Free-radical scavengers that are known to be effective protectors of proteins in solution are found to increase global radiation damage to protein crystals. Protective mechanisms may become deleterious in the protein-dense environment of a crystal.
Keywords: radiation damage, scavengers, radioprotection, global damage, site-specific damage
Abstract
Recent studies have defined a data-collection protocol and a metric that provide a robust measure of global radiation damage to protein crystals. Using this protocol and metric, 19 small-molecule compounds (introduced either by cocrystallization or soaking) were evaluated for their ability to protect lysozyme crystals from radiation damage. The compounds were selected based upon their ability to interact with radiolytic products (e.g. hydrated electrons, hydrogen, hydroxyl and perhydroxyl radicals) and/or their efficacy in protecting biological molecules from radiation damage in dilute aqueous solutions. At room temperature, 12 compounds had no effect and six had a sensitizing effect on global damage. Only one compound, sodium nitrate, appeared to extend crystal lifetimes, but not in all proteins and only by a factor of two or less. No compound provided protection at T = 100 K. Scavengers are ineffective in protecting protein crystals from global damage because a large fraction of primary X-ray-induced excitations are generated in and/or directly attack the protein and because the ratio of scavenger molecules to protein molecules is too small to provide appreciable competitive protection. The same reactivity that makes some scavengers effective radioprotectors in protein solutions may explain their sensitizing effect in the protein-dense environment of a crystal. A more productive focus for future efforts may be to identify and eliminate sensitizing compounds from crystallization solutions.
1. Introduction
Radiation damage to protein crystals remains a significant obstacle to structure determination by X-ray crystallography (Garman & Nave, 2002 ▶; Nave & Garman, 2005 ▶; Garman & Owen, 2006 ▶; Ravelli & Garman, 2006 ▶; Garman & McSweeney, 2007 ▶; Holton, 2009 ▶; Garman, 2010 ▶). This has become increasingly true as improvements in X-ray beam intensity/brilliance and in detector technology have enabled low-background measurements on smaller and smaller crystals (Coulibaly et al., 2007 ▶). At the temperatures typically used in cryocrystallography (∼100 K), the fraction of a complete data set that can be collected from a crystal before its diffraction degrades excessively depends upon the X-ray energy, the unit-cell size and symmetry, the number of unit cells within the illuminated volume and the atomic composition of the crystal (especially the concentrations of S, metals and other high-Z elements) as reflected in its mass–energy absorption coefficient (Haas & Rossmann, 1970 ▶; Matthews, 1977 ▶; Hope, 1988 ▶; Henderson, 1990 ▶; Young et al., 1993 ▶; Gonzalez & Nave, 1994 ▶; Teng & Moffat, 2002 ▶; Owen et al., 2006 ▶; Kmetko et al., 2006 ▶; Holton, 2009 ▶; Holton & Frankel, 2010 ▶).
Existing evidence indicates that all crystals are equally radiation sensitive at T ≃ 100 K: overall or ‘global’ damage depends only upon the radiation dose (Kmetko et al., 2006 ▶; Owen et al., 2006 ▶; Meents et al., 2007 ▶), although there can be significant variations in the extent of site-specific damage (Weik et al., 2000 ▶; Burmeister, 2000 ▶; Ravelli & McSweeney, 2000 ▶). All crystals are much more radiation sensitive at room temperature (Blake & Phillips, 1962 ▶; Southworth-Davies & Garman, 2007 ▶; Nave & Garman, 2005 ▶; Warkentin & Thorne, 2010 ▶). Unlike at low temperatures, anecdotal evidence suggests that crystal-to-crystal (protein-to-protein) variations in global sensitivity can be large.
Because radiation damage imposes limitations on data collection and interpretation, several approaches have been explored to mitigate it, including cooling to very low temperatures (Hope, 1988 ▶, 1990 ▶; Hanson et al., 2002 ▶; Teng & Moffat, 2002 ▶; Kmetko et al., 2006 ▶; Chinte et al., 2007 ▶; Meents et al., 2007 ▶, 2010 ▶; Warkentin & Thorne, 2010 ▶) and using crystals smaller than the mean free path of electrons generated following the initial X-ray absorption event (Nave & Hill, 2005 ▶; Stern et al., 2009 ▶; Finfrock et al., 2010 ▶).
Building upon extensive work on proteins in solution, cells and whole organisms, the use of scavengers and other ‘radioprotecting’ compounds has received particular attention. Early reports of successful protection employed styrene (Zaloga & Sarma, 1974 ▶) and polyethylene glycol (Cascio et al., 1984 ▶). Recent X-ray diffraction studies have screened a large number of potential radioprotectants (Murray & Garman, 2002 ▶; Kauffmann et al., 2006 ▶; Southworth-Davies & Garman, 2007 ▶; Borek et al., 2007 ▶; Macedo et al., 2009 ▶; Barker et al., 2009 ▶; Nowak et al., 2009 ▶). Radiation damage and its potential mitigation have also been examined using spectroscopic methods sensitive to radicals and to site-specific damage caused by, for example, photoreduction, including EXAFS (Yano et al., 2005 ▶), NEXAFS (Corbett et al., 2007 ▶), UV–Vis (McGeehan et al., 2009 ▶; Owen et al., 2009 ▶; Macedo et al., 2009 ▶), Raman (McGeehan et al., 2007 ▶; Carpentier et al., 2007 ▶) and EPR (Utschig et al., 2008 ▶) spectroscopies. Identification of radioprotectors and the mechanisms by which they provide protection would aid in the design of more effective compounds.
To date, no broadly applicable radioprotectors for protein crystals have been found. Several compounds have been reported to be effective, but often only for a single protein or in a particular temperature range. Protective effects found in one study are often not replicated in others. Protective effects evident using one damage metric may disappear when alternative metrics are used.
The difficulty of assessing radioprotection efficacy has several causes. Firstly, any protective effects in protein crystals are, for reasons to be discussed later, likely to be small. Secondly, metrics chosen to assess radiation damage may be unreliable. Damage has been monitored in reciprocal space via changes in total or individual reflection intensities, R factors, unit-cell volumes, mosaicity and absolute and relative B factors (Murray & Garman, 2002 ▶; Owen et al., 2006 ▶; Kmetko et al., 2006 ▶; Meents et al., 2007 ▶, 2010 ▶; Garman, 2010 ▶), in real space via changes in electron density at specific sites (Weik et al., 2000 ▶; Burmeister, 2000 ▶) and via changes in other metrics such as the pairwise decay factor R d (Sliz et al., 2003 ▶; Diederichs, 2006 ▶) and native structure factors R R (Borek et al., 2007 ▶). Damage should be assessed not versus time, frame number or incident X-ray flux but as a strict function of dose (energy deposited per unit mass; see, for example, Murray et al., 2004 ▶; Kmetko et al., 2006 ▶; Holton & Frankel, 2010 ▶). Calculating the dose requires knowledge of all of the atomic constituents within the unit cell, estimated from crystallography and quantified by chemical analysis (Murray et al., 2004 ▶; Kmetko et al., 2006 ▶).
Thirdly, errors in data collection can easily overwhelm expected effects. A non-square (e.g. Gaussian) beam-intensity profile will nonuniformly irradiate and damage the crystal. If the crystal is larger than the beam footprint, nonuniform irradiation will occur as the crystal is oscillated, especially if the center of rotation of the crystal does not remain aligned with the beam axis and/or if the angle through which the crystal is oscillated is large (Schulze-Briese et al., 2005 ▶; Kmetko et al., 2006 ▶). Crystal slippage and flutter, drift in beam position and intensity, and detector drift can all introduce substantial errors.
Here, we describe a systematic study of 19 small-molecule compounds as potential radioprotectors using our previously defined data-collection protocol and metric of global radiation damage (Kmetko et al., 2006 ▶; Warkentin & Thorne, 2010 ▶). No compound provides protection at cryogenic temperatures and only nitrate appears to provide protection at room temperature. We discuss why compounds that are known to be effective protectors in protein solutions and in vivo have negligible or negative effects on proteins in crystals.
2. Mechanisms of radiation damage and protection
As discussed in detail elsewhere (Teng & Moffat, 2000 ▶; Garman, 2010 ▶; Warkentin & Thorne, 2010 ▶), radiation damage can be roughly separated into primary and secondary damage. Primary damage results from X-ray absorption or inelastic scattering, producing a shower of secondary electrons and a cascade of radiochemical reactions on a timescale fast compared with that for diffusive atomic motions (Teng & Moffat, 2000 ▶). Secondary damage involves mostly diffusive motions of solvent, radicals, side chains, conformational subunits and entire molecules and largely freezes out at low temperatures (Warkentin & Thorne, 2010 ▶).
The damage to a given molecule can also crudely be distinguished as either direct or indirect (Dertinger & Jung, 1970a ▶). In direct damage, damage to a molecule is due to inelastic X-ray scattering by that molecule and/or to direct interaction with energetic primary and secondary electrons. In indirect damage, damage to a molecule is a consequence of interaction with other damaged molecules and their radiolytic products. This distinction is clearest for proteins in dilute solution, where indirect damage results from X-ray absorption by and/or primary excitation of the solvent; radicals produced in the solvent then diffuse to and react with the protein. The most important radiation-produced solvent radicals at room temperature include solvated electrons, hydroxyl, hydroperoxyl, oxygen and hydrogen radicals produced by radiolysis of water and hydrogen radicals from the protein itself.
Small-molecule compounds can reduce radiation damage in at least two ways. In competitive protection, the compound competes with the protein for diffusing radicals and scavenges reactive radiolysis products from the solvent. In restitutive protection, the compound helps to repair chemical damage to specific protein residues.
The radioprotective effects of small molecules on proteins in dilute aqueous solution and in the amorphous solid state and on living organisms has been a major focus of the radiation chemistry literature (Dertinger & Jung, 1970a ▶; Box, 1972 ▶; Livesay & Reed, 1987 ▶; Saha et al., 1995 ▶; Filali-Mouhim et al., 1997 ▶; Hategan et al., 2001 ▶; Kempner, 2001 ▶; Durchschlag et al., 2003 ▶; Shalaev et al., 2003 ▶; Audette-Stuart et al., 2005 ▶). At the molecular level, the goal is generally to protect conformational integrity and biological function. Reaction rate constants in dilute solutions can be determined by pulse radiolysis, spin trapping and ESR spectroscopy (Box, 1972 ▶; Garrison, 1987 ▶). Rates of inactivation of enzymatic function, denaturation and main-chain scission as well as analysis of radiolytic products (e.g. molecular fragments) can quantify protective effects (Saha et al., 1995 ▶). At the cellular or organismal level, the goal of protection is typically to increase survival rates after a given dose. These goals are different from that in macromolecular crystallography, which is to accurately preserve the local and global molecular structure and packing so as to maximize the resolution and fidelity of the resulting electron-density maps at a given dose. Consequently, the performance of a small-molecule compound in solution or in vivo may not be predictive of its performance in crystallography.
2.1. Selection of small-molecule compounds
Any potentially radioprotective compound introduced into a protein crystal by soaking or cocrystallization must be water soluble, compatible with the crystallization solvent and buffers, compatible with the protein and nonhazardous. These criteria eliminate many powerful free-radical traps and inhibitors that are normally soluble only in organic solvents. The X-ray absorption edges of the compound’s constituent elements should not interfere with the data-collection strategy: for example, a sulfur-containing compound could interfere with SAD phasing and cause additional damage. A radioprotective compound ideally should be universal, with protective effects that are not confined to a small subset of proteins.
Based upon these criteria, we selected the following 19 compounds for study.
Cysteamine, cysteine and cystamine. Examples of sulfur-containing aminothiols and their disulfides that are effective cellular protectants (Coggle, 1983 ▶). The generally accepted mechanism of cellular protection is via repair: H atoms from the sulfhydryl compounds can be transferred to the biological free radical. Cysteine can also stop chain reactions of organic peroxy radicals by reconstituting biological molecules (Coggle, 1983 ▶).
Thiourea. A sulfur-containing compound known to scavenge hydroxyl radicals.
1,4-Dithiothreitol (DTT). A thiol-containing molecule commonly used as a radioprotector.
β-Nicotinamide adenine dinucleotide (NADH). An electron donor.
Hydroquinone (HQ) and 4-methoxyphenol (MEHQ). Both readily quench radicals produced by photoinitiators and are often used as stabilizers, reducing agents, antioxidants and intermediates (Filali-Mouhim et al., 1997 ▶; Durchschlag et al., 2003 ▶; Shalaev et al., 2003 ▶; Audette-Stuart et al., 2005 ▶).
N-tert-Butyl-α-phenylnitrone (PBN). A commonly used free-radical spin trap.
Ascorbic acid. A powerful reducing agent (electron donor) previously tested in protein crystals (Murray & Garman, 2002 ▶; Barker et al., 2009 ▶; Macedo et al., 2009 ▶).
Glutathione. A good scavenger of hydroxyl radicals and singlet oxygen that can also play a repairing role.
Styrene, polyethylene glycol 200 (PEG 200) and hydroxyethyl methacrylate (HEMA). Undergo polymerization in the presence of radical initiators. Radiolytic products can be consumed by the condensation reaction of the polymer. Styrene (Zaloga & Sarma, 1974 ▶) and PEG (Cascio et al., 1984 ▶), a common crystallizing agent, have previously been tested in protein crystals.
Methimazole. Protects lysozyme in dilute solution (Taylor et al., 1984 ▶).
Sodium nitrate. Scavenges hydrated electrons (Audette-Stuart et al., 2005 ▶; Borek et al., 2007 ▶).
Sodium iodide and sodium bromide. Common additives used in halide-atom phasing (Dauter et al., 2000 ▶; Dauter & Dauter, 2001 ▶) but not expected to have scavenging action.
Deuterated water. Sometimes stabilizes macromolecules (Unno et al., 1989 ▶) and protects dilute solutions of enzymes from inactivation (Hategan et al., 2001 ▶).
Our selection of compounds is summarized in Table 1 ▶.
Table 1. Scavenger efficacy in lysozyme crystals at room temperature.
A change in s AD (ΔB rel/ΔD8π2) of at least 20% relative to native crystals is required for classification as a sensitizer or protector. Measurements at T = 100 K showed no protective or sensitizing effects for any molecule, with s AD values of 0.012 ± 0.0015.
| Compound | Cocrystallized (C) or soaked (S) | Concentration (mM) | Response | Coefficient of sensitivity, sAD (Å2 MGy−1) |
|---|---|---|---|---|
| None | — | Control | 0.57 | |
| Cysteamine, 1S | S | 100 | Null | 0.56 |
| Cysteine, 1S | C | 100 | Sensitizer | >3.5 |
| Cystamine, 2S | S | 200 | Null | 0.56 |
| Thiourea, 1S | C | 400 | Null | 0.57 |
| 1,4-Dithiothreitol, 2S | C | 100 | Null | 0.63 |
| β-Nicotinamide adenine dinucleotide (NADH), 2P | C | 50 | Null | 0.57 |
| Hydroquinone | C | 100 | Sensitizer | 0.75 |
| 4-Methoxyphenol (MEHQ) | 160 | Null | 0.58 | |
| N-tert-Butyl-α-phenylnitrone (PBN) | C | Sensitizer | 1.27 | |
| Ascorbic acid | C | 100 | Null | 0.57 |
| Glutathione (reduced), 1S | C | 100 | Null | 0.57 |
| Styrene | C | 100 | Sensitizer | 0.75 |
| Polyethylene glycol 200 | 100 | Sensitizer | 1.01 | |
| 2-Hydroxyethyl methacrylate (HEMA) | C | 10 | Sensitizer | 0.76 |
| Methimazole, 1S | C | 500 | Null | 0.60 |
| Sodium nitrate | S | 100 | Protector | 0.27 |
| Sodium iodide | S | 1000 | Null | 0.63 |
| Sodium bromide | S | 400 | Null | 0.68 |
| Deuterated water | C | (100%) | Null | 0.60 |
3. Methods
3.1. Crystal growth and scavenger soaks
All small-molecule compounds in this study (all ACS quality or better) and hen egg-white lysozyme (3× recrystallized; 14.4 kDa), thaumatin (22 kDa) and trypsin (23.8 kDa) were purchased from Sigma–Aldrich (St Louis, Missouri, USA) and used as received. Tetragonal lysozyme crystals (39% solvent content) were grown in Linbro plates by hanging-drop vapor diffusion. To allow the cleanest interpretation of scavenger experiments, lysozyme was dissolved in deionized water containing only 0.5 M sodium chloride and these buffer-free and cryoprotectant-free drops were suspended over wells containing 1 M sodium chloride solution. To maximize diffraction signal to noise, the lysozyme crystals selected for these experiments were 400 µm in size, as assessed by their fit into the aperture of a 400 µm MicroMount (Mitegen, Ithaca, New York, USA), and had regular shapes. Crystals of monoclinic lysozyme (33% solvent content), tetragonal thaumatin (56% solvent content) and orthorhombic trypsin (45% solvent content) were grown by hanging-drop vapor diffusion using 4%(w/v) sodium nitrate, 1 M sodium potassium tartrate and 0.2 M ammonium sulfate as precipitants, respectively (McPherson, 1999 ▶). The average size of these crystals was ∼100 µm. All small-molecule compounds were evaluated for their effects on tetragonal lysozyme; only nitrate was also evaluated using monoclinic lysozyme, thaumatin and trypsin.
Small-molecule compounds were introduced into the crystals either by direct addition to the initial crystallization medium or by post-growth soaking. For soaks, aqueous solutions of each small-molecule compound were prepared in a series of concentrations up to the aqueous solubility limit. To prevent osmotic shock, crystals were serially soaked in solutions of increasing concentration, remaining for roughly 3 min in each intermediate solution. To ensure consistent small-molecule uptake into the crystal channels, the final soaking drop had an initial volume of exactly 60 µl and the final soak lasted 3.0 min, ample time for diffusion to yield a uniform internal concentration. Crystals were then quickly (in 10 s or less) swiped through drops containing 15% glycerol in the final soaking solution and then flash-cooled in liquid nitrogen.
3.2. X-ray data collection
X-ray diffraction data were collected on beamlines A1 and F2 at the Cornell High-Energy Synchrotron Source (CHESS) using X-rays of energy 10 keV, away from any absorption edges of atoms within the crystals. The typical flux density was 1.6 × 1013 photons mm−2 s−1 on beamline A1 and 1.1 × 1012 photons mm−2 s−1 on beamline F2. The flux was continuously recorded during exposures using a standard ionization chamber calibrated against a solid-state detector. Crystals were illuminated through a D = 100 µm collimator, which produced a somewhat rounded ‘top-hat’ beam profile, and diffraction data were recorded using a Quantum 4 CCD detector (ADSC, Poway, California, USA). Radiation doses delivered to each crystal were determined using the incident flux density and the mass–energy absorption coefficient of the crystal, as calculated from the composition of the unit cell and the published absorption coefficients of each atomic constituent (Hubbell & Seltzer, 2004 ▶).
Room-temperature data collection was performed on crystals mounted on MicroMounts (Thorne et al., 2003 ▶) and then ‘covered’ with MicroRT capillaries (MiTeGen, Ithaca, New York, USA). Compared with conventional glass-capillary mounting, this mounting method minimized the liquid around the crystal and thus minimized crystal slippage. It also eliminated the optical distortion that occurs when crystals are held by liquid against capillary walls, making any sample motion much easier to optically detect (Kalinin et al., 2005 ▶). For data collection at cryogenic temperatures, crystals were mounted in MicroMounts, flash-cooled in liquid nitrogen and measured in a T = 100 K nitrogen-gas stream (Cryostream 700, Oxford Cryosystems, Devens, Massachusetts, USA).
As previously described (Kmetko et al., 2006 ▶; Diederichs, 2006 ▶; Warkentin & Thorne, 2010 ▶), any variations in illuminated crystal volume during data collection introduce substantial errors in damage measurements as more weakly damaged regions of the crystal move in and out of the beam. Our protocol for reliably and reproducibly evaluating radiation damage involves collecting data as 5° wedges and determining relative B factors from sequentially dosed data sets using the program SCALEIT (Howell & Smith, 1992 ▶) from the CCP4 crystallographic package (Winn et al., 2011 ▶) as a metric for radiation damage. Collecting only a wedge of data rather than a complete structural data set ensures uniform exposure, reproducible dose curves and expeditious collection. It requires a dose that is much smaller than the damaging dose delivered between data sets. Collecting only a wedge also minimizes rotation-induced crystal slippage during room-temperature data collection, which can be pronounced (i.e. 1 µm or larger) during the large rotations needed to collect a complete data set and which can lead to large errors in radiation sensitivity. The crystal position relative to the MicroMount and beam was carefully monitored through the beamline’s high-magnification telescope before and during data collection. Orientation matrices provided an additional check for sample motion and unit-cell parameters were monitored for any inadvertent dehydration (which produces much larger changes in cell volume than radiation damage).
We characterized the rate of global damage per unit dose using a coefficient of sensitivity to absorbed dose s AD = (ΔB rel/ΔD 8π2) (Kmetko et al., 2006 ▶; Warkentin & Thorne, 2010 ▶). This coefficient was determined from the slope of plots of relative B factor B rel versus dose D in the low-to-moderate dose regime where the relation is linear. Other metrics used to characterize global radiation damage versus dose in protein crystals include mosaicity, unit-cell expansion, intensity scale factors, intensity half-doses, the decay factor R d (Diederichs, 2006 ▶) and UV–Vis spectroscopy. Mosaicity is sensitive to the formation of cracks and other macroscopic disorder (Malkin & Thorne, 2004 ▶). Both mosaicity and cell expansion have yielded inconsistent results (Murray & Garman, 2002 ▶; Ravelli et al., 2002 ▶; Diederichs, 2006 ▶) and are not directly connected to the diffraction properties of greatest relevance in structure determination. Intensity scale factors are sensitive to flux-density variations in time and space and to sample motions, and intensity half-doses are related to the scale and B factors. Spectroscopy only probes the most optically active radiation-induced radicals (e.g. disulfides in McGeehan et al., 2009 ▶), which generally represent only a small fraction of the total radiation-induced radical population and an even smaller fraction of the chemical and structural disorder of relevance in crystallography. In our experience, which cumulatively has involved measurements on more than 200 crystals, relative B factors have provided the most consistent and reliable metric of global radiation damage.
Replacing solvent molecules with small-molecule compounds in general changes the mass–energy absorption coefficient of the crystal. This does not affect the results because damage is determined as a function of dose, not incident flux. In any case, for the small-molecule compounds studied here (excluding NaI and NaBr), the calculated changes in absorption coefficients are only a few percent.
In addition to collecting and analyzing data from 5° wedges to obtain relative B factors, for one small-molecule compound (nitrate) we collected and analyzed full structural data sets and refined these data to study the effect of the compound on molecular structure. The motivation and methods for these additional measurements are described together with the results in §4.4.
4. Results
4.1. Radiation sensitivity at cryogenic and room temperatures
As shown in Fig. 1 ▶, at room temperature plots of relative B factor B rel versus dose D deviate from linearity at large doses, with damage increasing more rapidly at large doses. This may indicate domino-like cascades of structural disordering once sufficient local damage has accumulated to destabilize the structure on larger scales (Blake & Phillips, 1962 ▶).
Figure 1.
Room-temperature data for relative B factor versus dose for lysozyme crystals soaked in six representative small-molecule compounds that had neither protecting nor sensitizing effects. See Table 1 ▶ for a full list of the compounds studied.
Previously reported increases in diffraction lifetime upon cooling to 100 K have varied widely from factors of ∼10 to ∼1000 depending upon the experimental setup and protein (Haas & Rossmann, 1970 ▶; Petsko, 1975 ▶; Hope, 1988 ▶; Henderson, 1990 ▶; Young et al., 1993 ▶; Gonzalez & Nave, 1994 ▶; Southworth-Davies et al., 2007 ▶). More recent work on several proteins (Kmetko et al., 2006 ▶; Warkentin & Thorne, 2010 ▶) has shown that dose curves at T = 100 K are linear up to doses of ∼15 MGy and that global radiation sensitivity is essentially independent of protein. The large range of cooling-related improvements in diffraction lifetimes thus arises from protein-to-protein variations in room-temperature sensitivity. Table 2 ▶ gives the ratios of T = 100 K to room-temperature radiation sensitivities that we have determined for several proteins. For the relatively well ordered, modest solvent content tetragonal lysozyme and tetragonal thaumatin crystals, the ratio is near 40. For crystals of the thiaminase enzyme TenA, which have a much higher solvent content (∼80%) and were relatively poorly ordered, our measured sensitivity ratio was roughly 1000.
Table 2. Ratio of room-temperature to low-temperature global radiation sensitivities s AD for selected proteins.
Preliminary data for urease (480 kDa, 51% solvent) gave a sensitivity ratio of 18.
| Protein | ||||
|---|---|---|---|---|
| Lysozyme | Thaumatin | Apoferritin | Bacillus subtilis TenA | |
| MW (kDa) | 14.4 | 22.2 | 476 | 27.3 |
| Solvent content (%) | 39 | 56 | 61 | ∼80 |
| Sensitivity ratio | 48 | 35 | 27 | ∼1000 |
4.2. Effect of dose rate
At T = 100 K, previous studies using global metrics of radiation damage found no dependence of damage versus dose on the dose rate (Teng & Moffat, 2002 ▶; Sliz et al., 2003 ▶; Leiros et al., 2006 ▶). At room temperature, one study reported a decrease in radiation sensitivity with increasing dose rate at very low dose rates of 6–10 Gy s−1 (Southworth-Davies et al., 2007 ▶).
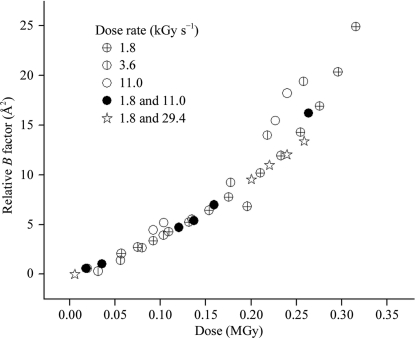
We have checked for dose-rate effects at room temperature, which is important in reliably evaluating possible radioprotective effects of small-molecule compounds. As shown in Fig. 2 ▶, we find no variation in sensitivity over a 16-fold dose-rate range corresponding to incident fluxes varying from 1.8 to 29 kGy s−1 both for native lysozyme (i.e. lysozyme without added small-molecule compounds) and for lysozyme soaked with small-molecule compounds (data not shown). Furthermore, at both room temperature and at T = 100 K we find no evidence of dark progression, i.e. of degradation of diffraction owing to, for example, post-irradiation chemistry continuing when the X-ray beam is turned off, on time scales of up to 10 h. Damage at both temperatures depends only upon dose, and not on how that dose is delivered, for the dose rates used. Consequently, flux variations owing to the gradual drop in synchrotron beam intensity during a fill and also owing to beam interruptions during fills do not affect measurements of damage, provided that integrated fluxes are carefully recorded.
Figure 2.
Room-temperature data for relative B factor versus dose for five native lysozyme crystals irradiated by a 10 keV X-ray beam at different dose rates. No variation of slope with dose rate is observed for dose rates differing by as much as 16; only the total dose is relevant. For the 11.0 kGy s−1 data set (the four circle-based symbols), the beam was turned off for 10 h between the first and second data points and between the third and fourth data points. No dark progression is observed: damage picks up where it left off prior to the break. The slight increase in B after the break arises from the small dose received in acquiring each data point.
4.3. Efficacy of small-molecule compounds
Table 1 ▶ gives the room-temperature radiation sensitivity of tetragonal lysozyme crystals soaked or cocrystallized with various small-molecule compounds, as well as of native small-molecule-free lysozyme crystals. Of the 19 additives, 12 (including deuterated water) showed no effect and six had a sensitizing effect on tetragonal lysozyme. Experiments were typically repeated on three crystals for each additive. Experiments using three of the sensitizers (styrene, HEMA and cysteine) were repeated on at least six crystals each to confirm the effect. Among the sensitizers, PEG 200 is most significant, as it is routinely added to protein crystallization solutions. Of the seven S-containing compounds, none had a protecting effect and one had a sensitizing effect. None of the compounds had any effect at cryogenic temperatures, with s AD values of 0.012 ± 0.0015 Å2 MGy−1. This is consistent with the freeze-out of radical mobility and the large drop in scavenger efficacy in dilute solution observed at low temperatures.
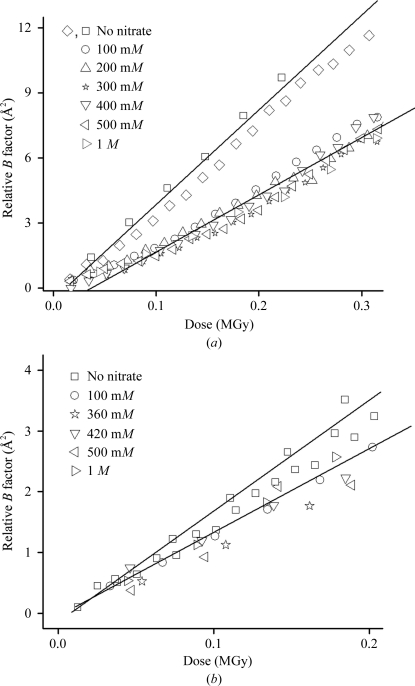
The only compound to show a statistically significant protecting effect at room temperature was sodium nitrate. To confirm this protecting effect, dose curves were measured for several lysozyme, thaumatin and trypsin crystals soaked with different nitrate concentrations. As shown in Fig. 3 ▶, a protective effect is observed at all concentrations that we examined for lysozyme and thaumatin. The magnitude of the effect (from the ratio of the slopes of the solid lines in Fig. 3 ▶) was a factor of 2 for tetragonal lysozyme, 1.6 for thaumatin and 1.6 for monoclinic lysozyme. However, no protection was observed for trypsin. Soaking and cocrystallization of nitrate were compared for tetragonal lysozyme and gave the same results. Site-specific preferential damage to nitrate ions in monoclinic lysozyme crystals has been observed by Borek et al. (2007 ▶).
Figure 3.
(a) Relative B factor versus dose for lysozyme crystals soaked in sodium nitrate and for two native crystals at room temperature. Experimental uncertainties are reflected in the difference between the native crystals. The solid lines are guides to the eye, indicating the overall trend of damage rates without and with nitrate. (b) Relative B factor versus dose for nitrate-soaked thaumatin crystals at room temperature.
4.4. Site-specific and structural changes
To explore differences in site-specific damage owing to sodium nitrate, full data sets were collected from tetragonal lysozyme crystals at both T = 100 K and room temperature before (‘fresh’) and after (‘dosed’) receiving a large radiation dose (1.9 MGy at T = 100 K and 0.12 MGy at room temperature) for both native nitrate-free crystals and crystals soaked in 100 mM nitrate. Frames were integrated using MOSFLM (Leslie, 1992 ▶) and integrated reflections were then truncated and scaled using TRUNCATE and SCALA (Winn et al., 2011 ▶). Structures were refined using BUSTER (Bricogne et al., 2011 ▶) with water positions adjusted, deleted and added as necessary, and using REFMAC5 (Winn et al., 1994 ▶) to a resolution of 1.9 Å with starting models derived from PDB entry 2w1l (Cianci et al., 2008 ▶) for sets at T = 100 K and at room temperature. Structure-factor amplitudes and phases for the refined models of the fresh and dosed sets from native (nitrate-free) crystals, the fresh and dosed sets from nitrate-soaked crystals, and the fresh sets from native and nitrate-soaked crystals were combined using the program CAD (included in CCP4). Data-collection and refinement statistics are provided in Tables 3 ▶ and 4 ▶.
Table 3. Data-processing statistics for eight tetragonal lysozyme crystals: four native (i.e. nitrate-free) crystals and four soaked in 100 mM nitrate.
Values in parentheses are for the highest resolution shell (2.0–1.9 Å). Indicated doses are typical values used at 100 and 295 K.
| Crystal | T (K) | Dose (MGy) | Unique reflections | Multiplicity | Completeness (%) | I/σ(I) | Rmerge (%) | Wilson B value (Å2) | Imean (SCALA) | I/I0 (SCALA) | Unit-cell parameters (Å) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Native | 100 | Fresh | 10042 | 6.8 (6.8) | 100.0 (100.0) | 5.0 (7.0) | 6.0 (8.3) | 14.4 | 6853 | 1.00 | a = 79.77, c = 37.54 |
| Native | 100 | 1.9 | 10063 | 6.8 (6.8) | 100.0 (100.0) | 4.9 (6.7) | 6.2 (8.7) | 15.8 | 5729 | 0.84 | a = 79.85, c = 37.56 |
| Native | 295 | Fresh | 10369 | 6.9 (7.0) | 100.0 (100.0) | 4.3 (4.3) | 5.9 (12.3) | 18.7 | 1458 | 1.00 | a = 80.15, c = 38.44 |
| Native | 295 | 0.12 | 10385 | 6.9 (7.0) | 100.0 (100.0) | 4.2 (3.6) | 6.3 (16.8) | 21.2 | 1117 | 0.77 | a = 80.19, c = 38.44 |
| Nitrate | 100 | Fresh | 10006 | 6.8 (6.9) | 100.0 (100.0) | 6.5 (3.7) | 7.3 (14.3) | 16.4 | 3685 | 1.00 | a = 79.56, c = 37.61 |
| Nitrate | 100 | 1.9 | 10046 | 6.8 (6.9) | 100.0 (100.0) | 6.7 (4.4) | 6.8 (13.6) | 17.1 | 2998 | 0.81 | a = 79.68, c = 37.66 |
| Nitrate | 295 | Fresh | 10397 | 6.9 (7.0) | 100.0 (100.0) | 4.1 (3.6) | 6.8 (16.3) | 19.7 | 1660 | 1.00 | a = 80.05, c = 38.64 |
| Nitrate | 295 | 0.12 | 10377 | 6.9 (7.0) | 100.0 (100.0) | 3.8 (3.2) | 7.3 (20.6) | 20.7 | 1466 | 0.88 | a = 79.88, c = 38.74 |
Table 4. Refinement statistics for each crystal in Table 3 ▶ .
Models for all data sets were refined to a resolution of 1.9 Å.
| Crystal | ||||||||
|---|---|---|---|---|---|---|---|---|
| Native | Nitrate | |||||||
| Fresh | Dosed | Fresh | Dosed | Fresh | Dosed | Fresh | Dosed | |
| T (K) | 100 | 100 | 295 | 295 | 100 | 100 | 295 | 295 |
| R factor (%) | 15.3 | 15.2 | 14.9 | 14.5 | 15.5 | 15.3 | 15.1 | 15.2 |
| Rfree (%) | 19.3 | 20.4 | 19.2 | 17.7 | 20.4 | 21.0 | 19.5 | 21.9 |
Using the refined structures, Fourier difference maps (|F fresh,obs − F dosed,obs|, ϕfresh) comparing fresh and dosed structures were calculated at each temperature for native (i.e. nitrate-free) crystals and nitrate-soaked crystals using FFT (part of CCP4). To identify structural changes arising from the nitrate soaks and to locate any ordered nitrate, structures of native fresh crystals and nitrate-soaked fresh crystals were compared using (|F nitrate,obs − F native,obs|, ϕnative). Sample results are shown in Fig. 4 ▶.
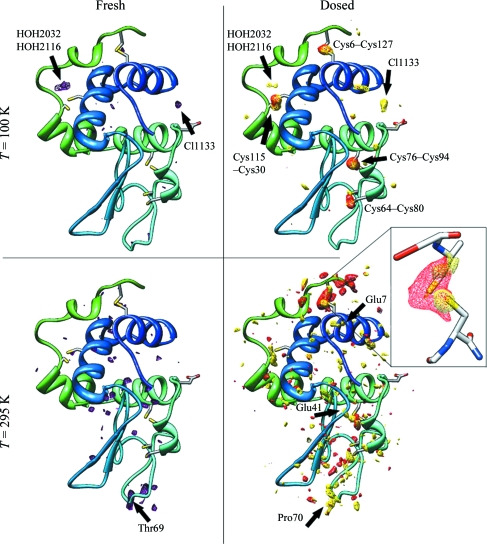
Figure 4.
Fourier difference maps comparing native tetragonal lysozyme crystals and crystals soaked in 100 mM nitrate. Top-row structures were acquired at T = 100 K and bottom-row structures at T = 295 K. Left column: difference maps (F nitrate,obs − F native,obs, ϕnative) acquired on ‘fresh’ crystals prior to delivery of a large dose. Purple contours correspond to levels of +4.4σ and +3.4σ at 100 and 295 K, respectively. At T = 100 K, two water molecules and the chloride have moved to new positions. At T = 295 K, the loop with Thr69 relocated to a new position. Right column: difference maps (F fresh,obs − F dosed,obs, ϕfresh) comparing ‘fresh’ and ‘dosed’ crystals, where damage arises from a large dose (0.12 MGy at room temperature and 1.9 MGy at T = 100 K) delivered between structural data sets. Red contours indicate damage to groups in native (nitrate-free) crystals, with levels of +4.3σ and +3.1σ at 100 and 295 K, respectively; yellow contours show damage in nitrate-soaked crystals, with levels of +4.7σ and +3.2σ at 100 and 295 K, respectively. At room temperature, the sulfur bridge Cys6–Cys127 appears to be protected by nitrate.
From the |F nitrate,obs − F native,obs| maps, nitrate soaking produces several changes in the ‘fresh’ (before dosing) structures both at room and cryogenic temperatures. Water molecules rearrange and side chains shift, but no changes attributable to ordered nitrate ions could be observed at either temperature. The calculated unit cell contracts slightly, by less than 0.3% (a result of questionable significance).
At T = 100 K, largely similar radiation-induced changes are observed after a dose of 1.9 MGy (corresponding to ΔB rel = 1.4 Å2) in both native and nitrate-soaked crystals and are consistent with previous measurements of site-specific damage in lysozyme (Weik et al., 2000 ▶; Ravelli & McSweeney, 2000 ▶; Nanao et al., 2005 ▶; Borek et al., 2007 ▶). All disulfide bridges are sensitive, with Cys76–Cys94 and Cys6–Cys127 being affected the most. Aspartic and glutamic acids suffer decarboxylation and several parts of the molecule undergo disorder via mobility, especially side chains on the surface of the protein. In nitrate-soaked crystals two sites show additional changes: one near waters HOH2032 and HOH2116 and another near chloride Cl1133. These two sites also show up in the difference map that compares native and nitrate-soaked crystals (|F nitrate,obs − F native,obs|, ϕnative; see Fig. 4 ▶, top left), so they may involve ordered nitrates. However, the likely partial occupancies of water, other ions and nitrates, disorder unique to each species, lack of an absolute scale for the map and the limited resolution of our data rule out any definitive identification.
At room temperature, after a dose of 0.12 MGy some differences in damage patterns are seen between native and nitrate-soaked crystals. For native crystals, this dose produced ΔB rel = 2.5 Å2. The most sensitive site was the Cys6–Cys127 bridge near the C-terminus. Some shifts of side chains (e.g. turns containing Gln41 and Thr69, Glu7 on helix 1) and minor damage to other typically radiation-sensitive sites are observed, but otherwise the damage appears to be uniformly distributed across the structure.
Nitrate-soaked crystals exposed at room temperature to the same dose show a smaller ΔB rel of 1.0 Å2. The previously sensitive Cys6–Cys127 disulfide bridge now suffers only minor damage and appears protected. Damage to Cys80–Cys64 and Cys94–Cys76 is slightly reduced. The C-terminal chain is more stable than in the native crystal, but the turn on the opposite side of the molecule at Pro70 remains quite mobile. Again, the damage is uniformly distributed across the structure, mostly owing to disorder stemming from mobility of side chains. Unlike at cryogenic temperatures, there is no additional damage in the nitrate-soaked crystal.
Despite these model differences in site-specific damage, no conclusion can be drawn as to their importance in determining the nitrate-related reduction in global radiation sensitivity as measured by s AD. As discussed by Kmetko et al. (2006 ▶), in crystallography ‘site-specific damage’ includes damage that is reproducible from cell to cell and in general this accounts for only a small fraction of the total damage to the crystal. The most important global damage-mitigating effects of nitrate may not be readily visible.
A manuscript that has appeared since our initial submission reports experiments on the efficacy of nitrate in protecting lysozyme crystals from damage at T = 100 K (De la Mora et al., 2011 ▶). In contrast to the present study, in which no protective effect was found at 100 K, De la Mora and coworkers (DLM) claimed a factor of two increase in dose tolerance. However, only one ‘native’ and three nitrate-soaked crystals were examined. Depending upon the damage metric used (overall intensity decay or decay R factor; Diederichs, 2006 ▶), two of the three nitrate-soaked crystals show the same sensitivity as the native crystal, suggesting that experimental uncertainties were at least as large as the claimed effect. Site-specific effects of nitrate, including the reduced sensitivity of the Cys6–Cys127 bridge seen here, are discussed. DLM also report four ordered nitrates in the T = 100 K structure based on 2F obs − F calc maps. As noted above, our refinement to comparable resolution (1.9 Å versus 2.0 Å by DLM) and using |F nitrate,obs − F native,obs| maps shows features at Cl1133 (the site of one of the nitrates in DLM) and near HOH2032 or HOH2116 (not reported by DLM) at 100 K, but no unambiguous evidence for ordered nitrates at either 100 or 300 K. The differences in the results may in part arise from differences in nitrate concentrations. The DLM data were collected from crystals soaked in 500 mM nitrate, whereas we only collected full structural data sets for crystals soaked in 100 mM nitrate. Using the same methods and criteria as were applied in analyzing our data, our analysis of DLM’s PDB data identifies one ordered nitrate site with high confidence.
5. Discussion
Of the 19 small-molecule compounds that we have screened, some are strongly sensitizing at room temperature. Only sodium nitrate appears to provide any protection against global radiation damage, but only to tetragonal and monoclinic lysozyme and to tetragonal thaumatin (but not trypsin) and only at room temperature. Nitrate also appears to protect against at least some site-specific damage. Although nitrate’s global protecting effect was reproduced in several crystals, the small magnitude of its effect on global damage together with some peculiar observations (e.g. the protecting effect in lysozyme does not increase with nitrate concentration beyond 100 mM) leave some residual doubt as to its validity. None of the compounds tested showed any sign of providing protection against global radiation damage, as quantified by s AD, at T = 100 K, where most structural data sets are acquired.
5.1. Comparison with previous studies
These results contrast with many previous reports of scavenger effectiveness, but are consistent with other studies that have found no protective effects. Styrene, chlorostyrene and methyl methacrylate have been reported to increase the room-temperature radiation lifetimes of IgG immunoglobin crystals by as much as a factor of ten (Zaloga & Sarma, 1974 ▶), but we and others (Murray & Garman, 2002 ▶) find that styrene has no protective effect with other proteins. PEG 4000 and PEG 20 000 (which may be too large to penetrate inside a crystal) have been reported to extend the lifetime of α-amylase and canavalin crystals at room temperature (Cascio et al., 1984 ▶), but we find that PEG 200 is a sensitizer. Kauffmann et al. (2006 ▶) reported that nicotinic acid, 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) and glutathione provided a roughly factor-of-two protection to lysoyzme, thaumatin and elastase crystals at cryogenic temperatures and observed some nonsystematic effects on site-specific damage. However, at cryogenic temperatures Nowak et al. (2009 ▶) found that diffraction from nicotinic acid-soaked trypsin crystals and native crystals were statistically indistinguishable and Macedo et al. (2009 ▶) found no effect of nicotinic acid on the increase in Wilson B factors with dose in azurin crystals; at both cryogenic and room temperature we find no effect of glutathione (in its reduced form) on lysozyme and thaumatin. Macedo et al. (2009 ▶) also found no effect of HEPES, 2-nitroimidazole, thiourea and l-cysteine at T = 100 K; the last two results are confirmed by the present work.
In a preliminary study on lysozyme at T = 100 K (Murray & Garman, 2002 ▶), ascorbic acid was found to reduce damage to disulfide bonds and to eliminate a 23% increase in Wilson B factor seen in native crystals after a dose of 10 MGy. A subsequent room-temperature study on lysozyme (Barker et al., 2009 ▶) found that ascorbic acid doubled the crystal dose tolerance, as reflected in the decay of summed diffraction peak intensities, and that 1,4-benzoquinone provided a factor of nine increase in dose tolerance. Macedo et al. (2009 ▶) found that at T = 100 K ascorbic acid and 2,3-dichloro-1,4-naphthoquinone (DNQ) reduced the increase in Wilson B factors with dose in azurin crystals, although the overall effect was comparable to the scatter in B factor versus dose for each crystal and to the scatter in B factors between native crystals. Ascorbic acid also reduced site-specific damage to disulfides, consistent with the results of Murray & Garman (2002 ▶). In contrast, we find that ascorbic acid has no effect on global radiation damage to lysozyme crystals at either room temperature or at T = 100 K.
Why have different studies obtained such different results? Reported protective effects could easily have resulted from issues in data collection and analysis. Early studies (Zaloga & Sarma, 1974 ▶; Cascio et al., 1984 ▶) were performed at room temperature and with very weak X-ray beams, most likely using capillary-mounted crystals. They did not record actual X-ray fluxes or use reliable metrics of damage. Crystal-to-crystal differences in slippage, dehydration and temperature history over the ∼20 or more hours required for data collection were likely to be significant. In both these early studies and in more recent studies data-collection protocols have often not been described in sufficient detail to determine whether the many potential sources of error described here and by Kmetko et al. (2006 ▶) have been adequately addressed, and sample-to-sample (or dose-to-dose) scatter in damage measurements has sometimes been comparable in size to claimed protective effects. At T = 100 K, non-uniform irradiation, arising from, for example, Gaussian rather than flat beam profiles, imprecise sample alignment in the X-ray beam and data collection over large rotation angles, can introduce errors comparable to or larger than reported protective effects. Differences in sample motion during data collection, owing to flutter in the cryostream at T = 100 K, owing to variable deflection of the crystal and nylon loop as the aerodynamic profile they present to the cryostream changes during rotation and owing to crystal slippage relative to the capillary or mount at room temperature, can introduce very large errors. Nowak et al. (2009 ▶) appear to have most thoroughly addressed the potential sources of error and verified their null results for nicotinic acid by repeating measurements on multiple crystals.
Evidence for data-collection issues in some studies is provided by large variations in reported room-temperature damage rates for native crystals of the same protein. For example, Barker et al. (2009 ▶) reported data for the summed intensity of reflections (their chosen damage metric) versus dose for six native lysozyme crystals at room temperature. The half-intensity dose (D 1/2) values (i.e. the dose at which the summed intensity is reduced to half its initial value) are ∼0.25, ∼0.25, ∼0.17, ∼0.13 and ∼0.15 MGy for five crystals irradiated on a home source, with dose rates of 4.6, 4.8, 5.7, 6.0 and 12.8 Gy s−1, respectively. The first two of these native crystals are less sensitive than either of the two ascorbate-soaked crystals that they report as showing protection. Furthermore, for a native crystal irradiated at the ESRF at a dose rate of 2800 Gy s−1, D 1/2 was measured to be ∼1 MGy s−1, a factor of 4–8 larger than on the home source.
The large difference between home and synchrotron source values was attributed to an ‘inverse’ dose-rate effect at low doses as reported by Southworth-Davies et al. (2007 ▶). This cannot be the case. Using the same damage metric as Barker et al. (2009), Southworth-Davies and coworkers reported a monotonic increase in D 1/2 for lysozyme crystals at room temperature from ∼0.38 MGy at 6.2 Gy s−1 to 1.63 MGy at 9.7 Gy s−1. Both the magnitudes and the trend of these data are inconsistent with those of Barker and coworkers in the same dose-rate range. At a dose rate of 6 Gy s−1, data collection to a dose of ∼1 MGy requires ∼50 h, with plenty of time for crystal motions owing to slippage and evaporation of surrounding mother liquor and perhaps also for X-ray beam drift relative to the sample
In the present study, HEMA initially showed significant protective effects, but at high concentrations it became strongly sensitizing. Additional experiments covering a wide range of concentrations showed a decrease in sensitivity with decreasing HEMA concentration, but no protective effect at any concentration. The initial protective effect was eventually traced to a crystal shift during data collection. These shifts can be very smooth and very hard to detect in diffraction, unless the position of the crystal is constantly monitored with a telescope and its alignment with the X-ray beam is checked at the start and end of data collection.
5.2. Radioprotectors and sensitizers
Based upon our data and the above discussion, we conclude that, with the possible exception of nitrate, none of the small-molecule compounds investigated to date has any unambiguous protective effect on global radiation damage to protein crystals at room temperature and none protect at T = 100 K. Some compounds may reduce site-specific damage to disulfide bonds, but this kind of damage is easily modelled and does not limit structural analysis (Ravelli et al., 2005 ▶). Reductions in other kinds of site-specific damage may also occur, but so far there is no evidence of the consistent and predictable effects necessary for broad utility. A study of 18 different radioprotectants showed useful effects on X-ray-induced reduction of metal centers in metalloproteins only for hexacyanoferrate III (Macedo et al., 2009 ▶).
Only nitrate appears to provide a modest protective effect and only at room temperature. Nitrate ions provide protection to, for example, T1 phages in solution (Dertinger & Jung, 1970a ▶). The size of the protective effect that we observe is consistent with the protection factor of ∼1.5–2 observed when much large scavenger concentrations are added to lyophilized protein [e.g. 50%(w/w) cystamine to ribonuclease; see Dertinger & Jung, 1970a ▶], but is orders of magnitude smaller than the maximum effect observed for proteins in dilute solution. Nitrate ions react effectively with hydrated electrons and may also affect radiation chemistry in other ways. However, nitrate could simply stabilize the protein conformation and crystal contacts so that for a given number of ‘hits’ they are less likely to be disrupted, as may occur when crystals are cross-linked. Our data and our reading of the literature do not allow us to speculate intelligently on the possible protective mechanism.
Given the lack of significant protective effects, a more relevant focus may be on screening compounds, especially those commonly found in crystallization solutions, to identify sensitizers. These can then be eliminated from growth solutions or via post-growth soaks. Polymers and polymerizing compounds such as styrene, HEMA and PEG (a common crystallization agent) appear to be sensitizing, as is cysteine, a sulfur-containing amino acid.
5.3. Why don’t known scavengers protect protein crystals against global damage?
Why are most of the small-molecule compounds studied here (and in some other studies as described in §5.1) and known to be effective radioprotectors in other contexts so ineffective in protecting protein crystals against global radiation damage? The absence of any effects, both protecting and sensitizing, on global damage to protein crystals at T = 100 K is not surprising. At low temperatures, crystals of all proteins, with their widely different sequences, solvent contents and structures and crystallized/soaked with an extremely wide array of compounds, apparently show similar radiation sensitivities (damage per dose). Were this not the case, the Henderson limit (Henderson, 1990 ▶; Owen et al., 2006 ▶) would have little predictive utility. One might therefore expect the impact of adding a few small organic molecules within the unit cell to be small.
At low temperatures (∼100 K and below), diffusion of essentially all radiolytic products except for electrons is frozen out. Small-molecule compounds and damaged protein are prevented from moving by the frozen solvent network, so that atomic positions are preserved. Small-molecule compounds could be effective in reducing damage if they somehow either prevent absorbed energy from reaching the protein molecules or siphon off energy absorbed by the protein before it does damage. We are not qualified to speculate on the nature and efficiency of the energy-transfer mechanisms remaining when translational atomic diffusion is no longer possible and vibrational motions become small, but they would appear to have little effect.
At room temperature, many small-molecule compounds have clear radioprotective effects on proteins in solution and in the amorphous solid state (Dertinger & Jung, 1970a ▶). In dilute aqueous solution most of the radiation is absorbed not by the protein but by the solvent, and most of the radicals produced by secondary electrons are generated in the solvent. Solvent radicals then diffuse to and attack the protein. Compounds that react with these radicals before they reach the protein and that are present in sufficiently large concentrations can thus provide significant competitive protection.
However, inside protein crystals there is comparatively little solvent and much of the X-ray dose is absorbed via ‘direct hits’ of X-rays and energetic primary and secondary electrons to the protein. This in part explains why proteins in crystals can survive much larger doses than proteins in dilute aqueous solution: for a given dose (energy deposited/volume) the number of hits per protein molecule in a crystal is smaller. Room-temperature half-dose values of ∼0.4 MGy (Barker et al., 2009 ▶) for lysozyme crystals can be compared with half doses for enzymatic inactivation of ribonuclease A of ∼0.3 MGy in the dry (lyophilized) state and 0.003 MGy in a 5 mg ml−1 (0.37 mM) aqueous solution (Smith & Adelstein, 1965 ▶; Dertinger & Jung, 1970b ▶).
Another reason why scavengers are so ineffective in protecting protein crystals is that the achievable ratio of scavenger molecules to protein molecules is generally much smaller than that required for significant protective effects in solution. As shown in Table 1 ▶, the concentrations of small-molecule compounds in our soaking solutions were typically 100 mM. Assuming an equal concentration in the roughly 50% of the unit-cell volume that is occupied by solvent (as suggested by studies of NaCl repartitioning and incorporation in lysozyme crystals; Vekilov et al., 1996 ▶), this corresponds to roughly one molecule per lysozyme molecule. For cysteine (MW = 128 Da), the mass ratio of scavenger:water:protein within the unit cell is then ∼1:82:99.
Shimazu & Tappel (1964 ▶) found that cysteine provided effective radioprotection of proteins in solution. For a protein concentration of 0.1%(w/w) and a cysteine concentration of 1 mM, the half-dose D 1/2 (corresponding to a 50% loss of enzymatic activity) was increased by a factor of seven for alcohol dehydrogenase (MW = 80 kDa) and 14.7 for ribonuclease A (MW = 13.7 kDa). The ratio of cysteine molecules to protein molecules in each solution was 80 and 13.7, respectively. The mass ratio of scavenger:water:protein was then 1:8300:8 for both proteins. Unlike in a protein crystal, most of the dose was absorbed by water, most of the radicals were generated in the water and most radicals first encountered scavenger molecules rather than protein molecules.
A more relevant comparison to protein crystals is provided by ‘dry’ mixtures of small-molecule radioprotectors and proteins produced by lyophilizing aqueous solutions. In mixtures of cystamine (MW = 152 Da) and ribonuclease A, no radioprotective effect was observed below a cystamine:protein mass ratio of 0.1 mg:1 mg, corresponding to nine cystamines per protein molecule (Dertinger & Jung, 1970c ▶). Unlike in dilute solution, the half-dose for enzymatic inactivation only increased by a factor of two before saturating at mass ratios beyond 1 mg:1 mg (corresponding to 90 cystamines per protein). This comparison suggests that the scope for protective effects in protein crystals is quite limited and will require small-molecule concentrations within the crystal one to two orders of magnitude larger than those that we have achieved by soaking.
Finally, good scavengers tend to be reactive, but in the protein-dense environment of a crystal this may in fact be deleterious. Reactions will occur not just with radicals in solvent channels but also (and with high probability) with the protein itself, possibly disrupting its conformation. This could explain why cysteine, a powerful free-radical scavenger and protector of proteins in solution, is such a strong radiation sensitizer in protein crystals and why only sodium nitrate, a relatively benign salt, appears to have any protective effect.
6. Conclusions
Many small-molecule compounds have been reported to protect protein crystals against radiation damage, but these reports have often not stood up on subsequent examination. We have evaluated 19 small-molecule compounds, most of which have shown protective effects either in crystals, in solution or in vivo, for their ability to reduce global radiation damage to lysozyme crystals. In carefully executed experiments using well defined metrics of irradiation (dose) and global damage (s AD), we find that no compound provides protective effects at T = 100 K, where most crystallographic data is acquired. At this temperature the diffusion of most radical species and thus of most radioprotective chemistry is frozen out.
At room temperature, nitrate appears to have a small but somewhat uncertain protective effect and several other compounds are sensitizers. Unlike in dilute solutions, in protein crystals most X-ray absorption is by the protein itself, not by the solvent, and the ratio of scavenger molecules to protein molecules is much smaller. These limit the scope for radioprotective effects. Scavengers may reduce some site-specific damage, but predictable and reliable benefits of a given scavenger for a wide variety of proteins have not been demonstrated.
Taken together with the results of previous studies, this suggests that alternative approaches, such as removing sensitizing compounds and large atomic number elements (Owen et al., 2006 ▶; Kmetko et al., 2006 ▶), collecting data at modestly reduced temperatures (Warkentin & Thorne, 2010 ▶), optimizing data-collection strategies (Holton, 2009 ▶) and using slightly larger crystals, are likely to be more useful than radioprotective compounds in reducing radiation damage to protein crystals.
Acknowledgments
The authors wish to thank the MacCHESS staff for extensive assistance in data processing, N. Husseini for assistance in data collection and Z. Otwinowski and J. Holton for suggesting the use of nitrate. This work was supported by the NIH (R01 GM65981). JK also acknowledges support from an HHMI Undergraduate Science Education Award to Kenyon College. X-ray diffraction measurements were performed at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under NSF award DMR-0225180, and using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health through its National Center for Research Resources. RET acknowledges a significant financial interest in MiTeGen, which provided some of the tools used in this work.
References
- Audette-Stuart, M., Houée-Levin, C. & Potier, M. (2005). Radiat. Phys. Chem. 72, 301–306.
- Barker, A. I., Southworth-Davies, R. J., Paithankar, K. S., Carmichael, I. & Garman, E. F. (2009). J. Synchrotron Rad. 16, 205–216. [DOI] [PubMed]
- Blake, C. & Phillips, D. C. (1962). Proceedings of the Symposium on the Biological Effects of Ionising Radiation at the Molecular Level, pp. 183–191. Vienna: International Atomic Energy Agency.
- Borek, D., Ginell, S. L., Cymborowski, M., Minor, W. & Otwinowski, Z. (2007). J. Synchrotron Rad. 14, 24–33. [DOI] [PubMed]
- Box, H. C. (1972). Annu. Rev. Nucl. Sci. 22, 355–382.
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER v.2.8.0. Cambridge: Global Phasing Ltd.
- Burmeister, W. P. (2000). Acta Cryst. D56, 328–341. [DOI] [PubMed]
- Carpentier, P., Royant, A., Ohana, J. & Bourgeois, D. (2007). J. Appl. Cryst. 40, 1113–1122.
- Cascio, D., Williams, R. & McPherson, A. (1984). J. Appl. Cryst. 17, 209–210.
- Chinte, U., Shah, B., Chen, Y.-S., Pinkerton, A. A., Schall, C. A. & Hanson, B. L. (2007). Acta Cryst. D63, 486–492. [DOI] [PubMed]
- Cianci, M., Helliwell, J. R. & Suzuki, A. (2008). Acta Cryst. D64, 1196–1209. [DOI] [PubMed]
- Coggle, J. E. (1983). Biological Effects of Radiation. London: Taylor & Francis.
- Corbett, M. C., Latimer, M. J., Poulos, T. L., Sevrioukova, I. F., Hodgson, K. O. & Hedman, B. (2007). Acta Cryst. D63, 951–960. [DOI] [PubMed]
- Coulibaly, F., Chiu, E., Ikeda, K., Gutmann, S., Haebel, P. W., Schulze-Briese, C., Mori, H. & Metcalf, P. (2007). Nature (London), 446, 97–101. [DOI] [PubMed]
- Dauter, Z. & Dauter, M. (2001). Structure, 9, R21–R26. [DOI] [PubMed]
- Dauter, Z., Dauter, M. & Rajashankar, K. R. (2000). Acta Cryst. D56, 232–237. [DOI] [PubMed]
- De la Mora, E., Carmichael, I. & Garman, E. F. (2011). J. Synchrotron Rad. 18, 346–357. [DOI] [PubMed]
- Dertinger, H. & Jung, H. (1970a). Molecular Radiation Biology New York: Springer-Verlag.
- Dertinger, H. & Jung, H. (1970b). Molecular Radiation Biology, p. 76. New York: Springer-Verlag.
- Dertinger, H. & Jung, H. (1970c). Molecular Radiation Biology, p. 85. New York: Springer-Verlag.
- Diederichs, K. (2006). Acta Cryst. D62, 96–101. [DOI] [PubMed]
- Durchschlag, H., Hefferle, T. & Zipper, P. (2003). Radiat. Phys. Chem. 67, 479–486.
- Filali-Mouhim, A., Audette, M., St-Louis, M., Thauvette, L., Denoroy, L., Penin, F., Chen, X., Rouleau, N., Le Caer, J. P., Rossier, J., Potier, M. & Le Maire, M. (1997). Int. J. Radiat. Biol. 72, 63–70. [DOI] [PubMed]
- Finfrock, Y. Z., Stern, E. A., Yacoby, Y., Alkire, R. W., Evans-Lutterodt, K., Stein, A., Isakovic, A. F., Kas, J. J. & Joachimiak, A. (2010). Acta Cryst. D66, 1287–1294. [DOI] [PubMed]
- Garman, E. F. (2010). Acta Cryst. D66, 339–351. [DOI] [PMC free article] [PubMed]
- Garman, E. F. & McSweeney, S. M. (2007). J. Synchrotron Rad. 14, 1–3. [DOI] [PubMed]
- Garman, E. & Nave, C. (2002). J. Synchrotron Rad. 9, 327–328. [DOI] [PubMed]
- Garman, E. F. & Owen, R. L. (2006). Acta Cryst. D62, 32–47. [DOI] [PubMed]
- Garrison, W. M. (1987). Chem. Rev. 87, 381–398.
- Gonzalez, A. & Nave, C. (1994). Acta Cryst. D50, 874–877. [DOI] [PubMed]
- Haas, D. J. & Rossmann, M. G. (1970). Acta Cryst. B26, 998–1004. [DOI] [PubMed]
- Hanson, B. L., Harp, J. M., Kirschbaum, K., Schall, C. A., DeWitt, K., Howard, A., Pinkerton, A. A. & Bunick, G. J. (2002). J. Synchrotron Rad. 9, 375–381. [DOI] [PubMed]
- Hategan, A., Martin, D., Popescu, L. M. & Butan, C. (2001). Bioelectrochemistry, 53, 193–197. [DOI] [PubMed]
- Henderson, R. (1990). Proc. R. Soc. Lond. B Biol. Sci. 241, 6–8.
- Holton, J. M. (2009). J. Synchrotron Rad. 16, 133–142.
- Holton, J. M. & Frankel, K. A. (2010). Acta Cryst. D66, 393–408. [DOI] [PMC free article] [PubMed]
- Hope, H. (1988). Acta Cryst. B44, 22–26. [DOI] [PubMed]
- Hope, H. (1990). Annu. Rev. Biophys. Biophys. Chem. 19, 107–126. [DOI] [PubMed]
- Howell, P. L. & Smith, G. D. (1992). J. Appl. Cryst. 25, 81–86.
- Hubbell, J. & Seltzer, S. (2004). Tables of X-ray Mass Attenuation Coefficients and Mass Energy Absorption Coefficients, v. 1.4. Gaithersburg: National Institute of Standards and Technology. http://physics.nist.gov/xaamdi.
- Kalinin, Y., Kmetko, J., Bartnik, A., Stewart, A., Gillilan, R., Lobkovsky, E. & Thorne, R. (2005). J. Appl. Cryst. 38, 333–339.
- Kauffmann, B., Weiss, M. S., Lamzin, V. S. & Schmidt, A. (2006). Structure, 14, 1099–1105. [DOI] [PubMed]
- Kempner, E. S. (2001). J. Pharm. Sci. 90, 1637–1646. [DOI] [PubMed]
- Kmetko, J., Husseini, N. S., Naides, M., Kalinin, Y. & Thorne, R. E. (2006). Acta Cryst. D62, 1030–1038. [DOI] [PubMed]
- Leiros, H.-K. S., Timmins, J., Ravelli, R. B. G. & McSweeney, S. M. (2006). Acta Cryst. D62, 125–132. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr. 26
- Livesay, J. C. & Reed, D. J. (1987). Adv. Radiat. Biol. 13, 285–340.
- Macedo, S., Pechlaner, M., Schmid, W., Weik, M., Sato, K., Dennison, C. & Djinović-Carugo, K. (2009). J. Synchrotron Rad. 16, 191–204. [DOI] [PubMed]
- Malkin, A. J. & Thorne, R. E. (2004). Methods, 34, 273–299. [DOI] [PubMed]
- Matthews, B. W. (1977). The Proteins, edited by H. Neurath & R. L. Hill, pp. 503–504. New York: Academic Press.
- McGeehan, J. E., Carpentier, P., Royant, A., Bourgeois, D. & Ravelli, R. B. G. (2007). J. Synchrotron Rad. 14, 99–108. [DOI] [PubMed]
- McGeehan, J., Ravelli, R. B. G., Murray, J. W., Owen, R. L., Cipriani, F., McSweeney, S., Weik, M. & Garman, E. F. (2009). J. Synchrotron Rad. 16, 163–172. [DOI] [PMC free article] [PubMed]
- McPherson, A. (1999). Crystallization of Biological Macromolecules. Cold Spring Harbor Laboratory Press.
- Meents, A., Gutmann, S., Wagner, A. & Schulze-Briese, C. (2010). Proc. Natl Acad. Sci. USA, 107, 1094–1099. [DOI] [PMC free article] [PubMed]
- Meents, A., Wagner, A., Schneider, R., Pradervand, C., Pohl, E. & Schulze-Briese, C. (2007). Acta Cryst. D63, 302–309. [DOI] [PubMed]
- Murray, J. & Garman, E. (2002). J. Synchrotron Rad. 9, 347–354. [DOI] [PubMed]
- Murray, J. W., Garman, E. F. & Ravelli, R. B. G. (2004). J. Appl. Cryst. 37, 513–522.
- Nanao, M. H., Sheldrick, G. M. & Ravelli, R. B. G. (2005). Acta Cryst. D61, 1227–1237. [DOI] [PubMed]
- Nave, C. & Garman, E. F. (2005). J. Synchrotron Rad. 12, 257–260. [DOI] [PubMed]
- Nave, C. & Hill, M. A. (2005). J. Synchrotron Rad. 12, 299–303. [DOI] [PubMed]
- Nowak, E., Brzuszkiewicz, A., Dauter, M., Dauter, Z. & Rosenbaum, G. (2009). Acta Cryst. D65, 1004–1006. [DOI] [PMC free article] [PubMed]
- Owen, R. L., Pearson, A. R., Meents, A., Boehler, P., Thominet, V. & Schulze-Briese, C. (2009). J. Synchrotron Rad. 16, 173–182. [DOI] [PMC free article] [PubMed]
- Owen, R. L., Rudiño-Piñera, E. & Garman, E. F. (2006). Proc. Natl Acad. Sci. USA, 103, 4912–4917. [DOI] [PMC free article] [PubMed]
- Petsko, G. A. (1975). J. Mol. Biol. 96, 381–392. [DOI] [PubMed]
- Ravelli, R. B. & Garman, E. F. (2006). Curr. Opin. Struct. Biol. 16, 624–629. [DOI] [PubMed]
- Ravelli, R. B. & McSweeney, S. M. (2000). Structure, 8, 315–328. [DOI] [PubMed]
- Ravelli, R. B. G., Nanao, M. H., Lovering, A., White, S. & McSweeney, S. (2005). J. Synchrotron Rad. 12, 276–284. [DOI] [PubMed]
- Ravelli, R. B. G., Theveneau, P., McSweeney, S. & Caffrey, M. (2002). J. Synchrotron Rad. 9, 355–360. [DOI] [PubMed]
- Saha, A., Mandal, P. C. & Bhattacharyya, S. N. (1995). Radiat. Phys. Chem. 46, 123–145.
- Schulze-Briese, C., Wagner, A., Tomizaki, T. & Oetiker, M. (2005). J. Synchrotron Rad. 12, 261–267. [DOI] [PubMed]
- Shalaev, E., Reddy, R., Kimball, R. N., Weinschenk, M. F., Guinn, M. & Margulis, L. (2003). Rad. Phys. Chem. 66, 237–245.
- Shimazu, F. & Tappel, A. L. (1964). Science, 143, 369–371. [DOI] [PubMed]
- Sliz, P., Harrison, S. C. & Rosenbaum, G. (2003). Structure, 11, 13–19. [DOI] [PubMed]
- Smith, T. W. & Adelstein, S. J. (1965). Radiat. Res. 24, 119–132. [PubMed]
- Southworth-Davies, R. J. & Garman, E. F. (2007). J. Synchrotron Rad. 14, 73–83. [DOI] [PubMed]
- Southworth-Davies, R. J., Medina, M. A., Carmichael, I. & Garman, E. F. (2007). Structure, 15, 1531–1541. [DOI] [PubMed]
- Stern, E. A., Yacoby, Y., Seidler, G. T., Nagle, K. P., Prange, M. P., Sorini, A. P., Rehr, J. J. & Joachimiak, A. (2009). Acta Cryst. D65, 366–374. [DOI] [PubMed]
- Taylor, J. J., Willson, R. L. & Kendalltaylor, P. (1984). FEBS Lett. 176, 337–340.
- Teng, T. & Moffat, K. (2000). J. Synchrotron Rad. 7, 313–317. [DOI] [PubMed]
- Teng, T.-Y. & Moffat, K. (2002). J. Synchrotron Rad. 9, 198–201. [DOI] [PubMed]
- Thorne, R. E., Stum, Z., Kmetko, J., O’Neill, K. & Gillilan, R. (2003). J. Appl. Cryst. 36, 1455–1460.
- Unno, K., Shimba, S. & Okada, S. (1989). Chem. Pharm. Bull. 37, 3047–3049. [DOI] [PubMed]
- Utschig, L. M., Chemerisov, S. D., Tiede, D. M. & Poluektov, O. G. (2008). Biochemistry, 47, 9251–9257. [DOI] [PubMed]
- Vekilov, P. G., Monaco, L. A., Thomas, B. R., Stojanoff, V. & Rosenberger, F. (1996). Acta Cryst. D52, 785–798. [DOI] [PubMed]
- Warkentin, M. & Thorne, R. E. (2010). Acta Cryst. D66, 1092–1100. [DOI] [PMC free article] [PubMed]
- Weik, M., Ravelli, R. B., Kryger, G., McSweeney, S., Raves, M. L., Harel, M., Gros, P., Silman, I., Kroon, J. & Sussman, J. L. (2000). Proc. Natl Acad. Sci. USA, 97, 623–628. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Yano, J., Kern, J., Irrgang, K. D., Latimer, M. J., Bergmann, U., Glatzel, P., Pushkar, Y., Biesiadka, J., Loll, B., Sauer, K., Messinger, J., Zouni, A. & Yachandra, V. K. (2005). Proc. Natl Acad. Sci. USA, 102, 12047–12052. [DOI] [PMC free article] [PubMed]
- Young, A. C. M., Dewan, J. C., Nave, C. & Tilton, R. F. (1993). J. Appl. Cryst. 26, 309–319.
- Zaloga, G. & Sarma, R. (1974). Nature (London), 251, 551–552. [DOI] [PubMed]