Summary
New structures of RNase J reported by Dorléans et al. and Newman et al. in this issue of Structure suggest how an enzyme whose identical subunits each contain a single, buried active site can function as both a 5’ exonuclease and an endonuclease.
RNase J, a prokaryotic ribonuclease present in about half of all bacterial species, including a number of important pathogens, plays a pivotal role in both RNA processing and degradation (Even et al., 2005; Britton et al., 2007; Mäder et al., 2008). It is unique among all known ribonucleases in that it can act as both an endonuclease and a processive 5’ exonuclease (Mathy et al., 2007; Li de la Sierra-Gallay et al., 2008; Richards et al., 2011). Its endonuclease activity, which is specific for single-stranded regions of RNA irrespective of their sequence or location (Daou-Chabo and Condon, 2009), enables it to cleave triphosphorylated primary transcripts internally, while its 5’ exonuclease activity, which is effective only on substrates bearing a single phosphate or a hydroxyl at the 5’ end (Mathy et al., 2007; Li de la Sierra-Gallay et al., 2008; Richards et al., 2011), allows it to rapidly degrade monophosphorylated intermediates produced either by internal cleavage or by the removal of 5’-terminal phosphates.
Many bacterial species contain a single form of RNase J, but some contain two, J1 and J2. In Bacillus subtilis, where RNase J has been best studied, these two paralogs have distinct properties. While both have endonuclease activity, only RNase J1 can function as a 5’ exonuclease (Mathy et al., 2010); furthermore, only RNase J1 is essential for cell growth (Even et al., 2005; Britton et al., 2007). The two paralogs exist in B. subtilis primarily as dimers or heterotetramers (2:2) (Mathy et al., 2010).
Three years ago, Harald Putzer and his coworkers reported the X-ray crystal structure of Thermus thermophilus RNase J bearing a mononucleotide ligand (Li de la Sierra-Gallay et al., 2008). The structure, which is unrelated to that of the eukaryotic 5’ exoribonuclease Xrn1 (Jinek et al., 2011), showed that each protein subunit comprises a hydrolytic metallo-β-lactamase domain flanked by a β-CASP domain and a distinctive C-terminal domain, and crystallographic symmetry suggested how the subunits assemble into dimers (Li de la Sierra-Gallay et al., 2008). As observed for other metallo-β-lactamases, the active site of RNase J was found to contain two zinc ions. Importantly, mutations in this region impaired both exonuclease and endonuclease activity, suggesting that the two hydrolytic activities share a single active site. The uridine monophosphate ligand was bound adjacent to the active site in an orientation suggesting structural mimicry of the 5’-terminal nucleotide of a monophosphorylated RNA molecule undergoing exonucleolytic degradation from the 5’ end. Oddly, however, the conformation of this form of the enzyme was “closed”, such that a polynucleotide ligand could not have been accommodated (Li de la Sierra-Gallay et al., 2008).
In this issue of Structure, two new X-ray crystal structures of RNase J are reported, one from the laboratories of Ciarán Condon and Harald Putzer (Dorléans et al., 2011) and the other from the laboratory of Richard Lewis (Newman et al., 2011). These new structures shed light on the interaction of RNase J with polynucleotide ligands and on its multimeric assembly and disassembly, and suggest how these properties govern its dual catalytic activities.
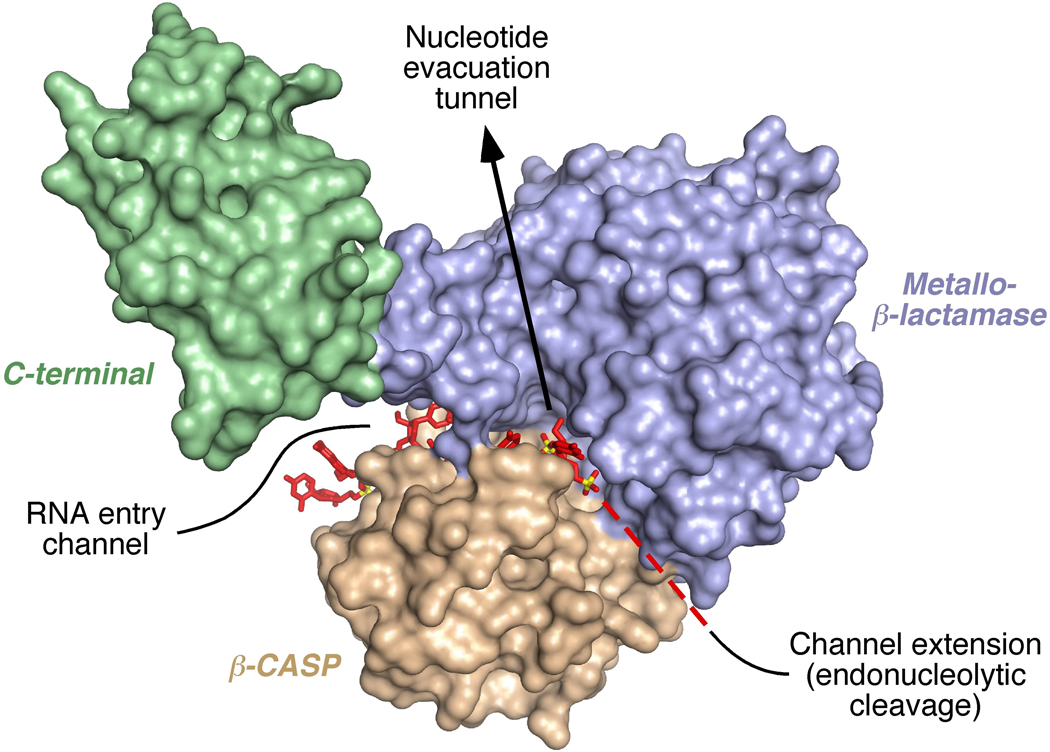
Condon, Putzer, and their coworkers have determined the structure of a catalytically inactive mutant of Thermus thermophilus RNase J bearing an oligonucleotide ligand seemingly poised to undergo 5’-exonucleolytic degradation (Dorléans et al., 2011). Unlike the RNase J structure described previously, the protein conformation is more open due to significant domain movements that create room for RNA binding in a channel between the β-CASP and metallo-β-lactamase domains, at the dimer interface (Figure 1). This RNA entry channel leads from the protein exterior just past the catalytic zinc ions to a 5’-end-binding site deep inside the protein core, the same site previously shown to bind a mononucleotide ligand. The channel is long enough to accommodate a 5’-terminal tetranucleotide and wide enough for single-stranded, but not double-stranded, RNA. Moreover, consistent with the enzyme’s negligible sequence specificity, most of its contacts with the ligand involve the sugar-phosphate backbone rather than the bases, which appear to be held less rigidly in place. A positively charged path on the exposed surface of the β-CASP domain, near the entry channel, may function as a second RNAbinding site that enables the enzyme to act processively when degrading longer RNA substrates exonucleolytically. A distinct, negatively charged tunnel from the buried 5’-end-binding site to the exterior is ideally situated to evacuate mononucleotides released during exonucleolytic degradation (Dorléans et al., 2011).
Figure 1. T. thermophilus RNase J monomer bearing an oligonucleotide ligand poised to undergo 5’-exonucleolytic degradation.
Although RNase J normally exists as a dimer or tetramer, the RNA entry channel, its unoccupied extension, and the putative nucleotide evacuation tunnel have been exposed in this image by omitting all but one protein subunit. In a dimer, the channels and tunnel would be sandwiched between the metallo-β-lactamase (blue) and β-CASP (tan) domains of the two subunits and hidden from view. The C-terminal domain (green) does not appear to contact the oligonucleotide, which is 2'-O-methylated and represented in stick format with all of its atoms colored red, except for phosphorus which is shown in yellow. To be degraded exonucleolytically, RNA must thread through the entry channel until its 5’ terminus reaches the 5’-end-binding site. To be cleaved endonucleolytically, Dorléans et al. and Newman et al. propose that RNA has to occupy both the entry channel and its extension (dashed red line).
Interestingly, the channel in which the RNA lies does not end at the 5’-end-binding site but continues beyond it, re-emerging on the other side of the protein. This suggests a possible mechanism by which RNase J could use the same active site to cleave RNA endonucleolytically. However, this model has a catch, as an RNA molecule could gain access to the whole channel and then get cut internally only if the protein subunits first undergo complete or partial dissociation and if the metallo-β-lactamase and β-CASP domains move apart to expose the channel along its entire length (Figure 1). An alternative model for endonucleolytic cleavage, in which the end of the RNA is threaded all the way through the channel without perturbing the protein subunits, is not consistent with the ability of this RNase J activity to differentiate between internal RNA segments that are intrinsically single- or double-stranded, cleaving only the former irrespective of their location (Dorléans et al., 2011).
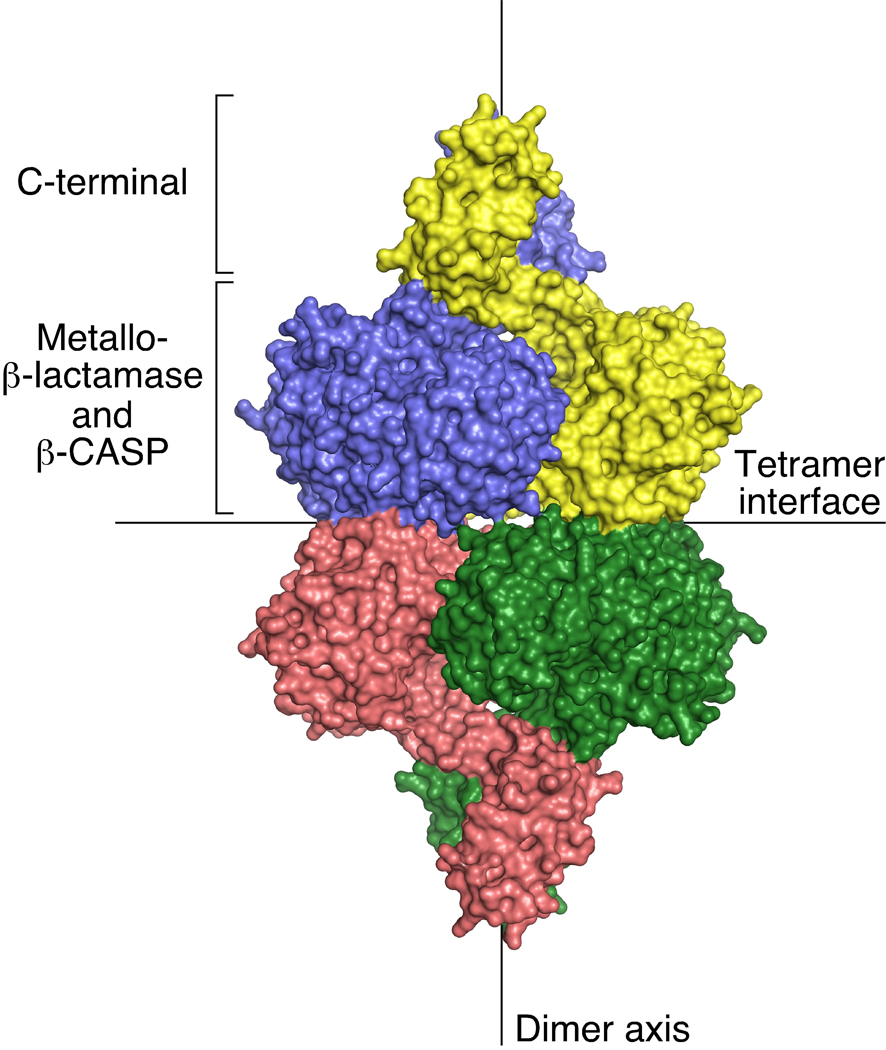
In a complementary study, Lewis and his coworkers have determined the structure of unliganded B. subtilis RNase J1 in an open conformation similar to that observed for the T. thermophilus enzyme bearing an oligonucleotide ligand (Newman et al., 2011). They have modeled how the protein would bind RNA on the basis of its homology to a member of the β-CASP family of metallo-β-lactamases (TTHA0252) whose structure with an oligonucleotide ligand is known. Their models for how RNase J1 could function as either an exonuclease or an endonuclease resemble those deduced from the structure of the T. thermophilus enzyme, and they too note that major structural changes would be needed for an endonucleolytic substrate to bind. In addition, they have carefully examined the dimer and tetramer interfaces in the asymmetric unit of their crystals and the multimeric state of purified RNase J1 and J2 in solution (Newman et al., 2011) (Figure 2). They conclude that the dimers are quite stable (buried surface area of 2500Å2 per subunit) and can assemble into tetramers (buried surface area of 1000Å2 per subunit) with a dissociation constant of about 0.1 µM. Whether dissociation to monomers can occur at a catalytically significant rate remains uncertain; alternatively, it is conceivable that the dimers may transiently undergo partial dissociation while maintaining contact via their C-terminal domains. Thus, depending on its concentration, there is the potential for RNase J to exist in cells as a mixture of tetramers, dimers, and either monomers or loosely associated dimers, all in dynamic equilibrium, consistent with the proposed requirement for complete or partial subunit dissociation prior to endonucleolytic substrate binding.
Figure 2. RNase J tetramer.
The four subunits of the tetramer (each distinctly colored) assemble as a dimer of dimers. All three domains of each subunit make intermolecular contact at the extensive dimer interface, whereas only the metallo-β-lactamase and β-CASP domains meet at the smaller tetramer interface. The figure depicts the enzyme from T. thermophilus; B. subtilis RNase J1 forms a similar tetramer.
In addition to providing important new insights into the function of RNase J, these structures suggest ways in which amino acid substitutions could be used to manipulate the ribonucleolytic activities of RNase J and thereby corroborate the authors’ functional inferences. For example, an amino acid substitution that occludes the putative nucleotide evacuation tunnel should specifically hinder exonucleolytic degradation without impairing endonucleolytic cleavage, whereas obstructing the portion of the RNA-binding channel that is thought to bind the 5’ (upstream) segment of endonucleolytic substrates should have the opposite effect. Similarly, if subunit dissociation must precede endonucleolytic cleavage, then the ratio of endonucleolytic to exonucleolytic activity might be expected to increase when RNase J is diluted or when the dimer and/or tetramer interface is disrupted by mutation. Other mutations could be used to test the importance of the positively charged path on the exposed surface of the β-CASP domain for exonucleolytic processivity.
Together with earlier studies, these new findings also raise some important questions that can now be addressed. For example, how is it that RNase J can cut RNA when the 5’ phosphate of the nucleotide in the 5’-end-binding site is joined to nothing (exonucleolytic degradation of monophosphorylated RNA) or to a polynucleotide chain (endonucleolytic cleavage), but not when it is joined to pyrophosphate (resistance of triphosphorylated RNA to exonucleolytic digestion)? Is RNase J1 essential in B. subtilis because it is the sole source of 5’-exoribonucleolytic activity, and what is the biological utility of a nonessential RNase J paralog (i.e. RNase J2) that is incapable of acting exonucleolytically? How is RNase J2 able to function as effectively as RNase J1 when cleaving RNA internally despite its exonucleolytic inactivity and substitutions for three zinc-coordinating residues? By elucidating these and other puzzles, future studies can be expected to clarify the mechanism and biological function of this important regulatory enzyme.
ACKNOWLEDGMENTS
Research in the Belasco laboratory is supported by Public Health Service grants GM35769 and GM79477 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, Tobin C, Daou R, Szyk J, Condon C. Mol. Microbiol. 2007;63:127–138. doi: 10.1111/j.1365-2958.2006.05499.x. [DOI] [PubMed] [Google Scholar]
- Daou-Chabo R, Condon C. RNA. 2009;15:1417–1425. doi: 10.1261/rna.1574309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorléans A, Li de la Sierra-Gallay I, Piton J, Zig L, Gilet L, Putzer H, Condon C. Structure. 2011;19 doi: 10.1016/j.str.2011.06.018. this issue. [DOI] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Coyle SM, Doudna JA. Mol. Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I, Zig L, Jamalli A, Putzer H. Nat. Struct. Mol. Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H. Mol. Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Mathy N, Hebert A, Mervelet P, Benard L, Dorleans A, Li de la Sierra-Gallay I, Noirot P, Putzer H, Condon C. Mol. Microbiol. 2010;75:489–498. doi: 10.1111/j.1365-2958.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- Newman JA, Hewitt L, Rodrigues C, Solovyova A, Harwood CR, Lewis RJ. Structure. 2011;19 doi: 10.1016/j.str.2011.06.017. this issue. [DOI] [PubMed] [Google Scholar]
- Richards J, Liu Q, Pellegrini O, Yao S, Bechhofer DH, Condon C, Belasco JG. Mol. Cell. 2011;43 doi: 10.1016/j.molcel.2011.07.023. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]