Abstract
Several methods have been developed to measure interactions between homologous chromosomes during meiosis in budding yeast. These include cytological analysis of fixed, spread nuclei using fluorescence in situ Hybridization (FISH) (1, 2), visualization of GFP-labeled chromosomal loci in living cells (3), and Chromosome-Conformation Capture (3C) (4). Here we describe a quantitative genetic assay that uses exogenous site-specific recombination to monitor the level of homolog associations between two defined loci in living cells of budding yeast (5). We have used the Cre/loxP assay to genetically dissect nuclear architecture and meiotic homolog pairing in budding yeast. Data obtained from this assay report on the relative spatial proximity or accessibility of two chromosomal loci located within the same strain and can be compared to measurements from different mutated strains.
Keywords: budding yeast, meiosis, homolog pairing, chromosome, collision assay, site-specific recombination, Cre/loxP
1.Introduction
The frequency of Cre-mediated recombination events (referred to here as “collisions”) between pairs of loxP sites engineered into the yeast genome provides a measure of the relative spatial proximity of those sites to one another in a living cell. A 13-fold level of induction of meiosis specific collision events in wild-type cells indicates a state of close, stable homolog juxtaposition (CSHJ) (6). From mutant analysis, we have shown that CSHJ is achieved primarily through the function of trans-acting factors required for early and intermediate stages of meiotic recombination (6–8).
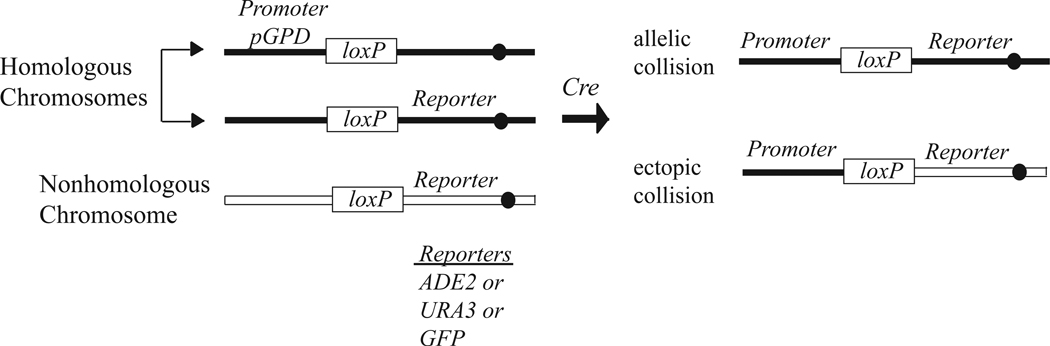
Cre recombinase is supplied under the control of galactose-inducible promoter (pGAL1). Cre-mediated crossing over between loxP sites engineered into specific chromosomal loci gives a genetically detectable product (Fig. 1) (6). One loxP insert bears an upstream constitutive promoter (pGPD) and the other loxP insert bears a downstream open reading frame from a reporter gene (e.g. ADE2, URA3 or GFP). Collisions between pairs of loxP sites can be measured at allelic positions on homologous chromosomes and at ectopic positions on nonhomologous chromosomes in the same strain by using two different reporters. While ectopic collisions are consistently low, allelic collisions increase as cells progress through meiosis (6). Depending on the type of reporter used for the assay, the output is either the frequency of prototroph formation upon return-to-growth (RTG) on media supporting vegetative growth or the frequency of fluorescent spores. For the protocol described here, pGPD1-loxP inserted in Chromosome V recombines with loxP:ura3 inserted at an allelic site, or with loxP:ade2 on Chromosome VIII at an ectopic position to generate Ura+ or Ade+ prototrophs, respectively. Strains listed in Table 1 are available upon request.
Figure 1.
Table1.
Yeast Strains
| Name | Genotype1 |
|---|---|
| SBY 1438 | MATα lys2::GAL1-Cre-LYS2 flo8::LEU2-pGPD1-loxP-lacZ ndt80::LEU2 |
| SBY 1160 | MATa lys2 flo8::LEU2-loxP-ura3 |
| SBY 1338 | MATa lys2 flo8::LEU2-loxP-ura3 ndt80::LEU2-loxP-ade2 |
| SBY 2877 | MATa lys2 arg4::LEU2-loxP-ade2 ndt80::LEU2 |
| SBY 2879 | MATα lys2::GAL1-Cre-LYS2 arg4::LEU2-pGPD-loxP ndt80::LEU2 |
| SBY 2865 | MATa lys2 flo8::LEU2-loxP-ade ndt80::LEU2 |
| SBY 2867 | MATa lys2 flo8::LEU2-loxP-ade |
| SBY 2021 | MATα lys2::GAL1-Cre-LYS2 flo8::LEU2-pGPD1-loxP-lacZ |
| SBY 2061 | MATα lys2 arg4::LEU2-loxP-ade2 |
| SBY 2063 | MATa lys2::GAL1-Cre-LYS2 arg4::LEU2-pGPD-loxP |
| SBY 1060 | MATa lys2 arg4::LEU2-loxP-ura3 |
| SBY 1780 | MATa lys2::GAL1-Cre-LYS2 arg4::LEU2-loxP-ura3 ndt80::LEU-loxP-ade2 |
| SBY 1755 | MATa lys2 arg4::LEU2-loxP-ura3 ndt80::LEU2-loxP-ade2 |
| SBY 1192 | MATα lys2 ndt80::LEU2-pGPD1-loxP-lacZ |
| SBY 1693 |
MATalys2::GAL1-Cre-LYS2 arg4::LEU2-act1-loxP-GFP ste7-1 cyh2-z MATα lys2 arg4::LEU2-GPD-act1-loxP-GFP ste7-1 CYH2 |
| SBY 1476 |
MATa lys2::GAL1-Cre-LYS2 flo8::LEU2-pGPD1-loxP-lacZ ndt80::LEU2 MATα lys2 flo8::LEU2-loxP-ura3 ndt80::LEU2-loxP-ade2 |
All strains are SK1 and contain the following genotype: ho::hisG ura3D::hisG leu2::hisG ade2D::hisG trp1::hisG GAL3. Wild type SK1 is gal3 (15). FLO8 is on Chromosome V and NDT80 is on Chromosome VIII.
2.Materials
2.1 Synchronization of cells for entry into meiosis
All media and solutions are made with deionized water.
Solid growth media: YPG (1% yeast extract, 2% peptone, 3% glycerol, 0.004% tryptophan, 0.01% adenine sulfate, 2% agar). YPD is the same as YPG except in place of glycerol, dextrose is added from a filter-sterilized 40% (w/v) stock to a final concentration of 2% (w/v) after autoclaving. For all media, tryptophan and adenine sulfate are added from filter-sterilized 1% (w/v) stock solutions to autoclaved media cooled to 65°C. Keep 1% tryptophan stock at 4°C and shield from light. Stored 1% adenine sulfate will form crystals that can be melted by heating the solution.
Liquid growth medium: Liquid YPD is prepared as above except agar is omitted. YPA is 1% yeast extract, 2% peptone, 1% potassium acetate, 0.004% tryptophan, 0.01% adenine sulfate.
Synthetic complete (SC) powder: mix together 0.8 g adenine sulfate, 0.8 g arginine, 2 g aspartic acid, 0.8 g histidine, 2.4 g leucine, 1.2 g lysine, 0.8 g methionine, 2 g phenylalanine, 8 g threonine, 0.8 g tryptophan, 1.2 g tyrosine, 0.8 g uracil and grind to a fine powder with a mortar and pestle. SC-URA powder is made similarly except that uracil is omitted.
Liquid sporulation medium (SPM): 1% potassium acetate, 0.02% raffinose, 0.009% SC powder.
18-cm and 25-cm test tubes
2.2 Cre induction and return-to-growth
SC-URA medium: 0.67% yeast nitrogen base without amino acids, 0.09 % SC-URA drop-out powder, 2% dextrose, 0.004% tryptophan, 0.01% adenine sulfate, 2% agar. Supplement with dextrose, tryptophan and adenine sulfate as described above.
YPD-ADE solid medium: same as YPD but without supplementation with adenine sulfate.
2% (w/v) galactose is made from a 20% (w/v) filter-sterilized stock solution, 2% (w/v) glucose from a 40% (w/v) filter-sterilized stock solution.
550 Sonic ZD-dismembrator (Fisher Scientific)
Glass pipets (1-mL and 0.2-mL)
Fixed volume or calibrated liquid dispenser
16-cm test tubes
3. Methods
Cells from three independent colonies are cultured and synchronized to enter meiosis upon transfer to sporulation medium (SPM). Transfer of cells to SPM culture marks t = 0 h of the meiotic time course. At t = 1 h (about the time of DNA replication), Cre recombinase expression is induced by the addition of galactose. Sample aliquots are removed from the culture at t = 1 (before induction), 2, 4, 6, 8, and 10 h after transfer to SPM and diluted appropriately for plating on selective and nonselective media for RTG. The ndt80Δ mutant arrests in pachytene with full-length synaptonemal complexes and unresolved double-Holliday junction intermediates (9, 10). The use of the ndt80Δ mutation allows for recovery of cells in most mutants via RTG that would otherwise be inviable if allowed to complete meiotic divisions and sporulate (11–13). Random spore analysis can be performed with strains that are NDT80. Expression of the GFP reporter can be visualized using fluorescence microscopy in individual spores contained in an ascus due to the spore-autonomous expression of the GPD promoter (14).The number of prototrophs per colony forming unit (C.F.U.) or the number of fluorescent tetrads per total tetrads for each time point indicates the frequency of collisions. Comparison of C.F.U. at each time point relative to t = 1 h reports on survivability of cells by RTG.
3.1 Synchronization of cells for entry into meiosis
From frozen stocks stored in 15% (v/v) glycerol at −80°C, patch cells onto YPG plates. Incubate less than 15 h at 30°C (see Note 1).
From YPG plate, streak for single well-spaced colonies on YPD plates. Incubate 2 d at 30°C.
Inoculate 5 mL YPD (18-cm test tube) with single colony from YPD plate. In a typical experiment, analysis of each strain is carried out in triplicate and is performed in conjunction with a wild-type control strain, also in triplicate (see Note 2). Incubate at 30°C on roller drum at 56 rpm for at least 30 h.
In a 25-cm test tube, inoculate 10 mL of YPA to a final OD600 of 0.23 (see Note 3). Incubate on roller drum at 30°C at 56 rpm for 14 h.
Examine cultures microscopically to determine whether vegetative growth has been arrested at G0/G1. Quantify the fraction of mitotic dividing cells, which are cells with small buds, for one culture of each strain. If more than 5% of cells are small-budded in any culture, consider incubating all of the cultures another 30 min. If more than 10% of cells have small buds, abort the experiment.
Pellet cells at 2,400g for 3 min.
Resuspend cells in 10 mL of sterile deionized water, pellet as described in Subheading 3.1, step 6.
Resuspend cells in 10 mL of SPM pre-warmed to 30°C and transfer to fresh 25-cm test tubes. Incubate on roller drum at 30°C at 56 rpm. The time at which the SPM cultures are placed back in the roller drum is designated as t = 0 h.
3.2 Cre induction and return-to-growth
Prepare one 16-cm test tube that contains 5 mL sterile deionized water and two 16-cm test tubes that contain 4.5 mL for each timepoint for each culture. These tubes will be used to perform the dilution series in Subheading 3.2, step 7. They can be prepared in advance using a fixed-volume water dispenser. We typically carry out analysis on four strains in triplicate so 36 test tubes are required for each time point.
At t = 1 h, remove 1 mL of uninduced cells from each culture and transfer to a 1.5 mL microcentrifuge tube.
After removing the first aliquot, induce Cre recombinase expression by adding 135 µL of 2% galactose into 9 mL of remaining culture (final concentration of 0.03% - see Note 4).
Place cultures back on rollerdrum to incubate at 30°C at 56 rpm. For GFP analysis, see Note 5.
Pellet the 1mL aliquot of cells for 1 min at 13,000g in a microcentrifuge and resuspend pellet in 1 mL of 2% glucose. For random spore analysis, see Note 6.
Briefly vortex resuspended cells.
To disperse cells, sonicate for 3 sec at setting 1.5 (15% maximum power) using the microtip of a 550 Sonic ZD-dismembrator.
Dilution series and plating: Add 25 µL of the sonicated cell suspension to a 16-cm test tube containing 5 mL of sterile deionized water. Vortex briefly. Transfer 0. 5 mL using a 1-mL pipet to a 16-cm test tube containing 4.5 mL water. This is the 10−3 dilution to be used for counting Ade+ prototrophs and Ura+ prototrophs. Transfer 0.5 mL from 10−3 dilution to 4.5 ml water. This is the 10−4 dilution to be used for determining C.F.U.s.
Briefly vortex the test tube before plating. Spread 0.2 mL from the 10−3 dilution onto YPD-ADE and SC-URA plates (see Note 7). From the 10−4 dilution, spread 0.2 mL onto YPD-ADE.
Incubate plates at 30°C.
Repeat Subheading 3.2, steps 5–10 at t = 2, 4, 6, 8, and 10 h.
Count the total number of colonies on 10−4 dilution plate after 2 d of growth (see Note 8).
Count white colonies (Ade+ prototrophs) on 10−3 dilution plate after 2 d of growth (see Note 9).
Count colonies (Ura+ prototrophs) on SC-URA plates after 4–5 d of growth.
3.3 Analysis
To calculate the frequency of ectopic collisions for each culture at each time point, divide the number of white colonies (Ade+ prototrophs) on the 10−3 plate by 10 times the total number of colonies on the 10−4 plate. To calculate the frequency of allelic collisions for each culture at each time point, divide the number of colonies (Ura+ prototrophs) on the 10−3 plate by 10 times the total number of colonies on the 10−4 plate. Viability of a strain is monitored by comparing colony-forming units (C.F.U.s) at each time point to C.F.U.s at the first time point. These calculations give the frequency of events per meiosis. If performing random spore analysis, the frequency is per chromatid.
Acknowledgements
The authors would like to give a special thanks to Tamara Peoples-Holst, Eric Dean, and Joshua Chang Mell for their contribution to the optimization of this protocol over the years. This work has been supported by the National Institutes of Health (NIH) grant NIH R01 GM075119 (S.M.B), the American Cancer Society RSG-01-053-01-CCG (S.M.B), and the NIH-Environmental Health Sciences training grant NIH T 32 ES07059 (D.Y.L).
Footnotes
YPG selects against growth of cells that have lost mitochrondrial function (i.e. are petite). Cells will sporulate if incubated on YPG longer than 15 h. Typically cells are incubated on YPG for 14 to 14.5 h.
Variation between cultures from independent colonies from the same strain is relatively low when performed in triplicate. Somewhat more variation is observed when comparing data obtained from experiments performed on different days. Thus, comparison of mutant phenotypes to wild type is more accurate when strains are processed in parallel. Relative differences between wild type and mutant are recapitulated in repeated experiments (6).
Strains that have slow vegetative growth can be inoculated here at higher densities to ensure proper density (OD600 2–2.5) prior to transfer to SPM in Subheading 3.1, step 8.
A final concentration of 0.03% galactose is sufficient to give high levels of induction of prototroph formation without affecting the kinetics or efficiency of the meiosis I divisions.
If performing Cre/loxP analysis with the GFP reporter strain, the protocol ends by using fluorescent microscopy to determine the number of tetrads that contain fluorescent spores per total tetrads. Tetrads are analyzed at t = 24 h.
If performing random spore analysis with NDT80 strains, resuspend cells in 1 mL of 20% sorbitol with 0.8 mg zymolyase. Incubate at 37°C for 1 h. Perform 4 rounds of sonication 15 sec at setting 3 (30% maximum power) using the microtip of a 550 Sonic ZD-dismembrator. Rest on ice between rounds. Use a microscope to inspect complete dispersal of spores. If not dispersed, repeat sonication. Continue with Subheading 3.2, step 8 but add 6.25 µL of sonicated cell suspension to 5 mL of water instead of 25 µL.
When plating, use a 0.2-mL glass pipet to spread the cell suspension evenly across the plate but take care to avoid the edges of the Petri dish. This is achieved by spinning a plate on a plating turntable and slowly releasing the cell suspension as one drags the pipet tip outwards. Leaving at least 0.5 cm of space between the edge of the plated suspension and the Petri dish will make counting colonies easier and more accurate.
Take care to distinguish between single round colonies and oddly shaped colonies that may represent two overlapping clones. If colonies are too small to count, slow growing strains can be left to grow for additional days.
Ade+ prototrophs are very distinctly pink or white; however, they may be hard to distinguish from petites or other slow growing colonies. The red color of Ade− auxotrophs does develop over time.
References
- 1.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 2.Loidl J, Scherthan H, Kaback DB. Physical association between nonhomologous chromosomes precedes distributive disjunction in yeast. Proc. Natl. Acad. Sci. USA. 1994;91:331–334. doi: 10.1073/pnas.91.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aragon-Alcaide L, Strunnikov AV. Functional dissection of in vivo interchromosome association in Saccharomyces cerevisiae. Nat. Cell Biol. 2000;2:812–818. doi: 10.1038/35041055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 5.Burgess SM, Kleckner N, Weiner BM. Somatic pairing of homologs in budding yeast: existence and modulation. Genes Dev. 1999;13:1627–1641. doi: 10.1101/gad.13.12.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peoples TL, Dean E, Gonzalez O, Lambourne L, Burgess SM. Close, stable homolog juxtaposition during meiosis in budding yeast is dependent on meiotic recombination, occurs independently of synapsis, and is distinct from DSB-independent pairing contacts. Genes Dev. 2002;16:1682–1695. doi: 10.1101/gad.983802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peoples-Holst TL, Burgess SM. Multiple branches of the meiotic recombination pathway contribute independently to homolog pairing and stable juxtaposition during meiosis in budding yeast. Genes Dev. 2005;19:863–874. doi: 10.1101/gad.1293605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lui DY, Peoples-Holst TL, Mell JC, Wu HY, Dean EW, Burgess SM. Analysis of close stable homolog juxtaposition during meiosis in mutants of Saccharomyces cerevisiae. Genetics. 2006;173:1207–1222. doi: 10.1534/genetics.105.050658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 11.Sherman F, Roman H. Evidence for two types of allelic recombination in yeast. Genetics. 1963;48:255–261. doi: 10.1093/genetics/48.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito RE, Esposito MS. Genetic recombination and commitment to meiosis in Saccharomyces. Proc. Natl. Acad. Sci. USA. 1974;71:3172–3176. doi: 10.1073/pnas.71.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zenvirth D, Loidl J, Klein S, Arbel A, Shemesh R, Simchen G. Switching yeast from meiosis to mitosis: double-strand break repair, recombination and synaptonemal complex. Genes to Cells. 1997;2:487–498. doi: 10.1046/j.1365-2443.1997.1370335.x. [DOI] [PubMed] [Google Scholar]
- 14.Mell JC, Weinhholz BL, Salem A, Burgess SM. Sites of recombination are local determinants of meiotic homolog pairing in Saccharomyces cerevisiae. Genetics. 2008;179:773–784. doi: 10.1534/genetics.107.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane SM, Roth R. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]