, nub, esg, Vsx-1, Lim1, disco, and OdsH. Direct inspection showed that many other such regions of <80 kb, which would not have been flagged in our analysis, also contained few if any MB insertions. However, not all PcG targets were coldspots; for example, there were many MB insertions in bru-3 (Figure 4C).
To investigate whether these PcG-regulated domains are coldspots for transposon insertion generally, we also examined whether P or piggyBac elements integrate normally into these same regions. As can be seen in the case of BX-C (Figure 4B), transposition of piggyBac and P elements is reduced in PcG-regulated domains as well (Figure 4B). However, some loci appeared to suppress transposon insertion selectively. For example, the region surrounding the PcG-regulated esg gene lacked Minos inserts, but contained many piggyBac and P-element insertions; indeed, esg is a P-element hotspot (Figure 4D). Interestingly, piggyBac insertions within many such PcG-target regions, including bru-3 and the esg region, were largely “f” class elements (PBac{WH}, Table 1, Table S3), suggesting that the engineered structure of the construct and not just the transposon type affects transposition or marker gene expression within such domains.
Coldspots for piggyBac frequently encode membrane proteins
We carried out a similar analysis of piggyBac insertions (Figure S1) and identified coldspots that account at least partly for the slower saturation curve of piggyBac relative to random integration or Minos integration (Table S4). Some were in PcG-target genes that corresponded to sites with reduced MB insertion, although the coldspots were not identical for the two elements. Most interesting, however, was a new class of sites that display normal levels of MB insertion, but exhibit strongly reduced levels of piggyBac insertion. These domains are not PcG targets, but are highly enriched in a class of genes with seemingly related function. For example, coldspots include clustered genes encoding acetylcholine receptors [nAcRα-96 (Figure S1A) and nAcRα-7E], olfactory or gustatory receptors (Or69A, Or92A, Or98A, Or22c, and Gr36a-d), neuropeptide receptors (DmsR, dpr10, and CG10418), GRHRII receptor (GRHRII), receptor protein tyrosine phosphatases [Lar (Figure S1B) and Ptp99A), dopamine receptors (D2R), and ryanodine receptor [Rya-r44F (Figure S1C)]. Many of the genes encode other putative membrane proteins, often members of the Ig superfamily [beat-IIIa, beat-Vc, dpr1, dpr2, dpr3, dpr5, and sns (Figure S1C)] and channels/transporters (Glut1, Rh50, Oatp58Da-c cluster, and Ir11a). We conclude that a group of genes with roles in neuronal function, signaling, and growth are coldspots for insertion by piggyBac elements.
Coldspots for P elements include many clustered specialized genes
P elements were absent from most of the PcG targets that were also low in MB or piggyBac insertion, including ANT-C and BX-C (Figure 3B). Some, but not all, of the domains refractory to piggyBac insertion were also low in P-element insertions (e.g., Figure S1, Table S4). However, the most frequent and quantitatively significant classes were intervals containing clusters of genes that were not targeted by P elements, but were hit by one or both of the other two transposons. For example, the 20-gene Osiris family (Dorer et al. 2003) represents one such cluster (Figure S2A). Other clusters, such as the 11-gene esterase complex in region 84D, are mostly refractory to insertion by P elements, except for alpha-Est10, which was hit 45 times (Figure S2B). In contrast, the 15 MB insertions and 10 piggyBac insertions in this region were spread more widely. The MB element in particular was able to insert in many clusters seemingly refractory to P-element insertion. For example, the MB screen included multiple insertions in eggshell protein genes, genes that have never been hit by P elements (Figure S2C).
Discussion
The current GDP collection
The GDP has now generated tools to help functionally analyze at least 9440 genes, approximately two-thirds of all annotated Drosophila protein-coding genes (Figure 1). Achieving this level of saturation using forward insertional mutagenesis required three different transposons and >200,000 independent transpositions. At the time the project began, the genome was not sequenced and relatively little was known about the physical organization of fly genes and regulatory elements. As the project progressed, Drosophila researchers and the fly genomics community increasingly documented multiple transcript isoforms, novel RNA genes, and key genomic regulatory elements. In response, the GDP project evolved beyond the concept of one disruption per gene, and now comprises >14,000 strains. New lines provide additional value by disrupting specific promoters or isoforms, and by providing access to unannotated genes, putative regulatory sequences, and still unknown aspects of genome function.
Recombination-based strategy for completing the GDP
The results reported here make clear that it would be extremely difficult to achieve 95% genome saturation by random insertional mutagenesis with P and piggyBac elements alone. Switching to the Minos element increased the yield of novel gene hits, but achieving 95% saturation of genes by Minos transposition would require an impractically large number of additional insertions to be generated and screened. Including attP sites in a Minos transposon will greatly enhance the general usefulness of insertions for manipulating the genome, since any DNA of interest could be subsequently added at the site of integration. Incorporating DNA that disrupts local chromatin structures might mutate nearby genes, but this approach would be similar to generating deletions from current insertions by imprecise excision. Homologous recombination (Rong et al. 2002) would provide the most attractive finishing strategy for the project, but it has not been technically and economically feasible to carry out on a large enough scale.
Our results suggest a hybrid strategy using attP-containing Minos insertions to provide access to the remaining genes. An attP site located near a target gene allows efficient homologous recombination by the SIRT method (Gao et al. 2008, 2009). A local duplication containing the mutation of interest is inserted at the attP site and then resolved by generating a local double-strand break (Gao et al. 2008, 2009; reviewed in Wesolowska and Rong 2010). Our results show that Minos could be used to tile the entire genome with attP-bearing insertions approximately every 40 kb, allowing the efficient application of homologous recombination to disrupt remaining unhit genes. Generating such a collection of elements represents a highly attractive finishing strategy for the GDP and would provide a powerful framework for future Drosophila genetic manipulation.
A dataset for deducing transposon–genome interactions
A further contribution of the GDP is the detailed knowledge it provides on how transposons interact with their genome. Many previous studies have demonstrated that specific transposons show a wide variety of nonrandom integration preferences (reviewed in Wu and Burgess 2004). Some elements are constrained to strongly preferred or invariant target sites by encoded nucleases; for example, piggyBac elements only insert at TTAA and Minos at TA motifs. In addition, chromatin structure further biases the spectrum of recovered insertion sites (Zhang and Spradling 1994; Wallrath and Elgin 1995; Yan et al. 2002; Bellen et al. 2004; Simons et al. 2006; Galvan et al. 2007; Babenko et al. 2010; Gangadharan et al. 2010; Grabundzija et al. 2010). However, in metazoans it has usually been difficult to separate site preferences from biases introduced by experimental design, by the loss of marker expression following insertion in suppressive chromatin, and by the failure to accurately map insertions in repetitive DNA.
The GDP datasets of MB and EY transpositions were largely free of bias, as every transposition event from the starting chromosomes that supports marker expression was recovered and analyzed. Quality flanking sequence data were obtained from both the 5′ and 3′ ends of most insertions, and automated alignments that failed to localize insertions uniquely were usually checked by a human annotator and frequently could be successfully mapped even within repeat-rich genomic regions. The number of insertions with repetitive flanking sequences that could not be mapped uniquely was relatively small (3–5% of total) and consisted of insertions within euchromatic transposons or within repetitive segments of centric heterochromatin and the Y chromosome. Thus, with respect to potential bias from both chromatin and repetitive genomic sequences, GDP data provide an accurate picture of transposon site selectivity within euchromatin, but an incomplete picture of transposition within centric heterochromatin.
Transposons avoid insertion within many Polycomb-regulated regions
Our data show how chromatin structure influences transposon insertion. In particular, many regions in the genome enriched in the repressive histone modification H3K27me3 were targeted much less frequently by all three transposons. Repressive domains frequently arise from the activity of Polycomb group genes (Schwartz et al. 2006, 2010). Many such regions contain clustered genes encoding key transcription factors such as Hox genes that regulate tissue differentiation and development. Each such cluster is repressed in some cells during development but active in others. Consequently, the roster of PcG-repressed domains depends on the cell type in question. In yeast, plants and Drosophila, H3K27me-rich centric heterochromatin is likely to be generated using other pathways, including the piRNA pathway (reviewed in Riddle and Elgin 2008). Our studies focused on germline transposition, which cluster size analysis places during premeiotic and meiotic adult germ cell development.
The observation that transposon insertions are recovered less frequently in PcG-regulated, “closed” domains has been reported previously (Bellen et al. 2004; Simons et al. 2006; Grabundzija et al. 2010). For example, Tol2 integrations are underrepresented in regions rich in H3K27me in human cells (Grabundzija et al. 2010); however, piggyBac insertions are not. However, in most cases it was difficult to determine whether the dearth of insertions was due to blocked transposition or reduced marker expression.
Our data suggest that PcG-regulated regions directly suppress transposition, but they likely also reduce marker gene expression. The yellow gene and the Pax6–GFP construct used to detect EY and MB transpositions are sufficiently robust to detect at least some insertions in centric heterochromatin. Hence these elements must actually transpose with reduced frequency into PcG domains because most such insertions would be detected. Consistent with this view, when suppressors of variegation were used to reveal the location of “suppressed” insertions they were only found in centric heterochromatin (Zhang and Spradling 1994; Yan et al. 2002). A direct effect on transposition is less certain in the case of piggyBac, because the elements studied carry the position-effect sensitive mini-white gene, and f insertions, which carry a chromatin insulator, were preferentially recovered in some PcG domains (Table S3). Our results suggest that as in pleuripotent mammalian cells (Boyer et al. 2006), Drosophila PcG domains are already established in premeiotic germ cells, where they can affect adult germline transposition. Functions of Polycomb genes in the early germline have been described in the male (Chen et al. 2005).
Transposon-free genomic regions
The genomes of many organisms, including humans, contain rare transposon-free regions (TFRs). Some of these encode clustered HOX genes such as the human HOXA4-11, HOXB4-6, and HOXD8-13 loci (Simons et al. 2006) that resemble the Drosophila BX-C and ANT-C complexes, which we observed to resist transposon insertion. We looked to see whether other human TFRs have Drosophila homologs that are also refractory to integration. For example, human DLX5 lies within a TFR, and its Drosophila homolog Distalless is a PcG-regulated gene that was not hit by any of the three transposons. Many TFRs did not show such a correlation, however. PAX6 lies within a TFR and the closely related Drosophila genes eyeless (ey) and sine oculis (so) both lie within PcG-regulated domains (Schwartz et al. 2010). However, ey received one piggyBac and two MB insertions in our experiments, while the so region was hit by two MB insertions. Finally, the region surrounding the NR2F1/COUP-TF1 gene is a TFR in at least six vertebrate genomes (Simons et al. 2006). Drosophila sevenup (svp) http://flybase.org/reports/FBgn0003651.html is a COUP-TF1 homolog; however, it does not lie within a PcG-regulated domain and was the target of 10 MB and three piggyBac insertions in our experiments. The domains that were refractory to insertion in our experiments frequently contain natural integrated transposons in some strains. Thus, in Drosophila suppression of transposon activity by PcG-regulated domains appears insufficient to sustain transposon-free regions. If PcG-mediated repression of transposon insertion is important in the genesis of mammalian TFRs, it may exert stronger effects, synergize with other regulatory mechanisms not present in Drosophila, and act on rates of germline transposition that are much lower than those in Drosophila.
Some transposons may evolve to benefit their host
Transposon insertions frequently disrupt vital genes; hence, the introduction and spread of transposable elements within a genome have the potential to be highly deleterious. Consequently, like viruses, transposons should evolve to minimize costs to host fitness. In addition, increasing evidence documents a major creative role for transposable elements in the evolution of new genes, regulatory elements, and on genome size itself (Sinzelle et al. 2009). A transposon that could generate useful variation within the genome of its host under conditions of stress, might contribute to the survival of both its host and itself (McClintock 1984). An element might minimize damage and maximize the chance of adaptive variation by avoiding insertion in evolutionarily stable genes and selectively targeting genes whose structure and/or regulation evolves rapidly.
We observed several examples of site specificity in our experiments that suggested such an adaptation. The gene cluster that encodes proteins with CHK-like kinase domains (Figure 2E) was one of few hotspots for Minos insertion. One of these genes, CHKov1, has been shown to harbor a Doc insertion in many wild Drosophila populations that confers enhanced insecticide resistance (Aminetzach et al. 2005). piggyBac elements rarely inserted in genes that encode a variety of membrane receptors for neurotransmitters and other ligands (Table S4). Conversely, many piggyBac hotspot loci contain genes affecting neural development and behavior (Table S2). Thus, of the three elements studied, piggyBac was the only one whose site preferences were suggestive of having evolved to minimize damage and to maximize changes in the regulation of potentially adaptive genes following insertion. There may be common transcription factors or chromatin configurations at these sites that allow such targeting.
Implications for other organisms utilizing transposon mutagenesis
Recently there has been growing interest in the application of insertional mutagenesis in a wide variety of experimental organisms both in the germline (Ding et al. 2005; Galváán et al. 2007; Sasakura et al. 2007; Sivasubbu et al. 2007; Bazopoulou and Tavernarakis 2009; de Wit et al. 2010; O’Malley and Ecker 2010) and in somatic cells (reviewed in Copeland and Jenkins 2010). Indeed, the potential application of transposons as human gene therapy vectors is currently undergoing clinical trials (Izsváák et al. 2010). The lessons learned in the GDP project regarding both the common and unique ways that transposons interact and evolve with the genome are certain to help these projects maximize the value of these exceptional tools for natural and human-guided genetic manipulation.
Acknowledgments
We thank Danqing Bei, Ying Fang, Adeel Jawaid, Jianping Li, Zhihua Wang, and Jin Yue at Baylor College of Medicine, Houston, TX, for generating and maintaining fly stocks. Vanessa Damm, Shelly Paterno, and Eric Chen assisted in the line maintenance and balancing at Carnegie Institution for Science, Baltimore, Maryland. We thank Soo Park and Kenneth Wan at Lawrence Berkeley National Laboratory, Berkeley, CA, for assistance with iPCR and sequencing of insertions. We are grateful to Exelixis and Aprogen (formerly GenExel) for providing lines and sequence data. We are grateful to researchers at Max Planck Institute, Göttingen, Germany, EMBL Heidelberg, Germany, and DeveloGen, Göttingen, Germany, for donating P-insertion lines to the public. We are grateful to Ulrich Schäfer and Herbert Jäckle for providing information and lines from the Göttingen X-linked insertion collection. We thank Peter Maroy for shipping lines to the project from the Szeged Stock Center. We thank Kathy Matthews, Kevin Cook, and Annette Parks for coordinating the transition of the lines to the Bloomington Drosophila Stock Center. We thank Koen Venken for useful suggestions. This work was supported by National Institute of General Medical Sciences (GM067858). Additional funds were provided through the support of the Spradling and Bellen labs from the Howard Hughes Medical Institute.
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Aminetzach Y. T., Macpherson J. M., Petrov D. A., 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309: 764–767 [DOI] [PubMed] [Google Scholar]
- Babenko V. N., Makunin I. V., Brusentsova I. V., Belyaeva E. S., Maksimov D. A., et al. , 2010. Paucity and preferential suppression of transgenes in late replication domains of the D. melanogaster genome. BMC Genomics 11: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Lee A. M., Wu C. T., 2006. Site-specific transformation of Drosophila via ϕC31 integrase-mediated cassette exchange. Genetics 173: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazopoulou D., Tavernarakis N., 2009. The NemaGENETAG initiative: large scale transposon insertion gene-tagging in Caenorhabditis elegans. Genetica 137: 39–46 [DOI] [PubMed] [Google Scholar]
- Beinert N., Werner M., Dowe G., Chung H.-R., Jäckle H., et al. , 2004. Systematic gene targeting on the X chromosome of Drosophila melanogaster. Chromosoma 113: 271–275 [DOI] [PubMed] [Google Scholar]
- Bellen H. J., O’Kane C. J., Wilson C., Grossniklaus U., Pearson R. K., et al. , 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3: 1288–1300 [DOI] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., et al. , 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Shepherd S., Lee K., McCall K., et al. , 1989. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 3: 1273–1287 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., et al. , 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., Cohen S. M., 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A. A., Stark A., Dus M., Kellis M., et al. , 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., et al. , 2007. The Carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175: 1505–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hiller M., Sancak Y., Fuller M. T., 2005. Tissue-specific TAFs counteract Polycomb to turn on terminal differentiation. Science 310: 869–872 [DOI] [PubMed] [Google Scholar]
- Cooley L., Kelley R., Spradling A., 1988. Insertional mutagenesis of the Drosophila genome with single P elements. Science 239: 1121–1128 [DOI] [PubMed] [Google Scholar]
- Copeland N. G., Jenkins N. A., 2010. Harnessing transposons for cancer gene discovery. Nat Rev Cancer 10: 696–706 [DOI] [PubMed] [Google Scholar]
- de Wit T., Dekker S., Maas A., Breedveld G., Knoch T. A., et al. , 2010. Tagged mutagenesis by efficient Minos-based germ line transposition. Mol. Cell Biol. 30: 68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., Han M., Zhuang Y., et al. , 2005. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122: 473–483 [DOI] [PubMed] [Google Scholar]
- Dorer D. R., Rudnick J. A., Moriyama E. N., Christensen A. C., 2003. A family of genes clustered at the Triplo-lethal locus of Drosophila melanogaster has an unusual evolutionary history and significant synteny with Anopheles gambiae. Genetics 165: 613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P., 1998. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194 [PubMed] [Google Scholar]
- Fujita P. A., Rhead B., Zweig A. S., Hinrichs A. S., Karolchik D., et al. , 2010. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 39: D876–D882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galván A., González-Ballester D., Fernández E., 2007. Insertional mutagenesis as a tool to study genes/functions in Chlamydomonas. Adv. Exp. Med. Biol. 616: 77–89 [DOI] [PubMed] [Google Scholar]
- Gangadharan S., Mularoni L., Fain-Thornton J., Wheelan S. J., Craig N. L., 2010. DNA transposon Hermes inserts into DNA in nucleosome-free regions in vivo. Proc. Natl. Acad. Sci. USA 107: 21966–21972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., McMahon C., Chen J., Rong Y. S., 2008. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc. Natl. Acad. Sci. USA 105: 13999–14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Wesolowska N., Rong Y. S., 2009. SIRT combines homologous recombination, site-specific integration, and bacterial recombineering for targeted mutagenesis in Drosophila. Cold Spring Harb. Protoc. doi:10.1101/pdb.prot5236 [DOI] [PubMed] [Google Scholar]
- Godfrey A. C., Kupsco J. M., Burch B. D., Zimmerman R. M., Dominski Z., et al. , 2006. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 12: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabundzija I., Irgang M., Mates L., Belay E., Matrai J., et al. , 2010. Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 18: 1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A. C., Fish M., Nusse R., Calos M. P., 2004. Construction of transgenic Drosophila by using the site-specific integrase from phage ϕC31. Genetics 166: 1775–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C., Jaunich B., Wimmer E. A., 2000. Highly sensitive, fluorescent transformation marker for Drosophila transgenesis. Dev. Genes. Evol. 210: 623–629 [DOI] [PubMed] [Google Scholar]
- Izsvák Z., Hackett P. B., Cooper L. J., Ivics Z., 2010. Translating Sleeping Beauty transposition into cellular therapies: victories and challenges. Bioessays 32: 756–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. I., Ryu T., Lee J., Heo Y. S., Ahnn J., et al. , 2010. A genetic screen for modifiers of Drosophila caspase Dcp-1 reveals caspase involvement in autophagy and novel caspase-related genes. BMC Cell. Biol. 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G. C., Rehm E. J., Rubin G. M., 2000. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97: 3347–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A., Oehler S., Klinakis A., Savakis C., 2005. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171: 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B., 1984. The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- Misra S., Crosby M. A., Mungall C. J., Matthews B. B., Campbell K. S., et al. , 2002. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 3: RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W., 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15050–15055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley R. C., Ecker J. R., 2010. Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61: 928–940 [DOI] [PubMed] [Google Scholar]
- Oh S. W., Kingsley T., Shin H. H., Zheng Z., Chen H. W., et al. , 2003. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics 163: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter A., Schöttler P., Werner M., Beinert N., Dowe G., et al. , 2002. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3: 34–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones-Coello A. T., Petrella L. N., Ayers L., Melillo A., Mazzalupo S., et al. , 2007. Exploring strategies for protein trapping in Drosophila. Genetics 175: 1089–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N. C., Elgin S. C., 2008. A role for RNAi in heterochromatin formation in Drosophila. Curr. Top. Microbiol. Immunol. 320: 185–209 [DOI] [PubMed] [Google Scholar]
- Ringrose L., Paro R., 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134: 223–232 [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., et al. , 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E., Blows F., Ashburner M., Bautista-Llacer R., Coulson D., et al. , 2004. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics 167: 797–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakura Y., Oogai Y., Matsuoka T., Satoh N., Awazu S., 2007. Transposon mediated transgenesis in a marine invertebrate chordate: Ciona intestinalis. Genome Biol. 8(Suppl 1): S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Nix D. A., Li X. Y., Bourgon R., et al. , 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Stenberg P., Ohno K., Bourgon R., et al. , 2010. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 6: e1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C., Pheasant M., Makunin I. V., Mattick J. S., 2006. Transposon-free regions in mammalian genomes. Genome Res. 16: 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinzelle L., Izsvák Z Z., Ivics Z., 2009. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell. Mol. Life. Sci. 66: 1073–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubbu S., Balciunas D., Amsterdam A., Ekker S. C., 2007. Insertional mutagenesis strategies in zebrafish. Genome Biol. 8(Suppl 1): S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Stern D. M., Kiss I., Roote J., Laverty T., et al. , 1995. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl. Acad. Sci. USA 92: 10824–10830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., et al. , 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153: 135–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt N., Molitor A., Somogyi K., Mata J., Curado S., et al. , 2005. Gain-of-function screen for genes that affect Drosophila muscle pattern formation. PLoS Genet. 1(e55): 499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., et al. , 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36: 283–287 [DOI] [PubMed] [Google Scholar]
- Tower J., Kurapati R., 1994. Preferential transposition of a Drosophila P element to the corresponding region of the homologous chromosome. Mol. Gen. Genet. 244: 484–490 [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., et al. , 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., He Y., Hoskins R. A., Bellen H. J., 2006. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2007. Transgenesis upgrades for Drosophila melanogaster. Development 134: 3571–3584 [DOI] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., et al. , 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6: 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L. L., Elgin S. C., 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263–1277 [DOI] [PubMed] [Google Scholar]
- Wesolowska N., Rong Y. S., 2010. The past, present and future of gene targeting in Drosophila. Fly 4: 53–59 [DOI] [PubMed] [Google Scholar]
- Wu X., Burgess S. M., 2004. Integration target site selection for retroviruses and transposable elements. Cell. Mol. Life Sci. 61: 2588–2596 [DOI] [PubMed] [Google Scholar]
- Yan C. M., Dobie K. W., Le H. D., Konev A. Y., Karpen G. H., 2002. Efficient recovery of centric heterochromatin P-element insertions in Drosophila melanogaster. Genetics 161: 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Spradling A. C., 1994. Insertional mutagenesis of Drosophila heterochromatin with single P elements. Proc. Natl. Acad. Sci. USA 91: 3539–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]