Abstract
Allopolyploidy has played a prominent role in organismal evolution, particularly in angiosperms. Allohexaploidization is a critical step leading to the formation of common wheat as a new species, Triticum aestivum, as well as for bestowing its remarkable adaptability. A recent study documented that the initial stages of wheat allohexaploidization was associated with rampant genetic and epigenetic instabilities at genomic regions flanking a retrotransposon family named Veju. Although this finding is in line with the prevailing opinion of rapid genomic instability associated with nascent plant allopolyploidy, its relevance to speciation of T. aestivum remains unclear. Here, we show that genetic instability at genomic regions flanking the Veju, flanking a more abundant retroelement BARE-1, as well as at a large number of randomly sampled genomic loci, is all extremely rare or nonexistent in preselected individuals representing three sets of independently formed nascent allohexaploid wheat lines, which had a transgenerationally stable genomic constitution analogous to that of T. aestivum. In contrast, extensive and transgenerationally heritable repatterning of DNA methylation at all three kinds of genomic loci were reproducibly detected. Thus, our results suggest that rampant genetic instability associated with nascent allohexaploidization in wheat likely represents incidental and anomalous phenomena that are confined to by-product individuals inconsequential to the establishment of the newly formed plants toward speciation of T. aestivum; instead, extensive and heritable epigenetic remodeling coupled with preponderant genetic stability is generally associated with nascent wheat allohexaploidy, and therefore, more likely a contributory factor to the speciation event(s).
ALLOPOLYPLOIDY, conditioned by interspecific hybridization prior to or followed by whole genome doubling (WGD), represents a pervasive force in speciation and evolution of many organismal taxa, particularly in the plant kingdom (Otto and Whitton 2000; Wendel 2000; Comai 2005; Otto 2007; Leitch and Leitch 2008; Soltis and Soltis 2009). Nonetheless, the genetic and molecular mechanism(s) whereby allopolyploidy facilitates the evolutionary process remains largely elusive (Otto 2007; Jones and Hegarty 2009). Findings made in the last two decades have revealed that the early stages of allopolyploidy are often associated with rapid and extensive genomic instabilities, and which are thought to be important for initial stabilization of the newly formed allopolyploids as well as contribute to their establishment as competitive new species (reviewed in Wendel 2000; Levy and Feldman 2004; Comai 2005; Chen 2007; Doyle et al. 2008; Feldman and Levy 2009; Jackson and Chen 2009; Jones and Hegarty 2009; Soltis and Soltis 2009).
Allohexaploid common or bread wheat (Triticum aestivum L.) is a classic example of speciation via allopolyploidy. The evolution of common wheat encompasses two allopolyploidization events: allotetraploidization to give rise to the allotetraploid wheat, T. turgidum (genome BBAA), and allohexaploidization to give rise to the allohexaploid common wheat, T. aestivum (genome BBAADD). Whereas the former event occurred ca. 0.5 million years ago between diploid wheat T. urartu (genome AA) and a goat-grass species belonging to the Sitopsis section of genus Aegilops with an S or S-like genome (Huang et al. 2002; Salamini et al. 2002), the later occurred <10,000 years ago (Feldman et al. 1995) between a free-threshing, cultivated form of T. turgidum (perhaps ssp. durum or the more primitive ssp. parvicoccum, genome BBAA) and another diploid Aegilops species, Aegilops tauschii (genome DD) (Kihara 1944; Mcfadden and Sears 1946; Feldman et al. 1995; Feldman 2000). Both allopolyploidization events can be readily reproduced in the laboratory, thus verifying authenticity of the evolutionary process and progenitors involved. Common wheat thus provides a unique example of evolution via allopolyploidy of a new species that instantaneously became arguably the human being's most important crop in a single step (no wild form of common wheat is known). Deciphering the process and elucidating its underlying mechanism of this speciation event is not only of evolutionary interest, but also might reveal novel clues toward more efficient crop genetic improvement.
Previous studies employing newly synthesized allopolyploid wheat lines have documented that the onset of both the tetra- and hexapolyploidization events is associated with rapid and extensive structural genomic instabilities including elimination of coding and noncoding sequences (Feldman et al. 1997; Liu et al. 1998b; Ozkan et al. 2001; Han et al. 2005) and changes in DNA methylation (Liu et al. 1998a; Shaked et al. 2001). Nonetheless, analysis at many unbiased loci from a genome-wide perspective was conducted thus far only at the tetraploid level (Shaked et al. 2001; Qi et al. 2010), while either only a few preselected loci known to be labile (chromosome- and genome-specific sequences) or a set of selected microsatellite loci have been investigated at the hexaploid level to detect allohexaploidy-associated rapid genomic changes (Feldman et al. 1997; Liu et al. 1998a,b; Ozkan et al. 2001; Mestiri et al. 2010).
More recently, Kraitshtein et al. (2010) have conducted a genome-wide analysis on genetic and DNA methylation changes at genomic loci flanking a high-copy number retrotransposon family called Veju (Sanmiguel et al. 2002) in a single newly synthesized allohexaploid wheat line. They found strikingly high frequencies (>50%) of both genetic change (including massive element loss followed by retrotranspositional burst) and methylation alteration in the studied line over the first five successive generations (Kraitshtein et al. 2010). Because only one line was used plus only euploidy was verified at the cytological level by conventional chromosome counting, it remained unclear whether the documented changes bear relevance to speciation of T. aestivum. This question is raised because it is well established that at the chromosomal level, apart from a few species-specific intergenomic translocations (Naranjo 1990; Jiang and Gill 1994), the three genomes (BB, AA, and DD) of T. aestivum are largely intact. Therefore, if the newly formed allohexaploid wheat plants possess many “anomalous” intergenomic translocations (i.e., absent from T. aestivum), then the genomic instabilities detected would represent accessory events accompanying the allopolyploidization process, which conceivably would have been rapidly purged out under natural selection, and hence, could not be among the original founders leading to speciation of T. aestivum.
To address this issue, we have focused our attention only on those plant individuals for each of three independently synthesized allohexaploid wheat lines, which have a transgenerationally stable chromosomal constitution, which is highly similar to the present-day natural common wheat, T. aestivum, on the basis of multicolor genomic in situ hybridization (GISH) analysis. It is conceivable that this subgroup of individuals with multigenerational chromosomal integrity likely represents a minority of the newly generated allohexaploid population under natural conditions, which however, was most parsimoniously to recapitulate the original founders of T. aestivum. We report here that two common features are associated with these carefully selected individuals from three newly synthesized allohexaploid wheat populations: (1) genetic instability at genomic regions flanking Veju (Sanmiguel et al. 2002), flanking a more abundant transposable element (TE), BARE-1 (Manninen and Schulman 1993), as well as at a large number of randomly sampled genomic loci is all extremely rare or nonexistent; and (2) extensive and transgenerationally heritable repatterning in DNA methylation of all three kinds of genomic regions was detected by DNA methylation-sensitive markers, and a subset of which was validated by bisulfite genomic sequencing. Both features are independent of genetic context and highly reproducible across the three independent lines. Thus, our results suggest that genome-wide rampant genetic instability associated with the initial stages of allohexaploidization in wheat is likely an attendant phenomenon occurring only in certain anomalous individuals. These plant individuals conceivably may have fitness disadvantages due to excessive chromosomal instability and therefore unlikely have been selected for under natural conditions to contribute to T. aestivum speciation. Instead, heritable epigenetic remodeling coupled with preponderant genetic stability is generally associated with wheat allohexaploidization, and hence, is more likely an important contributory factor to the speciation event(s).
Materials and Methods
Plant lines
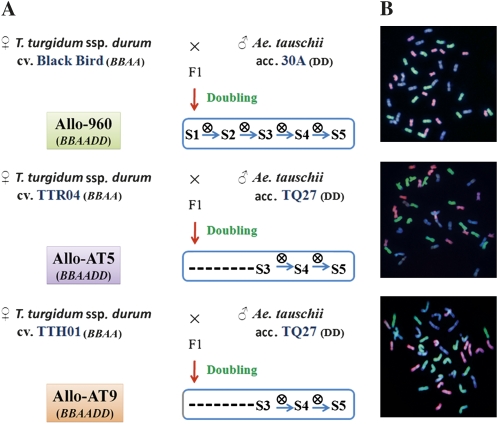
Three sets of synthetic allohexaploid lines (designated as Allo-960, Allo-AT5, and Allo-AT9) with identical genome constitution but different genotypes of one or both parental species were used (Figure 1A). Two of these lines were produced by crossing Triticum turgidum, ssp. durum, cv. TTR04 (for Allo-AT5) or ssp. carthlicum, cv. TTH01 (for Allo-AT9) with a common Ae. tauschii line (TQ27), followed by genome doubling with colchicine treatment (Ozkan et al. 2001). These two lines (at S1) were kindly provided by Professor Moshe Feldman at the Weizmann Institute of Science, Rehovot, Israel. The third line (Allo-960) was produced by crossing T. turgidum, ssp. durum, cv. Black-Bird with an Ae. tauschii line (30A), also followed by genome doubling with colchicine treatment. This line (at S1) was kindly provided by Dr. George Fedak of the Agriculture and Agri-Food Canada, Ottawa, Ontario, CA. Each of these lines was then self-pollinated for five consecutive generations in our hands, and one individual plant was selected from each generation for molecular analysis of genetic/epigenetic stability or changes. All plants including the corresponding parental lines were grown in controlled growth chambers at 22/20°C day/night of 12-h day length for genomic DNA isolation.
Figure 1.
(A) Diagrams showing pedigree of three independently formed allohexaploid wheat lines, designated as Allo-960, Allo-AT5, and Allo-AT9, each with three to five selfed generations available for this study. (B) Typical chromosomal constitutions for the selected individuals from each of the three allohexaploid wheat lines, as revealed by multicolor GISH. All plant individuals used for this study are with transgenerational chromosomal stability at the subgenomic level (euploidy and lack of any intergenomic rearrangements), as shown in B, which is highly similar to the natural common wheat, T. aestivum. The pink-, green-, and blue-colored chromosomes are of the BB, AA, and DD genomes, respectively.
Multicolor GISH
The protocol was essentially as described by Han et al. (2004) with minor modifications. Specifically, genomic DNA was isolated from young leaves of the three putative diploid progenitors of common wheat (T. aestivum), namely, T. urartu, Ae. speltoides, and Ae. tauschii, by a modified CTAB method. Genomic DNA of T. urartu and Ae. tauschii was labeled by nick translation with Chroma Tide Alexa Fluor 488-5-dUTP (Invitrogen; cat. no. C11397) and Texas Red-5-dCTP (Perkin Elmer; cat. no. NEL 426), respectively. Genomic DNA of Ae. speltoides was used as a blocker. Slide denaturation, hybridization, and washing conditions were carried out as described by the manufacturer's protocol (Invitrogen; cat. no. C11397). Slides were examined with an Olympus fluorescence microscope and digitally photographed.
Amplified fragment length polymorphism and methylation-sensitive amplified fragment length polymorphism
The amplified fragment length polymorphism (AFLP) procedure was performed essentially as originally described (Vos et al. 1995), with minor modifications for silver staining as detailed in Wang et al. (2005). The methylation-sensitive amplified fragment length polymorphism (MSAP) protocol, also employing silver staining, was exactly as described (Dong et al. 2006). For both markers, two technical replications (starting from independent DNA isolation) were performed and only clear and completely reproducible bands were scored, a subset of which was isolated for sequencing.
Transposon display and methylation-sensitive transposon display
The protocols for transposon display (TD) and methylation-sensitive transposon display (MSTD) were essentially as reported (Kraitshtein et al. 2010). Nested primers respectively specific to one terminus of each of the two retrotransposon families, Veju (Sanmiguel et al. 2002) and BARE-1 (Manninen and Schulman 1993), together with a primer matching the MseI adapter sequence, were used (supporting information, Table S1). Two technical replications were also included in the TD and MSTD analysis. Only clear and completely reproducible bands were scored, a subset of which was isolated for sequencing.
Bisulfite genomic sequencing
Genomic DNA was modified using the EZ DNA Methylation-Gold kit (Zymo Research, http://zymoresearch.com) to convert cytosine residues to uracil but leaves 5-methylcytosine residues unaffected by bisulfite. The amplification primer pairs for each of three isolated MSAP loci were designed using the Kismeth program (http://katahdin.mssm.edu/kismeth/ revpage.pl) and are given in Table S5. For each analyzed locus of each sample, 15 randomly chosen clones were sequenced, and the methylation levels presented as percentage (%) per site for each of the three types of cytosine residues, CG, CHG, and CHH, were calculated by dividing the number of nonconverted (methylated) cytosines by the total number of cytosines of each type within the sequenced regions.
Results
Defining chromosomal constitution of the nascent allohexaploid wheat plant individuals used for this study
Conventional chromosome number counting of 50–60 randomly chosen individuals of each of the three sets of synthetic allohexaploid wheat lines, Allo-960, Allo-AT5, and Allo-AT9, at each of the three (S3–S5) or five (S1–S5) selfed generations, indicated that the great majority (>95%) of the plants had the expected euploid chromosome number of 2n = 6X = 42, suggesting that general chromosomal numerical stability was characteristic of these newly formed allohexaploid lines harboring the functional Ph1 gene (Feldman et al. 1993; Mestiri et al. 2010). Further multicolor GISH analysis showed, however, that in all three sets of lines, a significant proportion (15–20%) of the individuals harbored intergenomic translocations primarily between the B-A genomes (N. Zhao, B. Zhu, M. Li, L. Wang, L. Xu, H. Zhang, S. Zheng, B. Qi, F. Han, and B. Liu, unpublished results), indicating that intergenomic rearrangements at the chromosomal level had frequently occurred in these newly formed allohexaploid lines, which is consistent with the results of meiosis analysis on an independent set of similar lines (Mestiri et al. 2010). These observations are supportive of the hypothesis that in nascent wheat allopolyploids, functionality of the Ph1 gene can be compromised and may entail additional or alternative mechanisms like homeologous chromosome differentiation via elimination of noncoding sequences to ensure rapid cytological diploidization (Feldman et al. 1997; Feldman and Levy 2005). Because the aim of this study was to explore the genomic status at the initial stages of wheat allohexaploidization in those individuals with a transgenerationally stable genome configuration most parsimoniously mimicking that of the present-day natural common wheat, T. aestivum, those plant individuals with discernibly aberrant translocations (i.e., different from T. aestivum) were excluded from further analysis. In fact, we set the strict criterion of choosing only those plants from each of the three lines, which showed complete transgenerational chromosomal stability at the subgenomic level (on the basis of the multicolor GISH analysis) across all the 3–5 generations examined (e.g., Figure 1B). That is, for example, if an intergenomic translocation event was detected in any of the derived progenies of a particular plant of an earlier generation, then the earlier-generation plant was not used for this study.
Genome-wide genetic stasis associated with nascent allohexaploidization in wheat
To investigate the structural genomic stasis or dynamics in the newly formed allohexaploid wheat individuals of the three independent lines that exhibited transgenerational chromosomal integrity at the subgenomic level described above, genomic regions flanking two high copy-number retrotransposon families, Veju (Sanmiguel et al. 2002) and BARE-1 (Manninen and Schulman 1993), revealed by the element-specific TD, as well as at a large number of unbiased genomic loci sampled by the AFLP maker, were analyzed. By using 16 pairs of selective primers for each of the TD markers (Table S1), 595, 622, and 632 bands by the Veju-TD, and 708, 711, and 717 bands by the BARE-1–TD, were scored for the three sets of allohexaploid wheat lines, Allo-960, Allo-AT5, and Allo-AT9, respectively. In parallel, by using 28 pairs of selective AFLP primers (Table S2), 1889, 1865, and 1887 bands were scored for the three sets of lines.
Previous studies have revealed two major types of genetic changes being often associated with nascent plant allopolyploidy: loss, disappearance of parental bands and gain, appearance of novel bands (Song et al. 1995; Liu et al. 1998a,b; Ozkan et al. 2001; Shaked et al. 2001). Whereas the detection of a loss event is dependent on the nature of the bands being polymorphic between the two parental species, a gain event will be detectable irrespective of parental polymorphism or monomorphism. With regard to the degree of parental polymorphism, we found that (1) between the two kinds of markers, TD and AFLP, the former is more polymorphic than the later (Figure S1), consistent with the more dynamic nature of TD-adjacent regions than other loci in the plant genomes (Parisod et al. 2009); and (2) between the two TD markers, Veju is more polymorphic than BARE-1, suggesting differential retrotranspositional activity of the two retroelements in the recent evolutionary past of one or both of the parental species of a given synthetic allohexaploid combination (Figure S1).
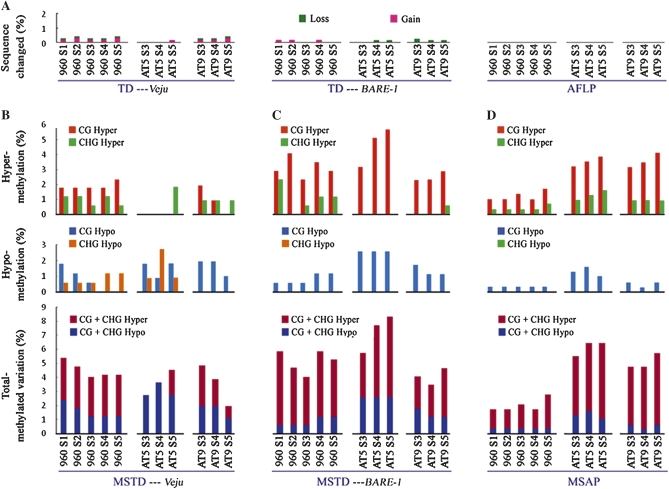
Totally beyond our expectation and in a sharp contrast with the recent report showing strikingly high levels of genetic dynamics including rampant element loss and retrotranspostional burst of the Veju retrotransposon family over the first five generations (S1–S5) in a newly formed allohexaploid wheat line (Kraitshtein et al. 2010), we detected near complete genetic stasis in all the plant individuals (n = 11) collected over 3–5 selfed generations of three independently synthesized allohexaploid wheat lines with different genotypic combinations (Figure 1A). Specifically, out of the 595, 622, and 632 bands scored for the three sets of allohexaploid wheat lines (Table S3), we detected from zero to three loss or gain events for the 11 individual plants; moreover, most of the losses were also detected in the corresponding parental DNA mixtures, indicating they were not “real” genetic changes associated with allohexaploidization, but resulting from PCR competition between the divergent parental species DNA (data not shown), consistent with the findings by a recent study monitoring microsatellite loci (Mestiri et al. 2010). Collectively, the allohexaploidization-induced genetic changes at genomic regions adjacent to the retrotransposon Veju are estimated as only in the range of 0–0.5% (Figure 2A).
Figure 2.
Genetic and DNA methylation changes in 11 studied individuals representing the three (S3–S5) or five (S1–S5) selfed generations of three independently formed allohexaploid wheat lines (Allo-960, Allo-AT5, and Allo-AT9). Two TD markers (Veju-TD and BARE-1–TD) and the AFLP marker were used to detect possible genetic changes, while their cytosine methylation-sensitive counterparts, two MSTD markers (Veju-MSTD and BARE-1–MSTD) and the MSAP marker, were used to detect possible alteration in cytosine methylation at the 5′-CCGG sites. The frequencies for the two major types of genetic changes, loss and gain, detected by the three kinds of markers, are depicted in A, while the frequencies of the four major patterns of cytosine methylation alteration as well as total, detected by Veju-MSTD, BARE-1–MSTD, and MSAP, respectively, are depicted in B–D. The detailed dataset for the scored variant bands is presented in Table S3 and Table S4.
Next, we analyzed the same allohexaploid plant individuals by another TD marker designed for a more abundant retrotransposon family, BARE-1 (Manninen and Schulman 1993), and at a large number of randomly sampled genomic loci by the AFLP marker. We detected even a greater degree of preponderance of genomic stasis by these two markers. Specifically, in BARE-1–TD, the number of variant bands excluding those due to parental DNA competition ranged from zero to two for the total numbers of 708, 711, and 717 bands scored for the three sets of independent lines, and in AFLP, no variant band due to “real” genetic changes was detected although 1889, 1865, and 1887 bands were scored for these lines (Figure 2A).
It should be emphasized again that for all three studied markers, some of the “variant” bands (particularly of the loss type) would have been scored as allopolyploidy-induced genetic changes if the experimentally mixed parental DNA was not included in the analysis (data not shown), underscoring essentiality of including this critical experimental control, as pointed out in a recent study (Mestiri et al. 2010).
Taking these results together, an important conclusion being reached is that all three markers targeting to different genomic regions point to the preponderance of genome-wide genetic stasis rather than dynamics in preselected individuals with a transgenerationally stable chromosomal constitution analogous to that of common wheat (T. aestivum); importantly, this holds true for all three independently formed lines with different parental genotypes.
Extensive and transgenerationally heritable repatterning of DNA methylation associated with nascent allohexaploidization in wheat
Previous investigations have documented that genome-wide alteration in cytosine methylation level and pattern is a major form of epigenetic dynamics associated with nascent plant allopolyploidy (Liu and Wendel 2003; Comai 2005; Chen 2007), including tetraploidization in wheat (Shaked et al. 2001; Qi et al. 2010). To explore whether similar epigenetic instability also occurred in the three sets of independently formed nascent allohexaploid wheat lines used in this study, we performed MSTD analysis, respectively targeting the genomic regions adjacent to the same two retrotransposons, Veju and BARE-1, and MSAP analysis, targeting a large number of randomly sampled loci across the genome. For all three kinds of markers, the methylation-sensitive isoschizomers, HpaII and MspI, were used, such that in all cases the 5′-CCGG sites were analyzed for possible methylation changes. Because HpaII will not cut if either of the cytosines at the 5′-CCGG sites is fully (double-strand) methylated, whereas MspI will not cut if the external cytosine is hemimethylated, the full methylation of the internal cytosine, or hemimethylation of the external cytosine at the assayed 5′-CCGG sites can be distinguished in the HpaII/MspI-based MSTD and MSAP fingerprinting profiles (Dong et al. 2006; Ngezahayo et al. 2009). For clarity, we hereby define these two major types of cytosine methylation as CG methylation (a band present in MspI digest but absent from HpaII digest) and CHG methylation (a band present in HpaII digest but absent from MspI digest), respectively. Accordingly, four patterns of methylation changes, namely, CG hypo, CHG hypo, CG hyper, and CHG hyper, could be assessed.
By using 16 pairs of selective primer pairs (Table S1), 604, 441, and 429 clear and reproducible bands in Veju-MSTD and 407, 421, and 414 such bands in BARE-1–MSTD were scored for the three sets of allohexaploid wheat lines, Allo-960, Allo-AT5, and Allo-AT9, respectively. In parallel, by using 20 pairs of selective primers (Table S2), 763, 728, and 745 clear and reproducible bands were scored by MSAP for these lines.
On the basis of the same rationale as for TD and AFLP, if no epigenetic remodeling occurred in these nascent allohexaploid wheat lines, we would expect complete additivity in the MSTD and MSAP patterns relative to their parental species and the corresponding parental DNA mixtures. Contrary to this expectation and in a sharp contrast to the preponderant genetic stasis, described above, substantial alteration in each of the four cytosine methylation patterns (CG hypo, CHG hypo, CG hyper, and CHG hyper) were detected by the three types of markers in all the studied individuals representing the three independent lines relative to the corresponding parental DNA mixtures (Figure 2, B–D). Specifically, the following results were obtained: (1) among the four patterns of methylation changes, a general trend revealed by all three types of markers is that the frequencies of the two kinds of hypermethylation (CG hyper and CHG hyper) were higher than those of hypomethylation in all studied plants except for allo-AT5 in Veju-MSTD (Figure 2B); (2) between the two types of methylation (CG vs. CHG), changes in CG methylation greatly surpassed those in CHG methylation for both hyper- and hypomethylation changes (Figure 2, B–D); (3) among the three kinds of markers, the two MSTDs showed higher frequencies of changes than MSAP in Allo-960, but the difference is not clearly discernible for the rest two lines, suggesting parental genotype-specific effects (Figure 2, B–D); (4) the collective frequencies for all four patterns of methylation changes across the 11 plant individuals representing the three independent lines ranged from 2 to 8.5% (Figure 2, B–D); (5) most of the detected methylation pattern changes are transgenerationally heritable, that is, a change that occurred in a particular individual representing a given generation was almost always detected in the later progenies descended from that particular individual (e.g., Figure 3).
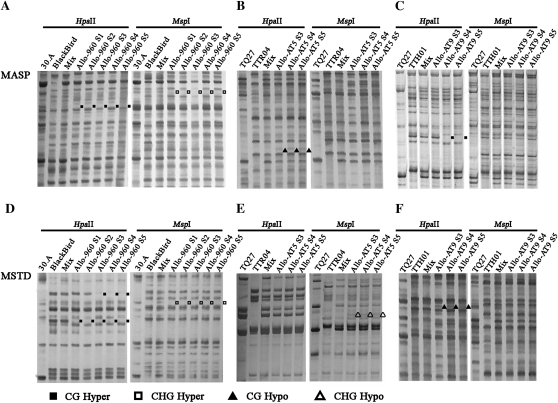
Figure 3.
Examples of typical MSAP and MSTD profiles for the same plants as in Figure 2 illustrating the four patterns of cytosine methylation alteration, CG hyper, CHG hyper, CG hypo, and CHG hypo, which can be unequivocally distinguished by these methylation-sensitive markers. A–F are produced by primer combinations of EcoRI-E + H/M-4, EcoRI-A + H/M-10, EcoRI-F + H/M-8, Veju + H/M-4, Veju + H/M-8, and BARE-1 + H/M-15, respectively.
Taken together, it is clear that, in sharp contrast to the predominant structural stasis in terms of genetic variation, extensive (detected by all three types of markers in all 11 analyzed plant individuals representing three independent lines) and transgenerationally heritable epigenetic remodeling in the form of cytosine methylation repatterning is a general occurrence associated with nascent allohexaploidization in wheat, which is highly reproducible among those individuals with a chromosomal constitution similar to the present-day natural allohexaploid common wheat, T. aestivum.
Validation of nascent allohexaploidization-induced DNA methylation repatterning in wheat by bisulfite genomic sequencing
To further investigate the cytosine methylation changes induced by nascent allohexaploidization in wheat, we selected several variant MSAP bands as representatives for bisulfite sequencing analysis. Three MSAP bands (MSAP19, MSAP27, and MSAP29), all from Allo-AT9, were chosen for the analysis, on the basis of the following two criteria: (1) they are low- or unique-copy on the basis of a blast analysis against the 5X nonannotated genomic sequence of common wheat cv. Chinese Spring (http://www.cerealsdb.uk.net/search_reads.htm); and (2) they are completely conserved at the nucleotide sequence level between the two parental species (Figure S2), such that no amplification bias would be expected when the allohexaploid synthetic line was subjected to the analysis. We obtained the following results for MSAP19, MSAP27, and MSAP29, respectively.
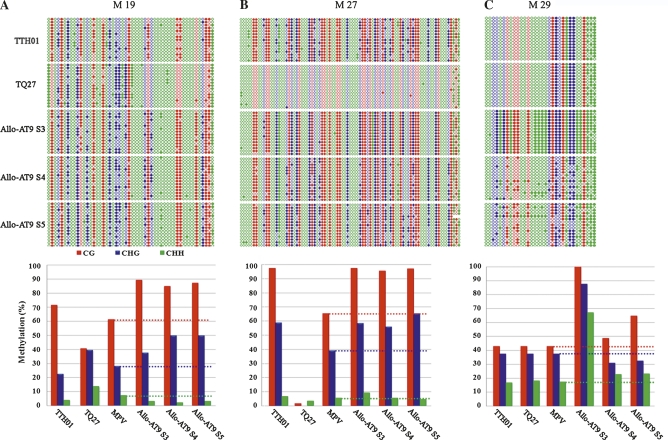
MSAP19 was moderately methylated in the diploid paternal species, Ae. tauschii (accession TQ27) for all three types of cytosine residues, CG (40.56%), CHG (39.39%), and CHH (3.56%), but more heavily methylated in the tetraploid maternal species, T. turgidum, ssp. durum (accession TTH01) at the CG residues (71.67%), whereas with CHG (22.42%) and CHH (3.91%) methylation levels being lower than or similar to those of the diploid species (Figure 4A). On the basis of these parental values, the expected middle-parental values (MPVs) for the three types of methylation levels were calculated according to the genomic contribution by the two parental species, i.e., 1/3 TQ27+2/3 TTH01. Compared with the calculated MPVs, Allo-AT9 showed marked increase in methylation for both CG (by 30–33%) and CHG (by 25–40%) across the three individual plants representing three generations (S3–S5), while with CHH, methylation was reduced by ∼50% (Figure 4A).
Figure 4.
The genomic bisulfite sequencing-based cytosine methylation maps and collective methylation levels (%) of the three types of cytosine residues, CG (red dots or columns), CHG (blue dots or columns), and CHH (green dots or columns), for each of the three variant MSAP loci [MSAP 19 (A), MSAP 27 (B), and MSAP 29 (C)] in three individual plants representing three successive selfed generations (S3–S5) of the nascent allohexaploid wheat line (AT9) and its parental species, T. turgidum, ssp. durum, cv. TTH01 and Ae. tauschii, line TQ27. The sequences for the studied loci are given in Figure S2, which are completely conserved between the two parental lines for AT9. For each analyzed locus, 15 clones were arbitrarily sequenced to reflect each biological sample. The calculated MPVs are included for comparison.
MSAP27 is unmethylated at all three types of residues (CG, CHG, and CHH) in the diploid parental species TQ27, but heavily methylated in the tetraploid parent TTH01 at CG and CHG residues (Figure 4B). Relative to the calculated MPVs, Allo-AT9 also showed marked increase in methylation of both CG (by ∼30%) and CHG (by 27–38%) across the three individual plants representing the three generations (S3–S5), with CHH methylation being increased by ∼80% in the S3 plant, but which were largely reverted back in the S4 and S5 plants (Figure 4B).
MSAP29 is similarly methylated at all three types of cytosine residues (∼42% for CG, ∼37% for CHG, and ∼16% for CHH) in both the diploid and tetraploid parents (TQ27 and TTH01) (Figure 4C). Relative to the calculated MPVs, the S3 plant showed conspicuous increase by 1–1.5 fold in methylation at all three types of cytosine residues, all of which however are reverting back toward the MPVs in the S4 and S5 plants (Figure 4C).
Thus, results of the bisulfite sequencing analysis of all three studied variant MSAP loci confirmed that extensive and, in two of the three loci, transgenerationally heritable methylation repatterning of all three types of cytosine residues (CG, CHG, and CHH) occurred in the studied nascent allohexaploid wheat line (Allo-AT9), with a clear trend toward hypermethylation. It is therefore reasonable to deduce that most of the methylation changes detected by MSAP and MSTD are real.
Sequences underlying nascent allohexaploidization-induced methylation repatterning in wheat
A subset of the variant bands in the nascent allohexaploid wheat lines detected by the MSTD and MSAP markers were isolated, cloned, and sequenced. A blast analysis of these variant bands at the National Center for Biotechnology Information (NCBI) Website (www.ncbi.nlm.nih.gov/) indicated that the great majority showed no meaningful similarity to the current database sequences, while the rest are with significant homology to those encoding for known function or predicted proteins, transposons, and retrotransposons (Table 1).
Table 1.
Functional classification of a subset of variant TD, MSTD, and MSAP bands that consistently occurred in all three independent allohexaploid wheat lines
| Category | No. and percentage (%) of variant MSTD bands | No. and percentage (%) of variant MSAP bands |
| Known-function genes | 4 (10.0) | 6 (15.4) |
| Predicted genes | 3 (7.5) | 8 (20.5) |
| TE-like sequences | 0 (0.0) | 0 (0.0) |
| No similarity | 33 (82.5) | 25 (64.1) |
| Total | 40 (100) | 39 (100) |
Discussion
Two natural allopolyploidization events at the tetraploid and hexaploid levels, respectively, between relevant Triticum and Aegilops species have fostered the speciation of allohexaploid common wheat (T. aestivum L.) that was to become arguably human beings’ most important crop (Feldman 2000; Dubcovsky and Dvorak 2007). Although the speciation routes and the three diploid progenitor species involved are well established (Kihara 1954; Kerby and Kuspira 1988; Dvorak et al. 1993; Feldman et al. 1995; Feldman 2000), the molecular and genetic bases of the speciation process leading to formation of T. aestivum remain largely mysterious (Feldman and Levy 2009).
Studies conducted over the last two decades have unveiled novel, non-Mendelian paradigms associated with the initial stages of allopolyploidization in plants. Among these, the most striking are rapid genetic, epigenetic, and gene expression dynamics associated with nascent allopolyploidy, which are proposed to have played important roles in the immediate stabilization and longer-term establishment of newly formed allopolyploids as new species (Wendel 2000; Liu and Wendel 2003; Levy and Feldman 2004; Ma et al. 2004; Pontes et al. 2004; Adams and Wendel 2005b; Feldman and Levy 2005; Chen and Ni 2006; Adams 2007; Chen 2007; Otto 2007; Hegarty and Hiscock 2008; Feldman and Levy 2009; Jones and Hegarty 2009). Paradoxically, not all newly formed plant allopolyploids are associated with rapid genomic changes, albeit they all represent established species (Hufton and Panopoulou 2009; Jackson and Chen 2009). For example, negligible genomic instability was detected in a set of newly formed cotton allopolyploids involving various parental combinations at different ploidy levels (Liu et al. 2001), and complete genetic stability at the microsatellite loci was found in a large number of newly synthesized allohexaploid wheat lines (Mestiri et al. 2010). On the other hand, alteration in gene expression appeared to be generally associated with nascent allopolyploidy in all studied cases involving diverse plant taxa, and which is proposed to result from an array of mechanisms, which play essential roles in the evolution of functional plasticity unique to allopolyploidy (Osborn et al. 2003; Riddle and Birchler 2003; Adams and Wendel 2005a,b; Comai 2005; Wang et al. 2006; Chen 2007; Doyle et al. 2008; Hegarty and Hiscock 2008; Flagel and Wendel 2009; Jackson and Chen 2009; Rapp et al. 2009; Soltis and Soltis 2009).
A hitherto unheeded but clearly important question concerning wheat evolution is what kind and to what extent the rapid genomic changes associated with the initial stages of allohexaploidization is likely consequential to speciation of T. aestivum rather than merely anomalous incidents occurring in byproduct individuals that did not contribute to the speciation process. We surmise that given the very short evolutionary history (<10,000 years) of T. aestivum, and the fact that its three constituent genomes (BB, AA, and DD) are largely distinct (Naranjo 1990; Jiang and Gill 1994), only those initial individuals that bear the same or highly similar chromosomal constitution as T. aestivum should be relevant. In this regard we note that almost all previous studies have not defined the chromosomal constitution of the newly formed allohexaploid lines at the subgenomic level, which entails multicolor GISH analysis to reveal possible intergenomic rearrangements. Consequently, if those initial individuals with unusual or anomalous intergenomic rearrangements and/or numerical changes (aneuploidy) were included in the analysis, then relevance of the detected changes to speciation of T. aestivum is questionable. This issue arises because it is inconceivable that the occurred chromosomal rearrangements would have been reverted back to the original, unchanged constitution over such a short evolutionary time span. Therefore, if the initial allohexaploidization-induced rapid genomic changes had played a role in the speciation and evolution of T. aestivum, then only those that occurred in individuals most parsimoniously recapitulating the original founders of the species are relevant.
It should be pointed out that some of the previously uncovered rapid genomic changes associated with wheat allopolyploidization such as elimination of coding and noncoding sequences have been documented as highly reproducible in randomly chosen individuals from multiple independently formed lines (Feldman et al. 1997; Liu et al. 1998a,b; Ozkan et al. 2001; Han et al. 2005). Therefore, this type of rapid genetic instability likely has played some roles in the initial establishment and eventual speciation of T. aestivum (Levy and Feldman 2004; Feldman and Levy 2005; Feldman and Levy 2009). Similarly, genome-wide changes in gene expression in the early generations of nascent allohexaploid wheat have also been documented as a general occurrence in multiple independent lines (Pumphrey et al. 2009; Akhunova et al. 2010; Chague et al. 2010; our unpublished data), so were in other studied plant taxa (reviewed in Osborn et al. 2003; Adams and Wendel 2005b; Comai 2005; Chen and Ni 2006; Adams 2007; Chen 2007; Doyle et al. 2008; Hegarty and Hiscock 2008; Flagel and Wendel 2009; Jackson and Chen 2009; Soltis and Soltis 2009).
A recent study by Kraitshtein et al. (2010) demonstrated that extraordinarily high frequencies of both genetic and DNA methylation changes occurred at genomic loci flanking a high copy-number retroelement Veju (Sanmiguel et al. 2002) across the initial generations (S1–S5) in wheat. In particular, the genetic changes were found to include rampant loss of >50% of the original element copies in the first three generations, and followed by element reinsertion, underscoring the stunning dynamics of the elements per se and/or their flanking sequences (Kraitshtein et al. 2010). In parallel, extensive DNA methylation changes were detected at the same Veju-adjacent genomic regions, and which were found to correlate with the genetic changes (Kraitshtein et al. 2010). Albeit interesting in its own right, the results are based on a single S1 plant individual and its derived progenies involving only one newly formed line; moreover, at the chromosomal level, only euploidy was verified by conventional cytological observations (Kraitshtein et al. 2010). Taken together, the relevance of the extremely high frequencies of rapid genomic changes detected in a single nascent individual to speciation of T. aestivum is debatable, because the chromosomal constitution of the studied S1 plant and its selfed progenies was not determined.
As an initial step to addressing the possible relevance of allohexaploidization-associated rapid genomic changes to speciation of T. aestivum, we conducted a parallel investigation on three independently formed allohexaploid wheat lines using combinatory studying approaches including cytological and molecular analyses. An important distinction of this investigation from all previous relevant studies is that we restricted the analysis to only those preselected individuals that bear a transgenerationally stable chromosomal constitution at the subgenomic level that is highly similar to T. aestivum. In a sharp contrast to the results of Kraitshtein et al. (2010), we found that the genomic regions flanking Veju showed very few or no genetic changes at all in these preselected individuals representing three independently formed allohexaploid lines across multiple (3–5) generations. Furthermore, even higher levels of genetic stability were detected by another TD marker targeting to genomic regions flanking a more prevalent retrotransposon (BARE-1) and complete stasis at a large number of genomic loci randomly sampled by the AFLP marker. In contrast, extensive DNA methylation repatterning occurred at all three kinds of genomic regions in all the sample plants subjected to the analysis; moreover, the altered methylation patterns are largely heritable across selfed generations, which is in agreement with the findings of Kraitshtein et al. (2010) and Yaakov and Kashkush (2011).
Taken in its entirety, our results have shown that preponderant structural genomic stasis but extensive and heritable repatterning of DNA methylation patterns are generally associated with the initial stages of allohexaploidization in wheat, as they were genotype independent and reproducibly seen in pre-selected individuals with a transgenerationally stable chromosomal constitution mimicking that of T. aestivum. In contrast, we also detected the occurrence of concomitant genetic and DNA methylation changes in a single plant individual of another newly formed allohexaploid wheat line (N. Zhao, B. Zhu, M. Li, L. Wang, L. Xu, H. Zhang, S. Zheng, B. Qi, F. Han, and B. Liu, unpublished results), which however exhibited chromosomal instability and with a chromosomal constitution substantially different from T. aestivum.
It should be mentioned that the feature of predominant genetic stasis, but extensive epigenetic remodeling being associated with the initial stages of allopolyploidy, has also been documented in several other plant taxa, including Spartina (Salmon et al. 2005), Brassica (Lukens et al. 2006; Gaeta et al. 2007), and Senecio (Hegarty et al. 2011). Moreover, the classic hybrid- and allopolyploid-associated phenomenon of “nucleolar dominance,” i.e., selectively silencing rRNA genes of one parental origin in a hybrid/allopolyploid genome, has been mechanistically established as being dictated by localized epigenetic difference (Chen and Pikaard 1997; Tucker et al. 2010). Taken together, these results have pointed to the intriguing possibility that epigenetic mechanisms may indeed have played a prominent role in allopolyploidy-mediated speciation and evolution in plants, which warrants further investigation.
Acknowledgments
We are grateful to Moshe Feldman and George Fedak for providing the initial plant lines used in this study. We are also indebted to two anonymous reviewers for their constructive comments to improve the manuscript. This work was supported by the National High-Tech Program of China (2010AA1000686001) and the Program for Introducing Talents to Universities (B07017).
Literature Cited
- Adams K. L., 2007. Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 98: 136–141 [DOI] [PubMed] [Google Scholar]
- Adams K. L., Wendel J. F., 2005a. Novel pattems of gene expression in polyploid plants. Trends Genet. 21: 539–543 [DOI] [PubMed] [Google Scholar]
- Adams K. L., Wendel J. F., 2005b. Polyploidy and genome evolution in plants. Curr. Opin. Plant. Biol. 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Akhunova A. R., Matniyazov R. T., Liang H., Akhunov E. D., 2010. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chague V., Just J., Mestiri I., Balzergue S., Tanguy A. M., et al. , 2010. Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 187: 1181–1194 [DOI] [PubMed] [Google Scholar]
- Chen Z. J., 2007. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Ni Z., 2006. Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. Bioessays 28: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Pikaard C. S., 1997. Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. USA 94: 3442–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Dong Z. Y., Wang Y. M., Zhang Z. J., Shen Y., Lin X. Y., et al. , 2006. Extent and pattern of DNA methylation alteration in rice lines derived from introgressive hybridization of rice and Zizania latifolia Griseb. Theor. Appl. Genet. 113: 196–205 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Flagel L. E., Paterson A. H., Rapp R. A., Soltis D. E., et al. , 2008. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Dubcovsky J., Dvorak J., 2007. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J., Terlizzi P., Zhang H. B., Resta P., 1993. The evolution of polyploid wheats: identification of the A genome donor species. Genome 36: 21–31 [DOI] [PubMed] [Google Scholar]
- Feldman M., 1993. Cytogenetic activity and mode of action of the pairing homoeologous (Ph1) gene of wheat. Crop science 33: 894–897 [Google Scholar]
- Feldman M., 2000. The origin of cultivated wheat, pp. 1–56 The Wheat Book, edited by A Bonjean, Angus W. Lavoisier Tech. & Doc, Paris [Google Scholar]
- Feldman M., Levy A. A., 2005. Allopolyploidy–a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 109: 250–258 [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A. A., 2009. Genome evolution in allopolyploid wheat–a revolutionary reprogramming followed by gradual changes. J. Genet. Genomics 36: 511–518 [DOI] [PubMed] [Google Scholar]
- Feldman M., Lupton F. G. H., Miller T. E., 1995. Wheats. Smartt J., N. W Simmonds.(eds.), Evolution of Crop Plants, 2nd Edition. Longman Scientific, London, pp. 184–192 [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A. A., et al. , 1997. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of because chromosomes. Genetics 147: 1381–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F., 2009. Gene duplication and evolutionary novelty in plants. New Phytol. 183: 557–564 [DOI] [PubMed] [Google Scholar]
- Gaeta R. T., Pires J. C., Iniguez-Luy F., Leon E., Osborn T. C., 2007. Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Fedak G., Guo W., Liu B., 2005. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R–1a) in newly synthesized wheat allopolyploids. Genetics 170: 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Liu B., Fedak G., Liu Z., 2004. Genomic constitution and variation in five partial amphiploids of wheat–Thinopyrum intermedium as revealed by GISH, multicolor GISH and seed storage protein analysis. Theor. Appl. Genet. 109: 1070–1076 [DOI] [PubMed] [Google Scholar]
- Hegarty M. J., Hiscock S. J., 2008. Genomic clues to the evolutionary success of polyploid plants. Curr. Biol. 18: R435–444 [DOI] [PubMed] [Google Scholar]
- Hegarty M. J., Batstone T., Barker G. L., Edwards K. J., Abbott R. J., et al. , 2011. Nonadditive changes to cytosine methylation as a consequence of hybridization and genome duplication in Senecio (Asteraceae). Mol. Ecol. 20: 105–113 [DOI] [PubMed] [Google Scholar]
- Huang S., Sirikhachornkit A., Su X., Faris J., Gill B., et al. , 2002. Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton A. L., Panopoulou G., 2009. Polyploidy and genome restructuring: a variety of outcomes. Curr. Opin. Genet. Dev. 19: 600–606 [DOI] [PubMed] [Google Scholar]
- Jackson S., Chen Z. J., 2009. Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 13: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Gill B. S., 1994. Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support the diphyletic origin of polyploid wheats. Chromosome Res. 2: 59–64 [DOI] [PubMed] [Google Scholar]
- Jones R. N., Hegarty M., 2009. Order out of chaos in the hybrid plant nucleus. Cytogenet. Genome Res. 126: 376–389 [DOI] [PubMed] [Google Scholar]
- Kerby K., Kuspira J., 1988. Cytological evidence bearing on the origin of the B genome in polyploid wheats. Genome 30: 36–43 [DOI] [PubMed] [Google Scholar]
- Kihara H., 1944. Discovery of the DD-analyzer, one of the ancestors of Triticum vulgare Agric. Hort. 19: 13–14 [Google Scholar]
- Kihara H., 1954. Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 19: 336–342 [Google Scholar]
- Kraitshtein Z., Yaakov B., Khasdan V., Kashkush K., 2010. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch A. R., Leitch I. J., 2008. Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483 [DOI] [PubMed] [Google Scholar]
- Levy A. A., Feldman M., 2004. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Lin. Soc. 82: 607–613 [Google Scholar]
- Liu B., Brubaker C. L., Mergeai G., Cronn R. C., Wendel J. F., 2001. Polyploid formation in cotton is not accompanied by rapid genomic changes. Genome 44: 321–330 [PubMed] [Google Scholar]
- Liu B., Vega J. M., Feldman M., 1998a. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41: 535–542 [DOI] [PubMed] [Google Scholar]
- Liu B., Vega J. M., Segal G., Abbo S., Rodova M., et al. , 1998b. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. I. Changes in low-copy non-coding DNA sequences. Genome 41: 272–277 [DOI] [PubMed] [Google Scholar]
- Liu B., Wendel J. F., 2003. Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogenet. Evol. 29: 365–379 [DOI] [PubMed] [Google Scholar]
- Lukens L. N., Pires J. C., Leon E., Vogelzang R., Oslach L., et al. , 2006. Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140: 336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. F., Fang P., Gustafson J. P., 2004. Polyploidization-induced genome variation in triticale. Genome 47: 839–848 [DOI] [PubMed] [Google Scholar]
- Manninen I., Schulman A. H., 1993. BARE-1, a copia-like retroelement in barley (Hordeum vulgare L.). Plant Mol. Biol. 22: 829–846 [DOI] [PubMed] [Google Scholar]
- McFadden E. S., Sears E. R., 1946. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37: 81–107 [DOI] [PubMed] [Google Scholar]
- Mestiri I., Chague V., Tanguy A. M., Huneau C., Huteau V., et al. , 2010. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 186: 86–101 [DOI] [PubMed] [Google Scholar]
- Naranjo T., 1990. Chromosome structure of durum wheat. Theor. Appl. Genet. 79: 397–400 [DOI] [PubMed] [Google Scholar]
- Ngezahayo F., Xu C., Wang H., Jiang L., Pang J., et al. , 2009. Tissue culture-induced transpositional activity of mPing is correlated with cytosine methylation in rice. BMC Plant Biol. 9: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn T. C., Pires J. C., Birchler J. A., Auger D. L., Chen Z. J., et al. , 2003. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147 [DOI] [PubMed] [Google Scholar]
- Otto S. P., 2007. The evolutionary consequences of polyploidy. Cell 131: 452–462 [DOI] [PubMed] [Google Scholar]
- Otto S. P., Whitton J., 2000. Polyploid incidence and evolution. Annu. Rev. Genet. 34: 401–437 [DOI] [PubMed] [Google Scholar]
- Ozkan H., Levy A. A., Feldman M., 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C., Salmon A., Zerjal T., Tenaillon M., Grandbastien M. A., et al. , 2009. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 184: 1003–1015 [DOI] [PubMed] [Google Scholar]
- Pontes O., Neves N., Silva M., Lewis M. S., Madlung A., et al. , 2004. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 101: 18240–18245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumphrey M., Bai J., Laudencia-Chingcuanco D., Anderson O., Gill B. S., 2009. Nonadditive expression of because genes is established upon polyploidization in hexaploid wheat. Genetics 181: 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B., Zhong X., Zhu B., Zhao N., Xu L., et al. , 2010. Generality and characteristics of genetic and epigenetic changes in newly synthesized allotetraploid wheat lines. J. Genet. Genomics 37: 737–748 [DOI] [PubMed] [Google Scholar]
- Rapp R. A., Udall J. A., Wendel J. F., 2009. Genomic expression dominance in allopolyploids. BMC Biol. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N. C., Birchler J. A., 2003. Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 19: 597–600 [DOI] [PubMed] [Google Scholar]
- Salamini F., Ozkan H., Brandolini A., Schafer-Pregl R., Martin W., 2002. Genetics and geography of wild cereal domestication in the near east. Nat. Rev. Genet. 3: 429–441 [DOI] [PubMed] [Google Scholar]
- Salmon A., Ainouche M. L., Wendel J. F., 2005. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14: 1163–1175 [DOI] [PubMed] [Google Scholar]
- SanMiguel P. J., Ramakrishna W., Bennetzen J. L., Busso C. S., Dubcovsky J., 2002. Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A(m). Funct. Integr. Genomics 2: 70–80 [DOI] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A., 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P. S., Soltis D. E., 2009. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60: 561–588 [DOI] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K., Osborn T. C., 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S., Vitins A., Pikaard C. S., 2010. Nucleolar dominance and ribosomal RNA gene silencing. Curr. Opin. Cell Biol. 22: 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., et al. , 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H. S., Wei N. E., Jiang H., et al. , 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. M., Dong Z. Y., Zhang Z. J., Lin X. Y., Shen Y., et al. , 2005. Extensive de Novo genomic variation in rice induced by introgression from wild rice (Zizania latifolia Griseb.). Genetics 170: 1945–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249 [PubMed] [Google Scholar]
- Yaakov B., Kashkush K., 2011. Massive alterations of the methylation patterns around DNA transposons in the first four generations of a newly formed wheat allohexaploid. Genome 54: 42–49 [DOI] [PubMed] [Google Scholar]