Abstract
The proneural transcription factor neurogenin 1 (neurog1) has been shown to be a key regulator of dorsal root ganglion (DRG) neuron development. Here we use a novel transgenic zebrafish line to demonstrate that the neural crest population that gives rise to DRG neurons becomes fate restricted to a neuronal/glial precursor before the onset of neurog1 function. We generated a stable transgenic zebrafish line that carries a modified bacterial artificial chromosome that expresses green fluorescent protein (GFP) under the control of the neurog1 promoter [Tg(neurog1:EGFP)]. In contrast to previously described neurog1 transgenic lines, Tg(neurog1:EGFP) expresses GFP in DRG neuronal precursors cells as they migrate ventrally and after their initial differentiation as neurons. Using this line, we are able to track the fate of DRG neuronal precursor cells during their specification. When Neurog1 function is blocked, either by neurog1 morpholino antisense oligonucleotide injection or in neurog1 mutants, GFP expression initiates in neural crest cells, although they fail to form DRG neurons. Rather, these cells take on a glial-like morphology, retain proliferative capacity, and express glial markers and become associated with the ventral motor root. These results suggest that, within the zebrafish neural crest, there is a fate-restricted lineage that is limited to form either sensory neurons or glia in the developing DRG. Neurog1 acts as the key factor in this lineage to direct the formation of sensory neurons.
Keywords: dorsal root ganglia, neural crest, fate restriction, neurogenin 1, sensory neuron, glia
Introduction
The neural crest has proved a rich system in which to study how an initially homogeneous population of cells is directed to adopt diverse fates (Le Douarin and Kalcheim, 1999). Although some neural crest cells (NCCs) are pleuripotent, other fate-restricted lineages give rise to specific cell types, such as neurons, glia, or melanocytes (Raible and Eisen, 1994; Henion and Weston, 1997; Henion et al., 2000; Selleck and Bronner-Fraser, 2000; Luo et al., 2003). Within the sublineage of neuronal and glial cells, there is a population of cells that form sensory neurons of the dorsal root ganglia (DRG) (Sieber-Blum, 1989; Greenwood et al., 1999). The DRG contain the sensory neurons of the peripheral nervous system that convey sensation from the body, such as touch, perception of pain (nociception), temperature, and sense of limb movement and position (proprioception) to the CNS. DRG neurons and associated glial cells arise from ventrally migrating trunk NCCs (Kalcheim, 1996; Anderson, 2000; Raible and Ungos, 2006).
In avian and mammalian embryos, the proneural transcription factors neurogenin 1 (neurog1) and neurogenin 2 (neurog2) are expressed in migrating NCCs that have not undergone neuronal differentiation but will form sensory neurons (Greenwood et al., 1999; Ma et al., 1999; Perez et al., 1999). In mouse embryos, when both neurogenin genes are functionally inactivated, DRG neurons are completely absent (Ma et al., 1999). In zebrafish, blocking the function of a single neurogenin gene, neurog1, with morpholino antisense oligonucleotides (MOs) leads to complete loss of DRG neurons as well as all neurons of the cranial ganglia (Andermann et al., 2002; Cornell and Eisen, 2002). Coupled with the fact that each DRG in the zebrafish arises from an initial population of two to five neurons (Raible et al., 1992), this genetic simplicity makes zebrafish an ideal system in which to study mechanisms of cell fate choice during neural crest specification.
Here we provide evidence that, in zebrafish, the sensory neuron-restricted lineage of neural crest in the absence of neurog1 gives rise to glial cells. We generated a transgenic zebrafish line that carries a bacterial artificial chromosome (BAC) clone modified to express the fluorescent reporter enhanced green fluorescent protein (EGFP) under the control of the regulatory elements of neurog1, Tg(neurog1:EGFP). Using this line, we visualize DRG precursor cells in living and fixed embryos. We see expression of Tg(neurog1:EGFP) in a subset of migrating trunk NCCs before overt formation of DRGs and later in DRG neurons. When we block neurog1 function, we continue to observe Tg(neurog1:EGFP)-positive migrating NCCs. These cells fail to differentiate as neurons but instead show a glial cell-like morphology and express glial-specific markers. We hypothesize that neurog1 expression directs this restricted lineage to make a binary fate choice: cells that express neurog1 form the sensory neurons of the DRG, but, in the absence of neurog1 function, these cells persist and take on a glial fate.
Materials and Methods
Generation of Tg(neurog1:EGFP) transgenic line.
We modified a neurog1-containing BAC clone by Escherichia coli-based homologous recom-bination (Zhang et al., 1998). BAC DanioKey clone DKEY-91L18 contains 17 kb of sequence upstream and 60 kb downstream of neurog1 (http://www.sanger.ac.uk/Projects/D_rerio/mapping.shtml). After recombination, the modified BAC clone contained an EGFP gene positioned at the endogenous start site of neurog1. The accuracy of recombination was evaluated by PCR, sequencing, and analysis of transient expression. To obtain a zebrafish line with germ-line incorporation, we microinjected 20 μg/ml linearized BAC DNA into zebrafish embryos, raised injected fish to adulthood, and screened their progeny for reporter gene expression. The germ-line transmission rate was 5%. The Tg(neurog1:EGFP)w61 strain has been outcrossed for six generations and transmits the transgene in a Mendelian manner.
Fish husbandry.
Embryos were obtained from natural spawning of adult zebrafish and staged according to Kimmel et al. (1995). Embryos were raised at 28.5°C in embryo medium (EM) (Westerfield, 1994). The neurog1 (neuroD3hi1089) mutant line (hereafter referred to as neurog1−/−), generated in a retroviral insertion screen (Golling et al., 2002), was subsequently crossed to the Tg(neurog1:EGFP) line and selected for neurog1+/− carriers of the Tg(neurog1:EGFP) transgene. The colorless (clst3) line contains a null mutation in the sox10 gene (Dutton et al., 2001); this line was also crossed with the Tg(neurog1:EGFP) line and selected for Tg(neurog1:EGFP); cls+/− carriers. The Tg(−3.1neurog1:GFP) transgenic line was described previously (Blader et al., 2003). The University of Washington Institutional Animal Care and Use Committee approved all procedures.
Whole-mount immunohistochemistry and RNA in situ hybridization.
For immunohistochemistry, embryos were collected at stages indicated, killed in MS-222 (3-amino benzoate methanesulfonic acid) (10 mg/ml in buffered embryo medium), and fixed in 4% paraformaldehyde in PBS for 2 h at room temperature. Antibody labeling was performed as described previously (Ungos et al., 2003). In brief, embryos were washed in PBS with 0.1% Triton X-100 (PBT) and blocked with the addition of 2% goat serum unless otherwise noted. Before blocking, embryos older than 48 h postfertilization (hpf) were made permeable with three 30-min washes in distilled water. Embryos were incubated in primary antibodies diluted in blocking solution overnight at room temperature (RT). Primary antibodies used were anti-GFP (1:700; rabbit or mouse anti-GFP; Invitrogen), anti-Elavl (1:700; mAB 16A11, also called anti-HuC/D; Invitrogen), anti-Sox10 (1:1000) (Park et al., 2005), anti-myelin basic protein (MBP) (1:50) (Lyons et al., 2005), anti-5-bromo-2-deoxyuridine (BrdU) (1:500; Becton Dickinson), anti-phosphohistone H3 (pH3,1:300; Upstate Cell Signaling Solutions), and anti-FoxD3 (1:1000 in blocking solution containing 20% goat serum) (Lister et al., 2006). Embryos were incubated in Alexa-488- or Alexa-568-conjugated secondary antibodies (Invitrogen) overnight at room temperature, rinsed in PBT, and then stored in 50% glycerol/PBS for imaging.
RNA in situ hybridization was preformed as described previously (Andermann et al., 2002). Digoxygenin-labeled antisense probe was generated for neurog1 by enzyme digestion with XhoI and synthesis with RNA polymerase T7 (Blader et al., 1997) and for neuroD probe by enzyme digestion using NotI and synthesis with T3 (Blader et al., 1997). Hybridization was performed overnight at 60°C. After development using 5-bromo-4-chloro-3-indolylphosphate and nitro blue tetrazolium, embryos were washed in PBT and then taken through immunohistochemistry with anti-GFP antibody to enhance the signal of Tg(neurog1:EGFP). Embryos were stored in 50% glycerol/PBS for imaging.
Data collection and time-lapse imaging.
Before imaging, embryos were deyolked and mounted on bridged coverslips in 50% glycerol/PBS. Images and cell counts were obtained using a Zeiss LSM-5.0 Pascal confocal microscope or a Zeiss Axioplan 2 microscope and a Spot CCD camera (Diagnostic Instruments), and whole images were processed for brightness and contrast using Adobe Photoshop (Adobe Systems). For individual cell counts, the five rostralmost DRG were analyzed in each condition. Non-neuronal cells were defined as associated with the DRG if they were in direct contact with labeled DRG neuronal cell bodies. Statistical significance was determined using a Student's t test or ANOVA depending on the experiment. To generate time-lapse movies, Tg(neurog1:EGFP) and Tg(neurog1:EGFP);neurog1−/− embryos were anesthetized in MS-222 (10 mg/ml in embryo medium), embedded in 1.5% agarose in EM, covered in EM containing MS-222, and imaged from 26 hpf to 36 or 39 hpf.
Morpholino injection.
MOs (Gene Tools) were diluted in RNase-free water, and 1–2 nl was pressure injected into embryos at the one- to two-cell stage. The neurog1 MO 5′-CCATATCGGAGTATACGATCTCCAT-3′ was designed against the neurog1 start codon and injected at 7 ng/nl, to block translation of native neurog1 message but not the Tg(neurog1:EGFP) transgene. This neurog1 MO showed a phenotype identical to that seen using previously reported neurog1 morpholinos and to the neurog1 mutant (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) (Andermann et al., 2002; Cornell and Eisen, 2002).
Cyclopamine treatment.
Tg(neurog1:EGFP) embryos were treated at 22 hpf with cyclopamine prepared as described by Ungos et al. (2003). In brief, cyclopamine (Toronto Research Chemicals) was diluted to 15 μg/ml in embryo medium from a stock of 4 mg/ml cyclopamine dissolved in 45% (w/v) 2-hydroxypropyl-β-cyclodextrin (HBC) (Sigma) and PBS. Embryos were incubated at 28.5°C in cyclopamine continuously beginning at indicated time points until they were collected for processing. Control embryos were treated with comparable concentrations of HBC in embryo medium and were indistinguishable from untreated embryos.
Cell cycle analysis.
To determine the number of cells in S-phase in the DRG, both Tg(neurog1:EGFP) control and Tg(neurog1:EGFP) neurog1 MO-injected embryos were exposed to an overnight pulse of 10 mm BrdU (Sigma) dissolved in 1% dimethylsulfoxide (DMSO) in EM beginning at 30 or 50 hpf and then collected for processing the following day. After fixation and immunolabeling for anti-GFP antibody, embryos were then refixed for 20 min at RT and washed three times for 10 min in PBS with 1% DMSO and 0.1% Tween 20 (PBDTw). BrdU incorporation was detected using a modified protocol developed in zebrafish (Harris et al., 2003). Embryos were dehydrated in a series of steps to 100% methanol, incubated a −20°C for 1 h, then rehydrated in a stepwise manner, and washed in PBDTw two times for 10 min. Embryos were permeabilized with 10 μl/mg Proteinase K (Invitrogen) in PBDTw for 20 min, refixed for 20 min in 4% paraformaldehyde, and washed three times for 10 min in PBDTw. Embryos were incubated in 1N HCl for 1 h at RT and then washed three times for 10 min in PBDTw. After washing, embryos were blocked in 10% goat serum in PBDTw for at least 1 h at RT and then incubated overnight at RT in mouse anti-BrdU IgG in blocking solution (1:100; Becton Dickinson). Embryos were washed three times for 30 min in PBDTw and incubated in Alexa-568 antibody for 6 h at RT, then washed in PBDTw, and stored in 50% glycerol/PBS until visualized as described above. Individual cells were counted in the five rostralmost DRG.
Cells in M-phase at 2 and 3 d postfertilization (dpf) were identified with the anti-pH3 antibody using the immunohistochemistry conditions described above. Because of the transient nature of pH3 expression, antibody labeling is infrequent. Therefore, all of the DRG along the length of the embryo were analyzed for pH3 labeling, and the percentage of DRG that contained at least one pH3-positive (pH3+) cell was quantified.
Results
Generation of a recombinant neurog1 BAC transgenic zebrafish line
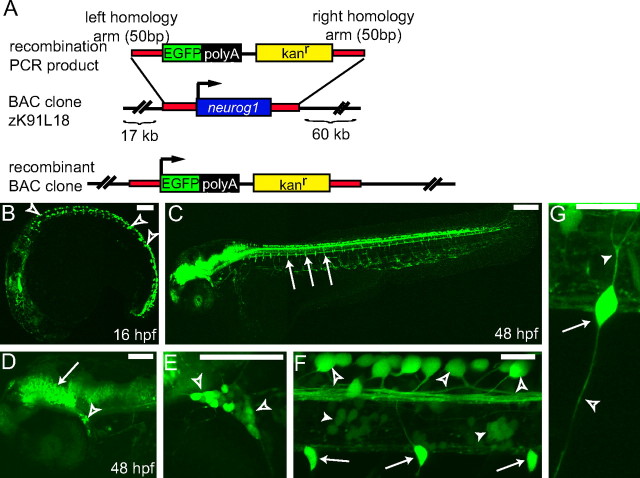
To generate a reporter line that allowed us to track DRG development, we screened the DanioKey BAC library (Koch et al., 2004) for a BAC that contained the coding sequence of neurog1; we identified the zebrafish BAC clone DKEY-91L18 (zK91L18). Mapping sequenced BAC ends onto the zebrafish genome reveals that the BAC contains 17 kb of sequence upstream and 60 kb downstream of the neurog1 coding sequence. Using bacterial recombination techniques (Zhang et al., 1998), the single exon neurog1 coding sequence in DKEY-91L18 was replaced with that of EGFP along with a kanamycin resistance cassette (Fig. 1A). The modified BAC was used to create a stable transgenic zebrafish line Tg(neurog1:EGFP). The expression pattern of GFP in this line is similar but not identical to that seen in previously reported neurog1 transgenic lines (Blader et al., 2003). In the Tg(neurog1:EGFP) line, GFP expression is visible in live embryos at 16 hpf in Rohon Beard (RB) sensory neurons and the developing CNS (Fig. 1B). At 48 hpf, GFP expression can be seen in the developing CNS in subsets of neurons in the spinal cord, the trigeminal ganglia, and RB cells (Fig. 1C–F), as well as DRG neurons arrayed along the trunk of the developing embryo (Fig. 1F,G). The central and peripheral axons of the DRG are also labeled with GFP (Fig. 1G). The BAC clone DKEY-91L18 does not contain all of the regulatory elements required for endogenous neurog1 expression; for example, with the exception of the trigeminal ganglion, all of the cranial ganglia that express neurog1 mRNA lack GFP expression (supplemental Fig. S2A, available at www.jneurosci.org as supplemental material) (Andermann et al., 2002). Ectopic GFP expression is also visible in areas in which neurog1 mRNA is not detected, including the vessels of the vascular system (supplemental Fig. S2B, available at www.jneurosci.org as supplemental material), indicating that this construct may lack some inhibitory as well as positive regulatory elements.
Figure 1.
Using BAC recombination to produce a Tg(neurog1:EGFP) transgenic line. A, Schematic showing the strategy for BAC recombination. In the BAC clone DKEY-91L18 (zK91L18) from the DanioKey library, the single coding exon of the neurog1 gene (blue bar) was replaced with an EGFP/kanamycin resistance cassette (green and black bars) by recombination using flanking homologous sequences (red bars). The EGFP gene is placed in-frame at the neurog1 start site. Schematic is not to scale. B–F, Confocal projections of live transgenic Tg(neurog1:EGFP) embryos. B, At 16 hpf, Tg(neurog1:EGFP) is expressed in developing brain, spinal neurons, and RB neurons (open arrowheads). C, By 48 hpf, Tg(neurog1:EGFP) is strongly expressed in the CNS, and DRG (arrows) neurons are visible ventral to the spinal cord along the trunk. D, In the head at 48 hpf, several regions in the brain are intensely GFP positive, including the midbrain–hindbrain boundary (arrow). A subset of cells in the trigeminal ganglia (open arrowhead) expresses Tg(neurog1:EGFP). E, GFP+ trigeminal ganglia neurons (open arrowheads). F, Close-up image showing GFP-expressing RB cells in the dorsal spinal cord and their associated axons (open arrowhead), cells in the spinal cord (arrowhead), and DRGs ventral to the spinal cord (arrow). G, DRGs containing a single GFP-expressing neuron (arrow) showing a centrally projecting axon (arrowhead) and a peripherally projecting axon (open arrowhead). Scale bars: B, D, 100 μm; C, 200 μm; E–G, 20 μm.
The Tg(neurog1:EGFP) line marks DRG precursors and nascent DRG neurons
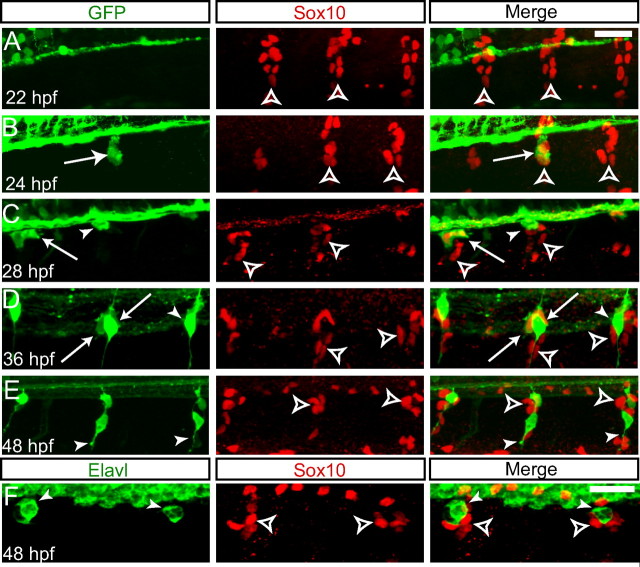
The Tg(neurog1:EGFP) transgenic line marks DRG precursors early in their development, in contrast to previously described neurog1 transgenic lines (Blader et al., 2003). At 22 hpf, segmental streams of neural crest cells are identifiable with anti-Sox10 antibody as they begin their ventral migration from the dorsal neural tube (Fig. 2A). Beginning at 24 hpf, a subset of the Sox10-positive neural crest cells begin to coexpress GFP, and we hypothesize that these GFP-positive cells are the precursors of the sensory neurons of the DRG (Fig. 2B). Beginning at 28 hpf, the GFP-positive cells begin coalesce to form rounded cell bodies (Fig. 2C), and, by 36 hpf, a subset of GFP-positive nascent DRG neurons have begun to send out axons centrally to the spinal cord and out to peripheral targets; at this stage, Sox10 has begun to be downregulated in some, but not all, GFP+ DRG cells (Fig. 2D). By 48 hpf, most GFP-positive cells have downregulated Sox10, whereas cells surrounding the DRG continue to express Sox10 (Fig. 2E). These Sox10-positive cells may represent undifferentiated neural crest cells or satellite glial cells; at the present time, no markers have been identified in zebrafish that allow definitive identification of these cells. At this stage, Sox10-negative (Sox10−)/GFP-positive cells express the Elavl antigen (Fig. 2F)
Figure 2.
Tg(neurog1:EGFP) is expressed in migrating DRG precursor cells. Formation of DRG anlagen after neural crest migration. A–E, Immunofluorescent detection of Sox10 in neural crest cells and glia (red) and GFP in developing DRG neurons (green). All images are confocal z-projections of the second to fourth rostralmost DRG. A, At 22 hpf, Sox10+ neural crest cells migrate ventrally in segmented streams (open arrowheads). B, A subset of Sox10+ neural crest cells begins to express Tg(neurog1:EGFP) at 24 hpf (arrow). C, By 28 hpf, GFP+ cells have begun to coalesce into rounded cells bodies, the majority of which coexpress Sox10 (arrows), although some GFP+ cells have downregulated Sox10 (arrowhead). Sox10+ glial cells surround the DRG neuronal precursor cells (open arrowhead). D, As the GFP+ cells differentiate as neurons at 36 hpf, they in turn downregulate Sox10 (arrowheads), whereas a subset of GFP+ cells continue to express Sox10 (arrow). E, By 48 hpf, DRG neurons have begun to send axons to both the periphery and centrally to the spinal cord (arrowheads). Sox10 is expressed in glial cells that surround the DRG neurons (open arrowheads). F, Anti-Elavl (green) and anti-Sox10 labeling in a 48 hpf Tg(neurog1:EGFP) embryo. DRG neurons are Elavl+ (arrowheads) but Sox10−, whereas glial cells continue to be labeled by Sox10 (open arrowheads). Scale bars, 20 μm.
To confirm that the GFP-positive cells form the neurons of the DRG, we used live time-lapse imaging of three Tg(neurog1:EGFP) transgenic embryos followed by immunohistochemical labeling with the anti-Elavl antibody to mark differentiated neurons. A representative Tg(neurog1:EGFP) embryo beginning at 26 hpf and continuing for 14 h is shown in Figure 3A (supplemental Movie 1, available at www.jneurosci.org as supplemental material). During this period GFP-positive cells can be seen to coalesce at the ventral edge of the spinal cord. During the later stages of development, these cells begin to extend ventral processes. After time-lapse image, these GFP-positive cells can be clearly identified by anti-Elavl antibody (Fig. 3B).
Figure 3.
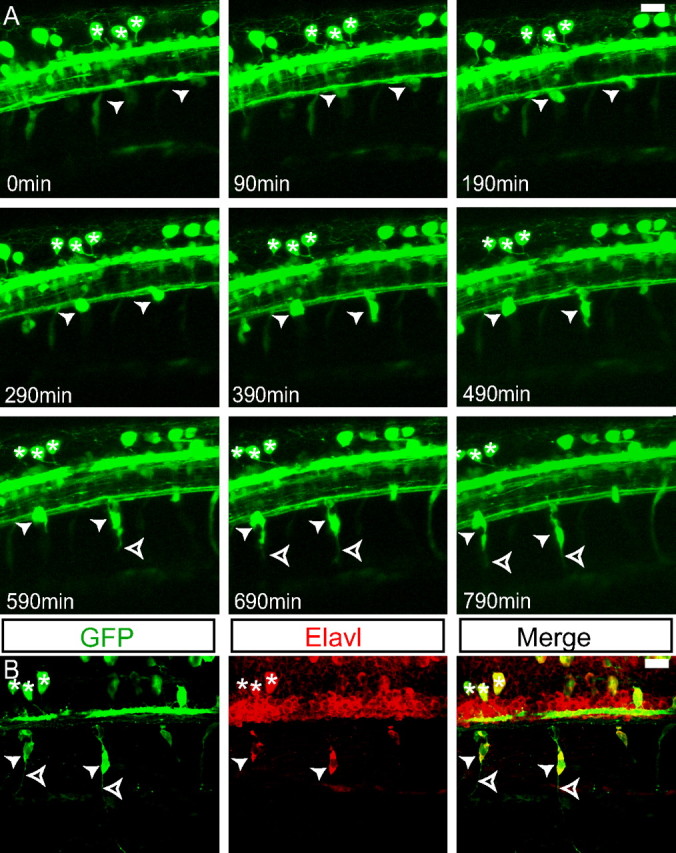
GFP-positive cells become DRG neurons. A, Individual frames of z-projections of a confocal time-lapse movie of a Tg(neurog1:EGFP) from 26 to 39 hpf. GFP-positive DRG precursor cells become visible at the ventral edge of the spinal cord (arrowheads). The same embryo is shown in supplemental Movie 1 (available at www.jneurosci.org as supplemental material). As the cells mature, they send out projections (open arrowheads). A group of RB neurons is maintained throughout the duration of the imaging (asterisks) and serves as a landmark for registration. B, Immunolabeling of the embryo at 40 hpf. The DRG cells from A are labeled with both anti-GFP and anti-Elavl antibodies, indicating that they have differentiated as neurons (arrowheads). Axons extend ventrally from the DRG neurons (open arrowheads). The group of RB neurons remains identifiable (asterisks). Scale bars, 20 μm.
Previous studies examining neurog1 expression during the development of the zebrafish DRG did not analyze time points before 30 hpf (Andermann et al., 2002; Cornell and Eisen, 2002; Ungos et al., 2003). As we see expression of the Tg(neurog1:EGFP) in the forming DRG before this period, we set out to correlate the Tg(neurog1:EGFP) with neurog1 RNA expression by in situ hybridization (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). The rostralmost DRG precursors showed neurog1 expression beginning at 26 hpf and continuing through 36 hpf. By 48 hpf, only occasional cells in the DRG express neurog1. Although transgene expression in combination with immunohistochemistry may allow visualization of DRG precursors slightly earlier than neurog1 in situ hybridization (24 vs 26 hpf), for the most part, Tg(neurog1:EGFP) shows reasonable fidelity to endogenous neurog1 expression.
Initial addition of neurons to the DRG
We followed the time course of neuronal addition in DRG during the first 4 d of development by monitoring expression of the pan-neuronal marker Elavl in the Tg(neurog1:EGFP) transgenic line. At 36 hpf, DRGs contain an average of two GFP-positive cells per ganglion, and, on average, one of these cells also expresses Elavl (Fig. 4A). Both GFP-positive and Elavl-positive neurons continue to be added to the DRG as the embryo develops (Fig. 4B); by 72 hpf, a subset of Elavl-positive cells has begun to downregulate Tg(neurog1:EGFP) expression (Fig. 4C). By 96 hpf, almost all GFP-positive cells also express Elavl, whereas the number of Elavl+ cells that have downregulated GFP expression has grown (Fig. 4D). A small number of cells express GFP alone, perhaps indicating that these are newly added cells that will give rise to neurons. Quantification of neuronal addition is shown in Figure 4E. Together, these results suggest that neural crest cells first turn on neurog1 and then downregulate Sox10 and express Elavl as they differentiate into DRG neurons, suggesting that neurog1 expression is the earliest step in sensory neuron specification. We note that GFP expression in our transgenic line is only a surrogate for endogenous Neurog1 expression, and we use it here to document the dynamics of DRG neuron addition.
Figure 4.
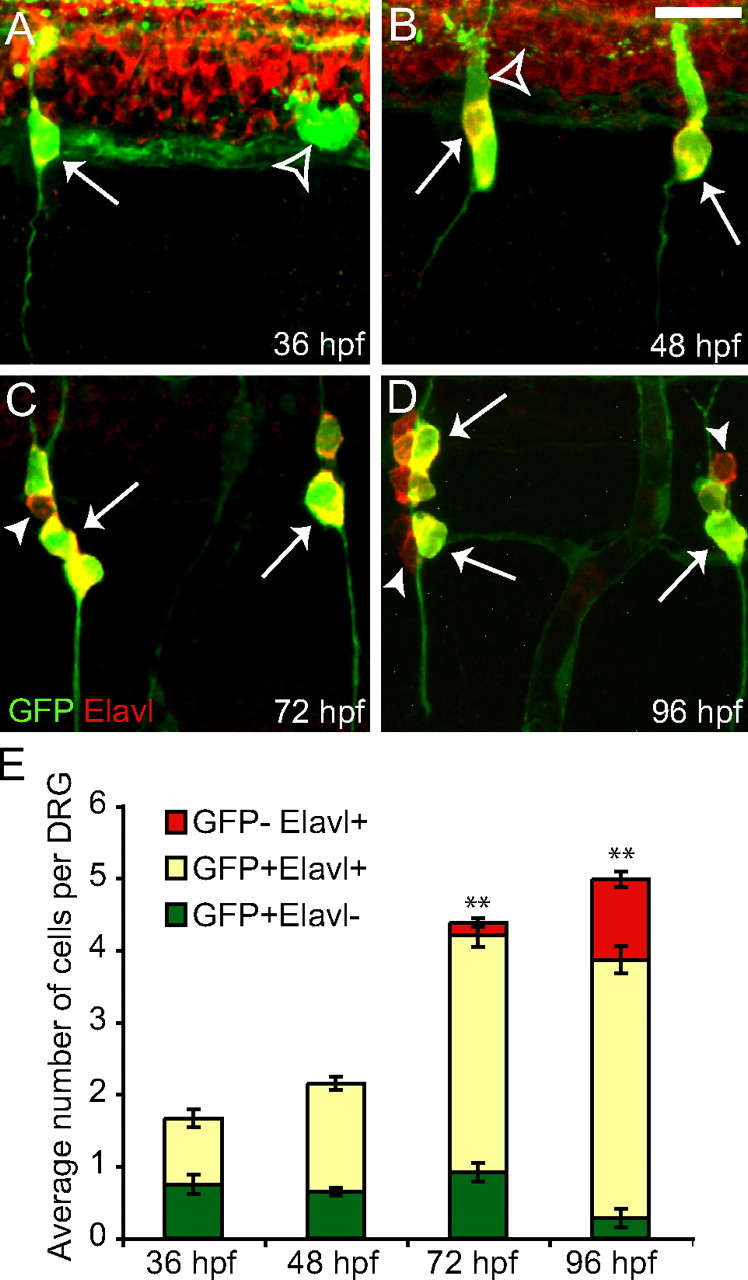
Tg(neurog1:EGFP) is transiently expressed in nascent DRG neurons. A–D, Immunofluorescent labeling of Tg(neurog1:EGFP) embryos between 36 and 96 hpf showing GFP (green) and Elavl (red). A, The DRGs initially form from one or two GFP+ cells per ganglion (open arrowhead), and a subset are also Elavl+ (arrow) at 36 hpf. B, At 48 hpf, all cells in the DRG are GFP+ (open arrowhead) and an increasing number of cells are also Elavl+ (arrow). C, By 72 hpf, a small number of Elavl+ DRG neurons have begun to downregulate GFP (arrowhead), although the majority of neurons colabel with GFP and Elavl (arrow), a trend that continues through 96 hpf as more cells become Elavl+ and GFP− (D). E, Quantification (mean ± SEM) of the contribution of GFP+ and Elavl+ cells to the DRG (cells from the 5 rostralmost DRGs were counted in each embryo; n = 12 embryos for 36 and 48 hpf; n = 11 for 72 and 96 hpf). The bar represent the average total number of cells in each DRG divided into those that only express GFP (green bars), those that express both GFP and Elavl (yellow bars), and those that only express Elavl (red bars). There is a significant increase in the total number of cells in the DRG (**p < 0.0001, ANOVA). The number of cells that express GFP alone significantly decrease over the same period (p=0.004, ANOVA). Scale bars, 20 μm.
The expression of the Tg(neurog1:EGFP) transgene in migrating neural crest cells contrasts with the expression of a previously described transgenic zebrafish line in which GFP is controlled by −3.1 kb of the upstream neurog1 regulatory sequence [Tg(−3.1neurog1:GFP)] (Blader et al., 2003). In the Tg(−3.1neurog1:GFP) line, GFP is not expressed in DRG neuronal precursor cells (supplemental Fig. S4A, available at www.jneurosci.org as supplemental material). GFP expression is not seen in the DRG until 36 hpf, at a stage when nascent DRG neurons have begun to express the transcription factor NeuroD (Ungos et al., 2003) and continues through 48 hpf (supplemental Fig S4C,E, available at www.jneurosci.org as supplemental material). GFP-positive cells are Sox10-negative, confirming that transgene expression commences with neuronal differentiation in the Tg(−3.1neurog1:gfp) line. Inactivation of neurog1 function using antisense morpholino oligonucleotides abolishes Tg(−3.1neurog1:gfp) expression in the DRG. These results suggest that there are positive autoregulatory elements in the neurog1 gene, as have been described previously for the related basic helix–loop–helix (bHLH) transcription factor atoh1 (Helms et al., 2000), although regulation might not be direct.
Following precursor formation in embryos with disrupted DRG development
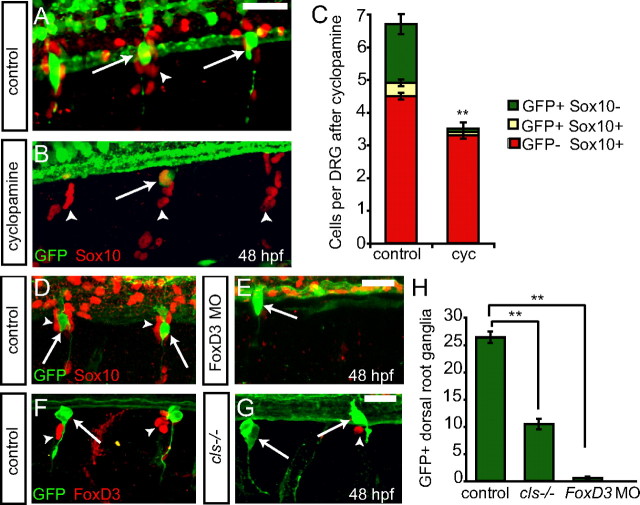
Previous work suggests that Hedgehog (Hh) signaling pathway is required for the formation of zebrafish DRG neurons and is required near the onset of neurog1 expression for proper sensory neuron development (Ungos et al., 2003). We treated Tg(neurog1:EGFP) embryos with the Hh antagonist cyclopamine or the vehicle HBC. Embryos treated with cyclopamine beginning at 22 hpf until 48 hpf lack the vast majority of GFP-positive DRG precursor cells, with only an occasional GFP-positive cell in the developing DRG (Fig. 5A,B). Although there is a significant reduction in the number of GFP+ cells after cyclopamine treatment, Sox10-positive cells remain present in treated embryos (Fig. 5C). These results support the idea that Hh signaling is required for the initiation of neurog1 expression and, by extension, specification of DRG neuronal precursors but acts downstream of neural crest specification and migration. Although Sox10+ cells remain in the position of the DRG, it is not possible to distinguish between undifferentiated Sox10-positive neural crest cells and Sox10-positive satellite glia. These results contrast with those seen after mutation of erbb3, in which no neural-crest-derived cells appear to remain in the position of the DRG (Honjo et al., 2008).
Figure 5.
Factors that act upstream of neurog1 block Tg(neurog1:EGFP) cells in the DRG. A, B, Tg(neurog1:EGFP) embryos at 48 hpf treated with cyclopamine or HBC at 22 hpf and labeled with anti-GFP (green) and anti-Sox10 (red). A, An HBC-treated control embryo showing GFP-labeled DRG neurons and Sox10+ glia. B, Cyclopamine treatment starting at 22 hpf leads to a loss of nearly all GFP expression in the DRG (arrow), although glial cells remain present (arrowhead). C, Quantification (±SEM) of the GFP+ neurons and Sox10+ glial cells in the DRG. There is a significant decrease in the number of GFP+ cells in cyclopamine-treated embryos. (n = 11; **p < 0.0001, Student's t test). D, E, Confocal z-projections of 48 hpf Tg(neurog1:EGFP) embryos injected with foxd3 MO and labeled with anti-GFP (green) and anti-Sox10 (red). D, Control uninjected embryo with GFP+ DRG neurons (arrows) and Sox10+ glia (arrowhead). E, foxd3 morphant embryo with a rare GFP+ DRG neuron (arrow). No Sox10+ glial cells are associated with the DRG. F, G, Confocal images of 48 hpf cls−/−; Tg(neurog1:EGFP) and control Tg(neurog1:EGFP) embryos labeled with anti-GFP (green) and anti-Foxd3 (red). F, Control embryo showing GFP+ DRG neurons (arrow) and FoxD3+ glia (red). G, The rostralmost GFP+ DRG neurons are present in cls−/− embryos (arrow), whereas the vast majority of Foxd3+ cells are absent (arrowhead). H, Comparison of the number of GFP+ DRG (±SEM) in control, foxd3 MO, and cls−/− embryos, showing the loss of most DRG in both experimental conditions (n = 10; **p < 0.0001, Student's t test). Scale bars, 20 μm.
The transcription factors FoxD3 and Sox10 have been described to act upstream of neurog1 expression in the developing DRG, with their loss of function leading to a loss of most but not all DRG neurons (Carney et al., 2006; Lister et al., 2006). Additionally, these factors are also required for the formation of neural-crest-derived glial cells and continue to be expressed in this lineage (Britsch et al., 2001; Dutton et al., 2001; Lister et al., 2006; Montero-Balaguer et al., 2006; Stewart et al., 2006). We monitored the development of DRG sensory neuron precursor cells after disruption of Foxd3 or Sox10 expression using the Tg(neurog1:EGFP) transgenic line. At 48 hpf, embryos lacking FoxD3 activity after foxd3 MO injection showed a loss of most DRG neurons as marked by Tg(neurog1:EGFP) as well as a loss of the vast majority of Sox10-positive cells (Fig. 5D,E,H). When DRG development is analyzed in the zebrafish sox10 mutant colorless (cls) carrying the Tg(neurog1:EGFP) transgene, we found that most DRG neurons were absent, and many that remain show disorganized process growth (Fig. 5F–H), probably because of the lack of glial cells described previously in sox10 mutants (Britsch et al., 2001; Dutton et al., 2001; Carney et al., 2006). Although we observed occasional Foxd3-positive cells in sox10 mutants, these cells were GFP negative. Together, these results provide additional support for the idea that Hedgehog signaling, Sox10, and Foxd3 all act upstream of neurog1 expression to specify DRG precursors.
Cell fate changes in the absence of neurog1 function
We reasoned that the early expression of the Tg(neurog1:EGFP) BAC transgene in migrating DRG neuron precursors might allow us to follow the fates of cells in the absence of neurog1 function. For these studies, we followed GFP expression in animals injected with neurog1 MO (Andermann et al., 2002; Cornell et al., 2002) or in animals with a retrovirus insertion into the neurog1 gene that disrupts neurog1 function (Golling et al., 2002). These neurog1 mutant animals have the same loss of cranial ganglia (supplemental Fig. S1A–C, available at www.jneurosci.org as supplemental material) and RB neuron phenotypes as neurog1 MO-injected animals (supplemental Fig. S1D–G, available at www.jneurosci.org as supplemental material). The loss of cranial ganglia neurons in the neurog1 mutant has been described in greater detail by Caron et al. (2008). Both the neurog1 MO and neurog1 mutant embryos fail to express neuroD at 36 hpf (supplemental Fig. S1H–J, available at www.jneurosci.org as supplemental material).
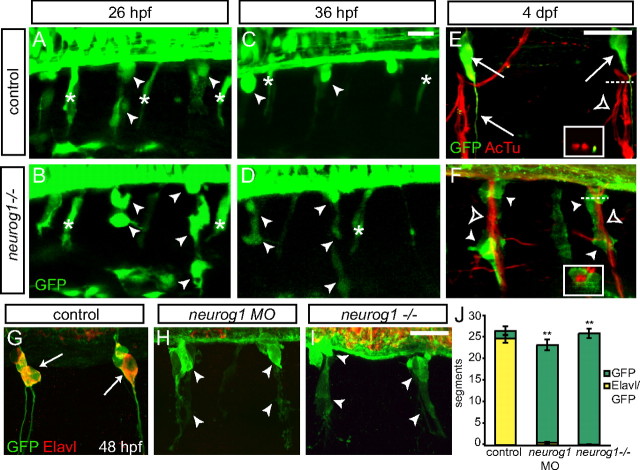
We used live time-lapse imaging of Tg(neurog1:EGFP) embryos to track the development of DRG neurons. Imaging began at 26 hpf during the period of late neural crest migration, after GFP has begun to be expressed in DRG precursor cells but before their coalescence at the ventral edge of the spinal cord (Fig. 2B). GFP-positive cells are found in similar locations with similar elongated morphologies of migrating neural crest cells in both control (Fig. 6A) and neurog1−/− embryos (Fig. 6B). By 36 hpf in control embryos, most of the GFP-positive neural crest cells have formed nascent DRGs (Fig. 6C). In contrast, in neurog1−/− embryos, GFP-positive cells are still visible, but rather than coalescing at the edge of the spinal cord, they remain elongated (Fig. 6D). By 48 hpf, most of the GFP-positive cells in the DRG of control embryos express the neuronal marker Elavl and have sent out central and peripheral axons (Fig. 6G). In contrast, GFP-positive cells in either neurog1 morphant or neurog1−/− embryos do not express the Elavl antigen and fail to send out axons (Fig. 6H,I). When the total number of segments that contain GFP-positive cells along the trunk of the embryo are quantified, the numbers in neurog1 MO and neurog1−/− do not differ significantly from controls. In the absence of neurog1 function, very few GFP+ cells express Elavl, indicating that they have not differentiated as neurons (Fig. 6J). In addition, GFP-positive neural crest cells also change their location in the absence of neurog1 function. Unlike the GFP-positive DRG neurons in control embryos that reside dorsolateral to the ventral motor roots of the spinal cord (Fig. 6E), GFP-positive cells in neurog1−/− embryos become associated with the ventral roots and in some instances enwrap them (Fig. 6F). These results lead us to hypothesize that these cells have switched to a glial fate.
Figure 6.
Tg(neurog1:EGFP)-expressing cells persist in the absence of neurog1 function. A–D, Live time-lapse imaging of the forming rostralmost DRG in Tg(neurog1:EGFP) embryos from 26 to 36 hpf in the presence and absence of neurog1 function. At 26 hpf, GFP+ DRG precursors migrate ventrally (arrowhead) in both control (A) and neurog1−/− (B) embryos. By 36 hpf, GFP+ DRG cells have coalesced at the ventral edge of the spinal cord (C). In contrast, GFP+ cells in the neurog1−/− embryo remain elongated (D). E, F, Confocal z-projections of 4 dpf Tg(neurog1:EGFP) and neurog1−/− embryos labeled with anti-GFP (green) to label DRG and anti-acetylated tubulin to label ventral motor roots (red). E, In control embryos, GFP+ DRG neurons and their axons (arrows) are located adjacent to the ventral roots (open arrowhead). Inset, Cross-section of the ventral root taken at the level denoted by the dashed line showing the proximity of DRG axons to the motor axons. F, In contrast, GFP+ cells (arrowheads) in neurog1−/− embryos become tightly associated with the ventral root (open arrowhead). Inset, Cross-section of the ventral root taken at the level denoted by the dashed line indicating that, in the neurog1 mutant, the GFP+ cells enwrap the ventral root. G, H, Confocal images of 48 hpf Tg(neurog1:EGFP) embryos in the presence or absence of neurog1 function label with anti-GFP (green) and anti-Elavl (red). G, In control embryos, the majority of GFP+ DRG neurons are also labeled with Elavl (arrow). In both neurog1 morphant embryos (H) and neurog1−/− embryos (I), GFP+ cells are present (arrowheads) but have an elongated morphology compared with control and are not labeled with Elavl. J, Quantification (±SEM) of the total number of segments along the trunk of the embryo that contain GFP+ (green bars) and Elavl+/GFP+ (yellow bars) clusters in the position of the DRG in control, neurog1 MO, and neurog1−/− embryos (n = 10 in all conditions). Although there is only a minor decrease in the number of GFP+ segments after loss of neurog1 function, there is a dramatic decrease in the number of Elavl+/GFP+ segments compared with control (**p < 0.0001, ANOVA). Scale bars, 20 μm.
To determine whether cells that would normally differentiate as DRG neurons have instead formed glial cells in the absence of neurog1 function, we examined whether Tg(neurog1:EGFP)-expressing cells displayed glial markers. Although Sox10 and FoxD3 are initially expressed in all trunk neural crest cells, including those that will give rise to neurons or glia, they are later expressed only in glial cells (Carney et al., 2006; Lister at al., 2006). Foxd3 is downregulated in neural crest cells before initial expression of Tg(neurog1:EGFP). FoxD3 continues to be expressed in cells surrounding the DRG, which are either unspecified neural crest cells or satellite glial cells (Fig. 7A,B). In neurog1 mutant animals, GFP-positive cells coexpress Foxd3 (Fig. 7B). Sox10 is coexpressed with GFP at the earliest stages of DRG formation (Fig. 2C–E) but is then downregulated in DRG neurons and expressed in cells surrounding the DRG (Fig. 7C). In contrast, Sox10 expression is maintained with loss of neurog1 function (Fig. 7D). Although the total number of cells that comprise the DRG shows a small but significant decrease in neurog1 MO and neurog1−/− embryos compared with controls, the number of cells that coexpress Sox10 and GFP is greatly increased in embryos lacking neurog1 function (Fig. 7E).
Figure 7.
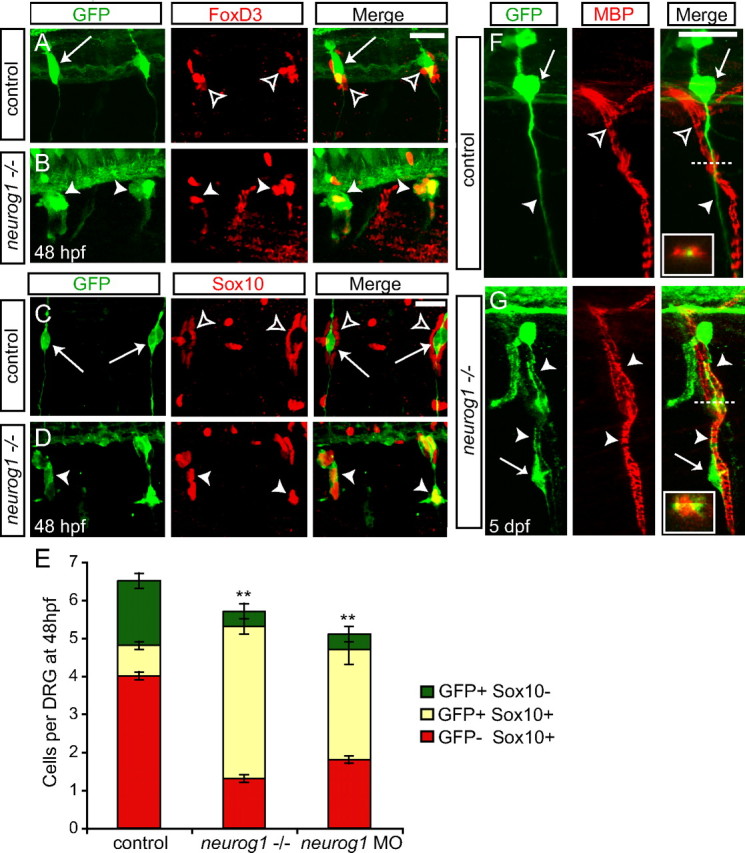
In the absence neurog1 function, GFP+ cells express glial markers. A, B, Confocal images of 48 hpf Tg(neurog1:EGFP) embryos with or without neurog1 MO injection and labeled with anti-GFP (green) and anti-Foxd3 (red). A, Foxd3 is expressed in glial cells associated with the DRG (open arrowhead) but is not coexpressed with GFP in DRG neurons in control embryos (arrow). B, In neurog1 morphant embryos, Foxd3 is coexpressed with GFP (arrowhead) in the DRGs. There remains a population of Foxd3+ glia that is not GFP+ (open arrowhead). C, D, Confocal images of a 48 hpf neurog1−/− embryo and a wild-type or heterozygous sibling labeled with anti-GFP (green) and anti-Sox10 (red). As with Foxd3, Sox10 marks glial cells in the DRG at 48 hpf. C, In control embryos, Sox10+ glia (open arrowheads) surround GFP+ DRG neurons (arrows). D, In contrast, GFP+ cells in neurog1−/− embryos coexpress Sox10 (arrowheads). E, Quantification (±SEM) of GFP + and Sox10+ cells in control embryos and in the absence of neurog1 function at 48 hpf. Each bar represents the number of GFP+ (green bars) cells in the DRG, plus the number of cells that are GFP+/Sox10+ (yellow bars) and the number of Sox10+ cells that are in direct contact with the DRG cells (red bars). On average, the total number of cells in the DRG decreases in neurog1 MO and neurog1−/− embryos compared with controls (p=0.04, ANOVA). There is not a significant difference in the total number of Sox10+ cells between neurog1 MO, neurog1−/−, and uninjected control embryos (control, neurog1 MO embryos, n = 10; neurog1−/−, n = 11). Significantly more cells are GFP+ and Sox10+ in the absence of neurog1 function compared with controls (**p < 0.0001, ANOVA). F, G, Images of 5 dpf neurog1 mutant and sibling embryos labeled with anti-GFP (green) and anti-MBP (red) to mark myelinating Schwann cells. Insets, Cross-sections of ventral roots taken at the level of the dashed line. F, A GFP+ DRG neuron (arrow) and its peripheral axon (arrowhead) are located adjacent to, but are not associated with, MBP+ Schwann cells surrounding the ventral motor root (open arrowhead). The inset shows the GFP+ axon in close proximity to MBP+ Schwann cells. G, In the neurog1 mutant, GFP+ cells (arrows) enwrap the ventral motor root and are MBP+ (arrowhead). The inset shows the coexpression of GFP and MBP in the neurog1−/− embryo. Scale bars, 20 μm.
Because both FoxD3 and Sox10 are markers of both neural crest cells and glial cells and it has not been established when the transition from neural crest to glia occurs in the zebrafish DRG, we used an additional glial marker. To determine whether GFP/Sox10-positive cells in embryos lacking neurog1 function simply remain undifferentiated or whether they instead become Schwann cells, we examined expression of MBP (Lyons et al., 2005). At 5 dpf, MBP marks the Schwann cells that are associated with the ventral motor roots (Brosamle and Halpern, 2002). In control embryos, the GFP-positive axons of the DRG run parallel to, but are not associated with, the ventral roots and are not labeled with MBP (Fig. 7F). In neurog1−/− embryos, GFP-positive cells that have become associated with the ventral roots coexpress MBP, suggesting that at least some of the GFP-positive cells have become Schwann cells (Fig. 7G).
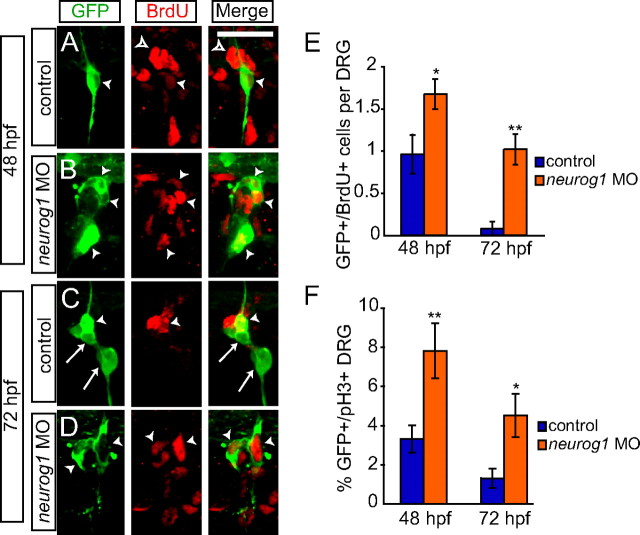
Changes in proliferation in the absence of neurog1 function
Differentiated sensory neurons in the zebrafish DRG show a very low level of cellular proliferation as the embryo develops (An et al., 2002). In contrast, zebrafish peripheral glial cells are derived from larger neural crest cell clones (Raible and Eisen, 1994), and, in other organisms, peripheral glial cells are typically highly proliferative (Jessen and Mirsky, 2005). To determine whether GFP-positive cells alter their rates of proliferation in embryos lacking neurog1 function, we assayed BrdU incorporation that marks cells in S-phase and expression of anti-pH3 antibody that marks cells in M-phase. We performed a BrdU pulse during two stages: during day 2 of development (30–48 hpf), beginning before most DRG precursor cells differentiate as neurons, and during day 3 (50–72 hpf), when most DRG neurons have begun to express the neuronal marker Elavl. In control embryos, some GFP+ cells were derived from cells that proliferate during the first period of BrdU treatment (Fig. 8A); an average of one cell of the approximately three that make up the DRG has incorporated BrdU (Fig. 8E). GFP+ cells had rarely incorporated BrdU during the later period (Fig. 8C,E). In contrast, embryos injected with the neurog1 MO show a significantly higher level of BrdU incorporation on both days (Fig. 8B,E), including robust incorporation on 3 dpf. These findings are supported by analysis using anti-pH3 antibody. Although pH3-positive cells are rare, there is still a significant increase in labeling when neurog1 function is blocked. We examined the total number of DRG that contained at least one pH3-labeled cell. Approximately 3% of DRG in controls show pH3 labeling, whereas 7.8% of DRG in neurog1 MO embryos are labeled with pH3 (p < 0.01). Considered together, these data indicate that the GFP-positive glial-like cells that form in the absence of neurog1 function act like satellite glia during early DRG development, and later at least a subset become associated with the ventral motor root, much like myelinating Schwann cells.
Figure 8.
GFP+ cells are mitotically active after neurog1 MO injection. A–D, Tg(neurog1:EGFP) embryos and neurog1 morphants labeled with anti-GFP (green) and anti-BrdU (red) to mark proliferating cells in S-phase. A, At 48 hpf, BrdU labels a GFP+ DRG neuron (arrowhead). Several BrdU+/GFP− cells can be seen surrounding the DRGs (open arrowhead). B, In a neurog1 MO embryo at 48 hpf, several GFP+ cells are also labeled with BrdU (arrows). C, At 72 hpf, a subset of GFP+ cells is also BrdU+ (arrowhead), although many GFP+ cells are not BrdU+ (arrow). D, At the same time point, GFP+/BrdU+ cells (arrowhead) can be seen in a neurog1 MO embryo. E, Quantification (±SEM) of GFP+ and BrdU+ cells 48 and 72 hpf. At 48 hpf, there are significantly more GFP+/BrdU+ cells in neurog1 MO-injected embryos (n = 10; orange bars) compared with controls (n = 12; blue bars; *p = 0.02, Student's t test). At 72 hpf, very few GFP+/BrdU+ cells are seen in control embryos (n = 9), whereas significantly more GFP/BrdU+ are still seen in neurog1 MO embryos (n = 10; **p = 0.0002, Student's t test). F, Comparison of anti-GFP antibody labeled DRG that contain at least one anti-pH3-labeled cell in the DRG of control (blue bars) and neurog1 MO (orange bars) embryos at 48 and 72 hpf (n = 15 embryos for all conditions). The percentage of pH3+/GFP+ DRG seen in neurog1 MO embryos compared with control is significantly high at both 48 hpf (**p < 0.005, Student's t test) and at 72 hpf (*p < 0.05, Student's t test). Scale bars, 20 μm.
Discussion
In this study, we show that, in zebrafish, a fate-restricted population of neural crest cells will form neurons of the DRG and that the proneural factor neurog1 is a key player in directing cell fate decisions within this population. We used our BAC transgenic zebrafish, Tg(neurog1:EGFP), to monitor the subset of migrating neural crest cells that give rise to the sensory neurons of the DRG and show that these precursor cells persist in the absence of neurog1 function to take on the attributes of myelinating glial cells. These cells also show an increase in proliferation compared with controls, which may indicate that neurog1 expression is required in the neural crest to restrict the fate of precursor cells to postmitotic neurons. Our data, together with the fact that loss of neurog1 function does not affect other neural-crest-derived cell types, suggest that there is a subpopulation of neural crest that is restricted to a sensory neuron/glial cell lineage before the expression of neurog1. These results are consistent with previous research documenting that, within the population of trunk neural crest, there exists cells that are fate restricted to give rise to specific classes of progeny (Raible and Eisen, 1994; Henion and Weston, 1997; Luo et al., 2003).
The expression of neurogenin appears to be the critical factor to direct neural crest cells preferentially toward sensory neuron fates. Our current study and work by others has shown that loss of zebrafish neurog1 function leads to a loss of DRG sensory neurons, whereas other neural-crest-derived neurons of the peripheral nervous system are unaffected (Andermann et al., 2002; Cornell and Eisen, 2002). A similar role for the neurogenins has been described in mouse and chick, indicating that neurog1 and neurog2 are required for the development of DRG sensory neurons from neural crest (Greenwood et al., 1999; Ma et al., 1999; Perez et al., 1999; Parras et al., 2002). Both neurog1 and neurog2 are expressed in a subset of migrating neural crest cells before their differentiation and later in nascent DRG neurons, suggesting that neurogenin expression may be acting to restrict these cells to a neuronal lineage (Ma et al., 1999; Perez et al., 1999). Overexpression studies suggest that neurogenin biases migrating neural crest to localize to the DRG and to express sensory neuron-specific markers (Perez et al., 1999). Moreover, ectopic expression of neurogenins in non-neural-crest-derived tissue elicited expression of sensory neuron-specific markers (Ma et al., 1996; Blader et al., 1997; Perez et al., 1999). In the mouse embryo, neurog1 and neurog2 are expressed in temporally distinct, but overlapping waves, and can be partially compensatory for one another; knockdown of both genes is required for complete loss of DRG neurons (Ma et al., 1999). Although alterations in cell fate were not investigated, an increase in terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling-positive cells was observed in neurog2−/− mice, confirming that some cells undergo apoptosis in the absence of proneural factors (Ma et al., 1999). In contrast to this previous study, we see little cell loss (Fig. 6E); rather, cells fated to become DRGs instead became glial cells in the absence of neurogenin function.
A role for neurogenin in directing binary cell fate decisions between neurons and glia has been suggested to occur in the CNS (Bertrand et al., 2002; Ross et al., 2003; Miller and Gauthier, 2007). In the developing cortex, neurons and then glia are born from multipotent precursors in a sequential manner (Qian et al., 2000; Shen et al., 2006). Expression of neurog1, neurog2, and mash1 play essential roles in directing cell fate decisions (Nieto et al., 2001; Britz et al., 2006). Using LacZ expression under the control of the neurog2 promoter to trace the fate of cortical cells in Mash1−/−, neurog2−/− mice, Nieto et al. (2001) found a significant decrease in the number of cortical neurons and an increase in the number of astrocytes, leading the authors to propose a model in which the proneural factors are acting to both promote a neuronal cell fate and to restrict a glial cell fate in cortical progenitor cells.
A mechanism underlying the dual function of neurog1 in promoting neural differentiation while inhibiting glia in multipotent progenitor cells has been described in detail for cultured rat cortical neurons (Sun et al., 2001). During neurogenesis, Neurog1 acts to turn on another proneural bHLH factor, neuroD, which in turn promotes neuronal differentiation (for review, see Bertrand et al., 2002). The coactivator proteins p300/CBP are recruited by Neurog1 to the neuroD promoter to active its expression (Sun et al., 2001). By sequestering p300/CBP, neurog1 also inhibits glia differentiation. The expression of glial fibrillary acid protein (GFAP) is activated by the cooperation of Smad1 and Stat1/3 proteins that in turn require p300/CBP as a coactivator. When Neurog1 sequesters available p300/CBP to the neuroD promoter, there is insufficient p300/CBP to activate the GFAP promoter. It is possible that a similar process may function during neuron versus glial cell fate decision during DRG development.
We found that loss of neurog1 resulted in increased cell division in addition to promotion of glial differentiation. Peripheral glial cells are typically highly proliferative (Jessen and Mirsky, 2005), so changes in proliferation rates in DRG precursors may simply reflect their acquisition of glial fates. Increased proliferation may also reflect the block in neuronal differentiation, because these cells become postmitotic. Expression of neurogenin genes in Xenopus primary neurons has been suggested to promote cell cycle exit through regulation of the cyclin-dependent kinase inhibitor p27 (Vernon et al., 2003) or its activating kinase (Souopgui et al., 2002), so loss of neurog1 would be predicted to allow cell cycle reentry. Alternatively, cells may retain characteristics of undifferentiated neural crest precursors. For example, the continued expression of Sox10 after loss of neurogenin function may maintain cells in a proliferative multipotent state (Kim et al., 2003). Our results suggest that, in the absence of neurog1 function, at least a subset of cells differentiate as Schwann cells, although these observations do not exclude the possibility that others remain undifferentiated.
Our results differ somewhat from those of Zirlinger et al. (2002), who followed the fates of mouse neural crest cells using an inducible Cre recombination system to permanently mark cells that express neurog2 at various stages during DRG formation. In that study, cells that expressed neurog2 were significantly more likely to be found in the DRGs than in sympathetic ganglia, indicating that neurog2 expression drives neural crest toward a sensory fate. However, cells that expressed neurog2 were equally likely to form DRG sensory neurons or satellite glia. These results suggest that, in mouse, there is a fate-restricted neural crest lineage that is biased toward a sensory ganglion fate rather than a neuronal fate. There are several possible explanations for the differences between this study and our findings. There may be differences in the sensitivity of the lineage tracing systems: the Cre–Lox marking system used in mice may be more sensitive and mark transiently expressing cells, whereas detectable GFP expression from the zebrafish transgene may require more prolonged gene transcription. Alternatively, expression of mouse neurog2 may not be sufficient to direct all cells to neuronal fates without expression of additional neurogenic factors, including neurog1, complementary functions that that are subsumed by the single zebrafish neurogenin gene. It is interesting to note however that, in both systems, the marked cells have fates limited to sensory neurons and glia. Together, these results suggest that signals upstream of neurogenin have limited neural crest cell fates to these lineages.
One crucial question that needs to be answered is how neurog1 expression is initiated in neural crest cells to direct them to become DRG precursors. Sox10 and FoxD3 have been shown to regulate neurog1 expression in the developing zebrafish DRG (Carney et al., 2006; Lister et al., 2006). Although these factors play important roles in mouse DRG development (Sonnenberg-Riethmacher et al., 2001; Teng et al., 2008), regulation of neurogenin genes has not been established. The receptor tyrosine kinases ErbB2 and ErbB3 have also been shown to be required for neurog1 expression and for the formation of both glial cells and DRG neurons (Honjo et al., 2008). It remains to be established whether these factors act directly to control neurog1 expression or whether they act by regulating other potential regulators. Additionally, all of these factors act in both neurons and glia. The Hh signaling pathway is also a good candidate for a key regulator of neurog1 during DRG development in the zebrafish and is not required for glial formation (Ungos et al., 2003). Additional research is needed to determine whether these regulatory pathways are required for sensory neurons as they are added later during DRG development. Because the zebrafish DRG continues to develop well into adulthood (An et al., 2002), it may prove to be a useful model in which to identify how neurog1 is directly regulated in the DRG.
Footnotes
This work was supported by National Institutes of Health Grants 2 T32 HDO7183 and 5 R01 NS057220. We thank Bruce Appel, Will Talbot, Nancy Hopkins, and Uwe Strähle for reagents and Branden Nelson for helpful comments on this manuscript. We thank David White for excellent fish care. We thank the University of Washington zebrafish community for their input and advice.
References
- An M, Luo R, Henion PD. Differentiation and maturation of zebrafish dorsal root and sympathetic ganglion neurons. J Comp Neurol. 2002;446:267–275. doi: 10.1002/cne.10214. [DOI] [PubMed] [Google Scholar]
- Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251:45–58. doi: 10.1006/dbio.2002.0820. [DOI] [PubMed] [Google Scholar]
- Anderson DJ. Genes, lineages and the neural crest: a speculative review. Philos Trans R Soc Lond B Biol Sci. 2000;355:953–964. doi: 10.1098/rstb.2000.0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Blader P, Fischer N, Gradwohl G, Guillemot F, Strähle U. The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development. 1997;124:4557–4569. doi: 10.1242/dev.124.22.4557. [DOI] [PubMed] [Google Scholar]
- Blader P, Plessy C, Strähle U. Multiple regulatory elements with spatially and temporally distinct activities control neurogenin1 expression in primary neurons of the zebrafish embryo. Mech Dev. 2003;120:211–218. doi: 10.1016/s0925-4773(02)00413-6. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz O, Mattar P, Nguyen L, Langevin LM, Zimmer C, Alam S, Guillemot F, Schuurmans C. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16(Suppl 1):i138–i151. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- Brösamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Carney TJ, Dutton KA, Greenhill E, Delfino-Machín M, Dufourcq P, Blader P, Kelsh RN. A direct role for Sox10 in specification of neural crest-derived sensory neurons. Development. 2006;133:4619–4630. doi: 10.1242/dev.02668. [DOI] [PubMed] [Google Scholar]
- Caron SJ, Prober D, Choy M, Schier AF. In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development. 2008;135:3259–3269. doi: 10.1242/dev.023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002;129:2639–2648. doi: 10.1242/dev.129.11.2639. [DOI] [PubMed] [Google Scholar]
- Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113–4125. doi: 10.1242/dev.128.21.4113. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Greenwood AL, Turner EE, Anderson DJ. Identification of dividing, determined sensory neuron precursors in the mammalian neural crest. Development. 1999;126:3545–3559. doi: 10.1242/dev.126.16.3545. [DOI] [PubMed] [Google Scholar]
- Harris JA, Cheng AG, Cunningham LL, MacDonald G, Raible DW, Rubel EW. Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio) J Assoc Res Otolaryngol. 2003;4:219–234. doi: 10.1007/s10162-002-3022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Henion PD, Weston JA. Timing and pattern of cell fate restrictions in the neural crest lineage. Development. 1997;124:4351–4359. doi: 10.1242/dev.124.21.4351. [DOI] [PubMed] [Google Scholar]
- Henion PD, Blyss GK, Luo R, An M, Maynard TM, Cole GJ, Weston JA. Avian transitin expression mirrors glial cell fate restrictions during neural crest development. Dev Dyn. 2000;218:150–159. doi: 10.1002/(SICI)1097-0177(200005)218:1<150::AID-DVDY13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Honjo Y, Kniss J, Eisen JS. Neuregulin-mediated ErbB3 signaling is required for formation of zebrafish dorsal root ganglion neurons. Development. 2008;135:2615–2625. doi: 10.1242/dev.022178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Kalcheim C. The role of neurotrophins in development of neural-crest cells that become sensory ganglia. Philos Trans R Soc Lond B Biol Sci. 1996;351:375–381. doi: 10.1098/rstb.1996.0031. [DOI] [PubMed] [Google Scholar]
- Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003;38:17–31. doi: 10.1016/s0896-6273(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Koch R, Rauch GJ, Humphray S, Geisler TR, Plasterk R. Bacterial artificial chromosome (BAC) clones and the current clone map of the zebrafish genome. Methods Cell Biol. 2004;77:295–304. doi: 10.1016/s0091-679x(04)77016-8. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The neural crest. Ed 2. Cambridge, UK: Cambridge UP; 1999. [Google Scholar]
- Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol. 2006;290:92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Luo R, Gao J, Wehrle-Haller B, Henion PD. Molecular identification of distinct neurogenic and melanogenic neural crest sublineages. Development. 2003;130:321–330. doi: 10.1242/dev.00213. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, Arana N, Jacobs J, Talbot WS. erbb3 and erbb2 are essential for Schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15:513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, Anderson DJ. Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller FD, Gauthier AS. Timing is everything: making neurons versus glia in the developing cortex. Neuron. 2007;54:357–369. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- Montero-Balaguer M, Lang MR, Sachdev SW, Knappmeyer C, Stewart RA, De La Guardia A, Hatzopoulos AK, Knapik EW. The mother superior mutation ablates foxd3 activity in neural crest progenitor cells and depletes neural crest derivatives in zebrafish. Dev Dyn. 2006;235:3199–3212. doi: 10.1002/dvdy.20959. [DOI] [PubMed] [Google Scholar]
- Nieto M, Schuurmans C, Britz O, Guillemot F. Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron. 2001;29:401–413. doi: 10.1016/s0896-6273(01)00214-8. [DOI] [PubMed] [Google Scholar]
- Park HC, Boyce J, Shin J, Appel B. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J Neurosci. 2005;25:6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez SE, Rebelo S, Anderson DJ. Early specification of sensory neuron fate revealed by expression and function of neurogenins in the chick embryo. Development. 1999;126:1715–1728. doi: 10.1242/dev.126.8.1715. [DOI] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Raible DW, Eisen JS. Restriction of neural crest cell fate in the trunk of the embryonic zebrafish. Development. 1994;120:495–503. doi: 10.1242/dev.120.3.495. [DOI] [PubMed] [Google Scholar]
- Raible DW, Ungos JM. Specification of sensory neuron cell fate from the neural crest. Adv Exp Med Biol. 2006;589:170–180. doi: 10.1007/978-0-387-46954-6_10. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodsdon W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cells in the embryonic zebrafish. Dev Dyn. 1992;195:29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Bronner-Fraser M. Avian neural crest cell fate decisions: a diffusible signal mediates induction of neural crest by the ectoderm. Int J Dev Neurosci. 2000;18:621–627. doi: 10.1016/s0736-5748(00)00037-x. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Sieber-Blum M. Commitment of neural crest cells to the sensory neuron lineage. Science. 1989;243:1608–1611. doi: 10.1126/science.2564699. [DOI] [PubMed] [Google Scholar]
- Sonnenberg-Riethmacher E, Miehe M, Stolt CC, Goerich DE, Wegner M, Riethmacher D. Development and degeneration of dorsal root ganglia in the absence of the HMG-domain transcription factor Sox10. Mech Dev. 2001;109:253–265. doi: 10.1016/s0925-4773(01)00547-0. [DOI] [PubMed] [Google Scholar]
- Souopgui J, Sölter M, Pieler T. XPak3 promotes cell cycle withdrawal during primary neurogenesis in Xenopus laevis. EMBO J. 2002;21:6429–6439. doi: 10.1093/emboj/cdf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174–188. doi: 10.1016/j.ydbio.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungos JM, Karlstrom RO, Raible DW. Hedgehog signaling is directly required for the development of zebrafish dorsal root ganglia neurons. Development. 2003;130:5351–5362. doi: 10.1242/dev.00722. [DOI] [PubMed] [Google Scholar]
- Vernon AE, Devine C, Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development. 2003;130:85–92. doi: 10.1242/dev.00193. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene, OR: University of Oregon; 1994. [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- Zirlinger M, Lo L, McMahon J, McMahon AP, Anderson DJ. Transient expression of the bHLH factor neurogenin-2 marks a subpopulation of neural crest cells biased for a sensory but not a neuronal fate. Proc Natl Acad Sci U S A. 2002;99:8084–8089. doi: 10.1073/pnas.122231199. [DOI] [PMC free article] [PubMed] [Google Scholar]