Abstract
Premature ovarian failure in the autosomal dominant disorder blepharophimosis-ptosis-epicanthus inversus is due to mutations in the gene encoding Forkhead L2 (FOXL2), producing putative truncated proteins. We previously demonstrated that FOXL2 is a transcriptional repressor of the steroidogenic acute regulatory (StAR), P450SCC (CYP11A), P450aromatase (CYP19), and cyclin D2 (CCND2) genes, markers of ovarian follicle proliferation and differentiation. Furthermore, we found that mutations of FOXL2 may regulate wild-type FOXL2, leading to loss of transcriptional repression of CYP19, similar to StAR. However, the regulatory mechanisms underlying these premature ovarian failure-associated mutations remain largely unknown. Therefore, we examined the effects of a FOXL2 mutant protein on the transcriptional repression of the CYP19 promoter by the full-length protein. We found that mutant FOXL2 exerts a dominant-negative effect on the repression of CYP19 by wild-type FOXL2. Both wild-type and mutant FOXL2 and can form homo- and heterodimers. We identified a minimal −57-bp human CYP19 promoter containing two potential FOXL2-binding regions and found that both wild-type and mutant FOXL2 can bind to either of these regions. Mutational analysis revealed that either site is sufficient for transcriptional repression by wild-type FOXL2, and the dominant-negative effect of mutant FOXL2, but these are eliminated when both sites are mutated. These findings confirm that mutant FOXL2 exerts a dominant-negative effect on wild-type FOXL2's activity as a transcriptional repressor of key genes in ovarian follicle differentiation and suggest that this is likely due to heterodimer formation and possibly also competition for DNA binding.
Blepharophimosis-ptosis-epicanthus inversus (BPES) syndrome is an autosomal dominant disorder in which patients exhibit a characteristic eyelid dysplasia, blepharophimosis, ptosis, and epicanthus inversus (1), and is due to heterozygous mutations in the forkhead transcription factor Forkhead L2 (FOXL2) gene (2). There are two types of BPES: type 1, in which patients exhibit characteristic eyelid dysplasia in association with premature ovarian failure (POF) and infertility (in affected females only); and type 2, in which the characteristic eyelid dysplasia is not associated with POF, and both affected males and females are fertile (1). Ovaries from patients with BPES type 1 are variable in their histological appearance, ranging from the presence of some primordial follicles with atretic follicles to complete absence of follicles and scarring of the ovaries (3, 4).
FOXL2 is a member of the forkhead/hepatocyte nuclear factor 3 (FKH/HNF3) family of transcription factors (2), members of which are expressed in all eukaryotes and play important roles in the establishment of the body axis and the development of embryonic tissue. Heterozygous mutations of FOXL2 in individuals with BPES type 1 create premature stop codons, which are predicted to result in truncated proteins lacking the carboxyl-terminal alanine/proline-rich domain, which is involved in transcriptional repression (2, 5–8) and therefore plays a pivotal role in the function of this transcription factor. Unlike the FOXL2 mutations that commonly occur in individuals with BPES type 1, the FOXL2 mutations that occur in individuals with BPES type 2 result in elongation of the alanine/proline-rich domain (2, 5–7). These differing mutations may underlie the difference in ovarian phenotypes between BPES type 1 and type 2.
FOXL2 is selectively expressed in the eyelids of developing mice (2) and in the ovarian follicles of adult mice (2, 8, 9). Specifically, it is expressed in the undifferentiated granulosa cells of small and medium follicles in the ovary (8), where it likely functions as a transcriptional repressor of granulosa cell proliferation and differentiation. Steroidogenic acute regulatory protein (StAR) translocates cholesterol from the outer to the inner membrane of mitochondria, which is the rate-limiting step in steroidogenesis (10–12). We have previously shown that FOXL2 binds to the human StAR promoter and suppresses its activity (8). We subsequently demonstrated that FOXL2 also functions as a transcriptional repressor of the P450aromatase (CYP19) promoter (13), which is a key enzyme expressed in granulosa cells and is the rate-limiting step in the conversion of androgens to estrogens. Two other genes that play central roles in ovarian steroidogenesis and granulosa cell proliferation, P450scc (CYP11A) and cyclin D2 (CCND2), are also regulated by FOXL2 (13). Furthermore, we have shown that the alanine/proline-rich C-terminal region of FOXL2 functions as a transrepression domain for the StAR (8) and CYP19 (13) genes. A human FOXL2 mutation (Q219X) that is associated with POF in BPES type 1 (2) produces a truncated FOXL2 protein, FOXL2 [amino acids (aa) 1–218], that lacks the entire alanine/proline-rich region, with resultant loss of repressor activity of the StAR (8) and CYP19 (13) promoters. Interestingly, the FOXL2 (aa 1–218) protein retains its ability to repress the CYP11A and CCND2 promoters (13), suggesting that there are complex mechanisms regulating the transcriptional activity of FOXL2. In this study, we characterized the regulation of the CYP19 promoter by FOXL2, to better understand the mechanisms underlying its role as a transcriptional repressor during follicle development. In addition, because the heterozygous FOXL2 mutation (Q219X) associated with POF in BPES type 1 (2) leads to selective loss of repressor activity of the CYP19 promoter (13), and this is a key enzyme signifying granulosa cell differentiation, we set out to determine the effects of this heterozygous mutation on transcriptional repression of the CYP19 promoter by wild-type FOXL2, to identify its potential role in POF in patients with BPES type 1.
Materials and Methods
Plasmids
Wild-type and mutant human FOXL2 pcDNA3 expression vectors were prepared as described previously (8). These two cDNA sequences were released from a pcDNA3 backbone using the restriction enzymes HindIII and XbaI and subcloned into pFLAG-CMV-2 (Sigma Chemical Co., St. Louis, MO) and phCMV (Genlantis, San Diego, CA) to generate FLAG-tagged and hemagglutinin (HA)-tagged cDNA. The human CYP11A promoter (−1430 bp, GenBank accession no. M60421) and human CDKN1B promoter (−1899-bp, sequence obtained from UCSC Genome Browser) constructs were described previously (13). Ovary-specific human CYP19 promoter fragments (GenBank accession no. S85356) spanning from −633, −313,−114, and −57 bp to the translational start site were amplified from genomic DNA and Hotstar Taq polymerase (QIAGEN, Valencia, CA) according to the manufacturer's protocol. PCR products for each of these promoter fragments were electrophoresed, extracted, and subcloned into the pGL2 luciferase reporter vector backbone (Promega, Madison, WI). Three mutants for the −57-bp ovary-specific human CYP19 promoter, 5′-cacggcacacacacgacaggactctaaattgccccctctgaggtcaaggaacacaag-3′ (mt1), 5′-gtccctttgatttccacaggactctaaattgccccctctgagctcacggcacacacg-3′ (mt2), and 5′-cacggcacacacacgacaggactctaaattgccccctctgagctcacggcacacacg-3′ (mt1/2) were synthesized and subcloned into the pGL2 luciferase reporter vector backbone (Promega). The human CCND2 (−1624 bp) promoter was a generous gift from Dr. Masataka Nakamura.
Transfection and reporter assays
CHO cells (1 × 105 per well in a 24-well plate) were cultured in DMEM/F12 supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine. The following day, cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with the CYP19, CYP11A, CCND2, or CDKN1B promoter luciferase reporter vectors (500 ng/well), or empty pGL2 backbone (as a control) and wild-type and/or mutant FOXL2 (0, 10, or 20 ng/well). The total DNA concentration in each well was maintained at 1 μg by adding the empty pcDNA3 expression vector, including 50 ng indicator plasmid, pCMV-β-galactosidase. Transfections were performed in Opti-MEM (Invitrogen) for 4 h, after which cells were cultured in fresh medium with 10% fetal bovine serum for 24 h. Twenty-four hours after transfection, the cells were washed with PBS and lysed in 100 μl 1× reporter lysis buffer (Promega), followed by freeze/thawing to ensure complete lysis. Twenty microliters of each sample were used to measure luciferase activity using a FLUOstar OPTIMA plate reader (BMG Labtech, Offenberg, Germany). β-Galactosidase activity was measured using the same plate reader, after the addition of 50 μl β-galactosidase substrate (200 mm sodium phosphate buffer, 2 mm magnesium chloride, 100 mm β-mercaptoethanol, and 1.33 mg/ml ortho-nitrophenyl-β-galactoside) to 5 μl cell lysate from each sample in a 96-well plate. Results were first normalized using the β-galactosidase activity. In each case, the normalized luciferase activity was then divided by the normalized luciferase activity obtained for the empty pGL2 vector backbone under the same experimental conditions, and the results are reported as the fold change relative to the luciferase activity of the empty vector backbone. Each condition was tested in quadruplicate, and each experiment was repeated at least three times.
Immunoprecipitation
CHO cells were transfected with FLAG-tagged wild-type or mutant FOXL2 for 24 h and then lysed in an immunoprecipitation (IP) buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, and 0.2 mm phenylmethylsulfonyl fluoride. The cell lysates were incubated with anti-FLAG M2 affinity gel (Sigma) at 4 C for 4 h. The gel was then washed with Tris-buffered saline [50 mm Tris-HCl (pH 7.4) and 150 mm NaCl] to eliminate nonspecific binding and incubated with 100 μg/ml FLAG peptide (Sigma) in Tris-buffered saline at 4 C for 30 min to elute the bound proteins. The eluted samples or cell lysates were added to 4× sodium dodecyl sulfate (SDS) sample buffer and heated at 95 C for 5 min to denature the proteins. The proteins of interest in the eluted samples or cell lysates were then analyzed by Western blotting.
Western blotting
The nuclear and cytoplasmic extracts, cell lysates, and IP products described in the previous sections were added to 4× SDS sample buffer and heated at 95 C for 5 min to denature the proteins. The cell lysates were then separated on 7.5% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. The membranes were incubated with primary antibodies to FLAG or HA (Sigma), washed, and then incubated with horseradish peroxidase-conjugated secondary antibodies. Chemiluminescent detection was performed using enhanced chemiluminescence Western blotting detection reagents (Amersham Biosciences, Piscataway, NJ).
Electrophoretic mobility shift assay
FLAG-tagged wild-type FOXL2 and mutant FOXL2 were immunoprecipitated from transfected CHO cells as described above. EMSA was then performed using a LightShift chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL). The IP products were mixed for 20 min with 20 fmol of a 5′ biotin-labeled DNA probe for the wild-type or mutant −57-bp CYP19 promoter fragment, either alone or with 2 pmol of the unlabeled −57-bp DNA probe as a competitor. The mixtures were then electrophoresed through 6% polyacrylamide gels and transferred to nylon membranes, and the bound biotin-labeled DNA fragments were detected by chemiluminescence.
Chromatin IP (ChIP)
KGN cells were grown in DMEM/F12 supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mm glutamine in a humidified atmosphere with 5% CO2 at 37 C. After the cells reached 90% confluency, they were transfected with either empty pFLAG-CMV-2, wild-type FOXL2 pFLAG-CMV-2, or mutant FOXL2 pFLAG-CMV-2 using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, formaldehyde (Fisher Scientific, Pittsburgh, PA) was added directly to cell culture media at a final concentration of 1%. Fixation was allowed to proceed for 10 min and then stopped by the addition of glycine to a final concentration of 0.125 m. The cells were then washed twice with ice-cold PBS and harvested in lysis buffer [1% Triton X-100, 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 0.5 mm phenylmethylsulfonyl fluoride]. The cell lysates were sonicated on ice to obtain DNA fragments with an average length of 200-1000 bp. The sonicated lysates were precleared by adding protein G-agarose (Millipore, Billerica, MA) and rotated at 4 C for 1 h. The supernatants were then incubated with mouse IgG or M2 FLAG antibody (Sigma-Aldrich, St. Louis, MO) at 4 C overnight, and then protein G-agarose was added for another 2-h incubation. The agarose was then washed twice with buffer containing 250 mm LiCl, 1% Triton X-100, 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 0.5 mm phenylmethylsulfonyl fluoride and twice more with buffer containing 50 mm Tris-HCl (pH 7.4) and 150 mm NaCl. FLAG-tagged proteins were then eluted using 100 μg/ml FLAG peptide in 50 mm Tris-HCl (pH 7.4) and 150 mm NaCl. The DNA-protein complexes were de-cross-linked at 65 C overnight. The eluates were treated with ribonuclease at 37 C for 1 h and proteinase K at 45 C for 2 h. The DNA fragments in the eluates were then concentrated using a PCR cleanup kit (QIAGEN). The final products were subjected to real-time PCR using the primers 5′-CCTCTGAAGCAACAGGAGCTAT-3′ and 5′-AAAACCATCTTGTGTTCCTTGAC-3′, which correspond to the −113- to +8-bp region of the human CYP19 promoter.
Real-time PCR was performed on a MyiQ thermal cycler (Bio-Rad Laboratories, Hercules, CA) using iQ SYBR green supermix (Bio-Rad) for 50 cycles in two-step reactions: 95 C for 20 sec and 58 C for 1 min. The cycle threshold (Ct) counts for IP with IgG and M2 FLAG antibodies were first normalized against the Ct counts for 1% input, to give the percentage of total input. The percentages of total input from IP using the M2 FLAG antibody were then divided by the percentages of total input from IP using the IgG antibody. The results from ChIP with cells transfected with wild-type FOXL2 and mutant FOXL2 were further normalized against the results from ChIP with cells transfected with empty pFLAG-CMV-2 vector (control).
Statistical analysis
For promoter assay studies, experiments were performed in quadruplicate and the se reported. For the ChIP assay study, the results shown are the average of three individual experiments and the sd are as indicated. Statistical analysis was performed using one-way ANOVA, and significance was at P < 0.05.
Results
Dominant-negative effect of mutant FOXL2
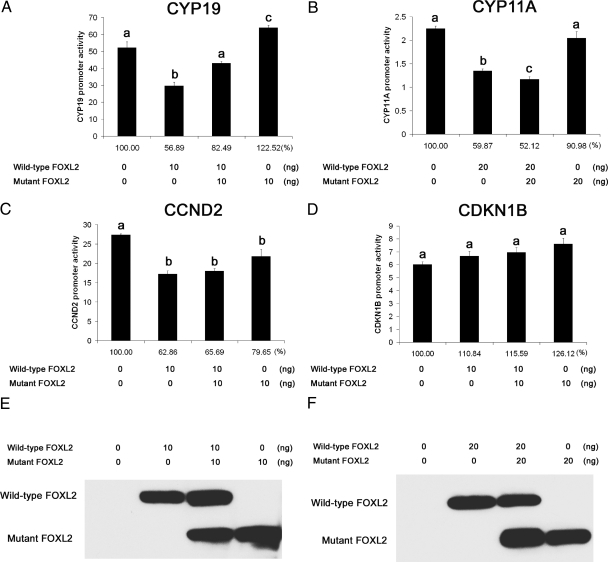
Ovarian failure in BPES type 1 has been attributed to either haploinsufficiency or a dominant-negative effect whereby the mutant protein interferes with the action of wild-type FOXL2 (2, 5, 14). Our previous studies have shown that the FOXL2 (aa 1–218) mutant protein exerts a dominant-negative effect on wild-type FOXL2's activity as a transcriptional repressor of the StAR promoter (8). To determine whether mutant FOXL2 also adversely affects the transcriptional repression of other promoters by FOXL2, mutant FOXL2 was cotransfected with wild-type FOXL2 and CYP19, CYP11A, CCND2, or CDKN1B promoter constructs. Equal concentrations of mutant and wild-type FOXL2 were used for these experiments (Fig. 1, A–F). Wild-type FOXL2's activity as a transcriptional repressor of the CYP19 promoter diminished when coexpressed with mutant FOXL2 (Fig. 1A). However, this effect was not observed for the CYP11A or CCND2 promoters (Fig. 1, B and C). Furthermore, mutant FOXL2 alone slightly induced transcription of the CYP19A promoter (Fig. 1A) and slightly repressed transcription of the CCND2 promoter (Fig. 1C). Neither mutant nor wild-type FOXL2 had a repressive effect on CDKN1B promoter activity (Fig. 1D).
Fig. 1.
Dominant-negative effect of mutant FOXL2. CHO cells were transiently transfected with wild-type FOXL2 and/or the FOXL2 (aa 1–218) mutant expression constructs, together with reporter constructs for CYP19 (A), CYP11A (B), CCND2 (C), or the CDKN1B control promoter (D). Twenty-four hours after transfection, the cells were lysed and luciferase activity measured. Results were first normalized against β-galactosidase activity and reported as the fold change relative to the luciferase activity of the empty pGL2 vector backbone under the same experimental conditions. Results were also calculated as the percentage of control (no FOXL2 transfected, 100%), shown below the columns. Coexpression with mutant FOXL2 led to a decrease in wild-type FOXL2's activity as a transcriptional repressor of the CYP19 promoter (A) but did not affect transcriptional repression of the CYP11A (B) or CCND2 (C) promoters. Mutant FOXL2 alone slightly induced transcription of the CYP19A promoter (A) and slightly repressed transcription of the CCND2 promoter (C). Each condition was tested in quadruplicate. One-way ANOVA was performed between samples, and different letters (a, b, or c) denote significant differences (P < 0.05) between samples. E and F, Western blots showing the relative amounts of wild-type and mutant FOXL2 protein expressed after transient transfection.
Dimerization of wild-type and mutant FOXL2
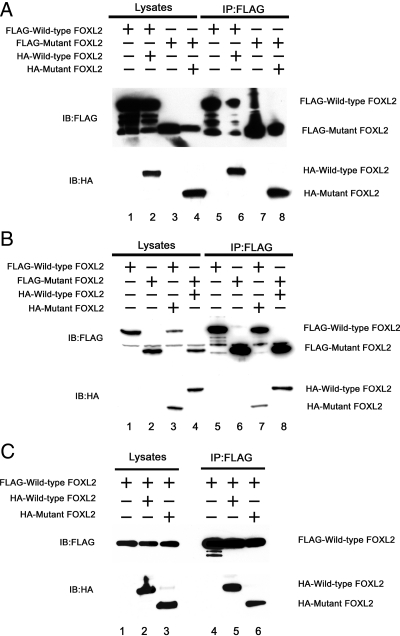
The forkhead recognition motif is structurally similar to helix-turn-helix proteins (15, 16) that are associated with dimerization (17–19). Recently, Lamba et al. (20) demonstrated that FOXL2 can self-associate in cells, and other forkhead family members exist as monomers and dimers (21). As dominant-negative effects may occur as a result of dimerization, we determined whether wild-type and mutant FOXL2 can exist as dimers. CHO cells were transiently cotransfected with FLAG-tagged and HA-tagged FOXL2 (wild-type or mutant). FLAG-tagged FOXL2 (wild-type or mutant) was then immunoprecipitated using an anti-FLAG antibody, and the immunoprecipitates were analyzed by Western blotting with anti-FLAG and anti-HA antibodies. As shown in Fig. 2A, when CHO cells coexpressed FLAG-tagged and HA-tagged wild-type FOXL2 (lane 2), HA-tagged wild-type FOXL2 was present in the FLAG immunoprecipitates [lane 6, immunoblot (IB) HA]. When CHO cells only expressed FLAG-tagged wild-type FOXL2 (lane 1), HA-tagged FOXL2 was absent from the FLAG immunoprecipitates (lane 5, IB HA). Similarly, when CHO cells coexpressed FLAG-tagged and HA-tagged mutant FOXL2 (lane 4), HA-tagged mutant FOXL2 was present in the FLAG immunoprecipitates (lane 8, IB HA). When CHO cells expressed only FLAG-tagged wild-type FOXL2 (lane 3), HA-tagged FOXL2 was absent from the FLAG immunoprecipitates (lane 7, IB HA). Thus, both wild-type and mutant FOXL2 appear to form homodimers.
Fig. 2.
Dimerization of wild-type and mutant FOXL2. A, To test for the ability to form homodimers, CHO cells were transiently cotransfected with FLAG-tagged wild-type FOXL2 alone (lane 1), with FLAG-tagged and HA-tagged wild-type FOXL2 (lane 2), with FLAG-tagged mutant FOXL2 alone (lane 3), or with FLAG-tagged and HA-tagged mutant FOXL2 (lane 4). The cell lysates were immunoprecipitated using an anti-FLAG antibody (lanes 5–8) and analyzed by Western blotting with antibodies to FLAG and HA. FLAG-wild-type FOXL2 coimmunoprecipitates with HA-wild-type FOXL2. Furthermore, FLAG-mutant FOXL2 coimmunoprecipitates with HA-mutant FOXL2. B, To test the ability to form heterodimers, CHO cells were transiently cotransfected with FLAG-tagged wild-type FOXL2 alone (lane 1), FLAG-tagged mutant FOXL2 alone (lane 2), FLAG-tagged wild-type FOXL2 and HA-tagged mutant FOXL2 (lane 3), or with FLAG-tagged mutant FOXL2 and HA-tagged wild-type FOXL2 (lane 4). The cell lysates were immunoprecipitated using an anti-FLAG antibody (lanes 5–8) and analyzed by Western blotting with antibodies to FLAG and HA. FLAG-wild-type FOXL2 and HA-mutant FOXL2 coimmunoprecipitate, as do HA-wild-type FOXL2 and FLAG-mutant FOXL2. C, To examine whether wild-type FOXL2 has different affinity for wild-type vs. mutant FOXL2, CHO cells were transiently cotransfected with FLAG-tagged wild-type FOXL2 alone (lane 1), FLAG-tagged wild-type FOXL2 and HA-tagged wild-type FOXL2 (lane 2), or FLAG-tagged wild-type FOXL2 and HA-tagged mutant FOXL2 (lane 3). The cell lysates were immunoprecipitated using an anti-FLAG antibody (lanes 4–6) and analyzed by Western blotting with antibodies to FLAG and HA. FLAG-tagged wild-type FOXL2 had affinity for HA-tagged mutant FOXL2 but slightly less than for HA-tagged wild-type FOXL2.
We then tested whether wild-type and mutant FOXL2 could form heterodimers, which might explain the dominant-negative effects of mutant FOXL2 on the wild-type protein. As shown in Fig. 2B, when CHO cells coexpressed FLAG-tagged wild-type FOXL2 and HA-tagged mutant FOXL2 (lane 3), HA-tagged mutant FOXL2 was present in the FLAG immunoprecipitates (lane 7, IB HA). When CHO cells coexpressed FLAG-tagged mutant FOXL2 and HA-tagged wild-type FOXL2 (lane 4), HA-tagged wild-type FOXL2 was present in FLAG immunoprecipitates (lane 8, IB HA). When CHO cells expressed only FLAG-tagged wild-type or mutant FOXL2 (lanes 1 and 2), no HA-tagged FOXL2 was detected in the immunoprecipitates (lanes 5 and 6). Thus, in addition to homodimer formation, the wild-type and mutant FOXL2 proteins also appear to form heterodimers.
To examine whether wild-type FOXL2 has greater affinity for wild-type or mutant FOXL2 (i.e. whether it will preferentially form homodimers or heterodimers), FLAG-tagged wild-type FOXL2 was cotransfected with HA-tagged wild-type or mutant FOXL2 (Fig. 2C, lanes 1–3). The cell lysates were immunoprecipitated using an anti-FLAG antibody (lanes 4–6). HA-tagged wild-type and mutant FOXL2 were both present in immunoprecipitates (lanes 5 and 6). The amount of HA-tagged mutant FOXL2 was slightly less than that of HA-tagged wild-type FOXL2, suggesting that FLAG-tagged wild-type FOXL2 binds to mutant FOXL2 with high affinity but slightly less than its affinity for wild-type FOXL2.
Effects of mutant FOXL2 on repression of the minimal CYP19 promoter by wild-type FOXL2
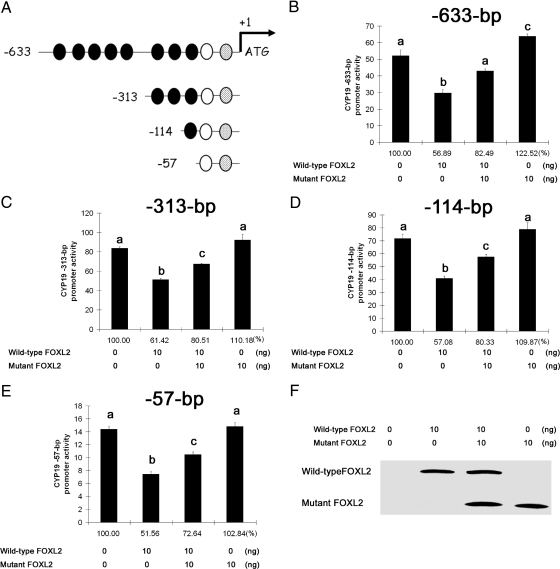
As our results to date indicate that FOXL2 functions as a transcriptional repressor of the CYP19 promoter (13), we set out to determine the minimal CYP19 promoter region responsive to FOXL2 regulation. We first generated a series of luciferase reporter vectors containing truncated CYP19 promoter fragments (Fig. 3A). We performed an in silico analysis of the CYP19 promoter and found that the −663-, −313-, −114-, and −57-bp promoter fragments contain eight, three, one, and zero traditional forkhead consensus sites, 5′-(G/A)(T/C)(C/A)AA(C/T)A-3′, respectively. All of these promoter fragments also contain a sequence similar to the core of the nonconsensus FOXL2 binding element identified by Lamba et al. (22), 5′-TGTTTA-3′. The region surrounding this binding element also has sequence similarity to the reverse complement of the FOXL2 binding site identified by Benayoun et al. (23), 5′-GTCAAGG(T/C)-3′. An additional binding site identified by Benayoun et al. (23) is located downstream, closer to the start site, in all of the truncated promoter fragments (Fig. 3A). Wild-type FOXL2 and/or mutant FOXL2 and each of these CYP19 promoter-luciferase constructs were transiently cotransfected into CHO cells. As demonstrated in Fig. 3, B–E, wild-type FOXL2 functions as a transcriptional repressor of all of the CYP19 promoter fragments, including the smallest −57-bp region, whereas mutant FOXL2 slightly induced the −633-bp region but had no effect on the other CYP19 promoter fragments. Furthermore, wild-type FOXL2's activity as a transcriptional repressor of these CYP19 promoter fragments was diminished in all cases when coexpressed with mutant FOXL2 (Fig. 3, B–E).
Fig. 3.
Minimal aromatase promoter region responsive to repression by FOXL2. A, Schematic representation of the human ovary-specific CYP19 promoter fragments used in this study. The black ovals represent forkhead consensus sites. The white oval represents a region that contains a sequence similar to the core of the nonconsensus FOXL2 binding element identified by Lamba et al. (22), 5′-TGTTTA-3′ and also has sequence similarity to the reverse complement of the FOXL2-binding site identified by Benayoun et al. (23), 5′-GTCAAGG(T/C)-3′. The gray oval represents a region containing the binding sequence identified by Benayoun et al. (23). B–E, CHO cells were transiently transfected with wild-type FOXL2 and/or mutant FOXL2 as well as the reporter constructs for the −633-bp (B), −313-bp (C), −114-bp (D), or −57-bp (E) CYP19 promoter fragments. Twenty-four hours after transfection, luciferase activity was measured. Results were normalized against β-galactosidase activity and reported as fold change relative to the luciferase activity of the empty pGL2 vector backbone under the same experimental conditions. Results were also calculated as the percentage of control (no FOXL2 transfected, 100%), shown below the columns. Wild-type FOXL2 represses the activities of all of the CYP19 promoter fragments (by 40–50%), but none were repressed by mutant FOXL2. Cotransfection with both wild-type and mutant FOXL2 results in diminished repressor activity. Each condition was tested in quadruplicate. One-way ANOVA was performed between samples, and different letters (a, b, c, or d) denote significant differences (P < 0.05) between samples. F, Western blot showing the relative amounts of wild-type and mutant FOXL2 protein expressed after transient transfection.
Wild-type and mutant FOXL2 bind to the −57-bp region of the CYP19 promoter
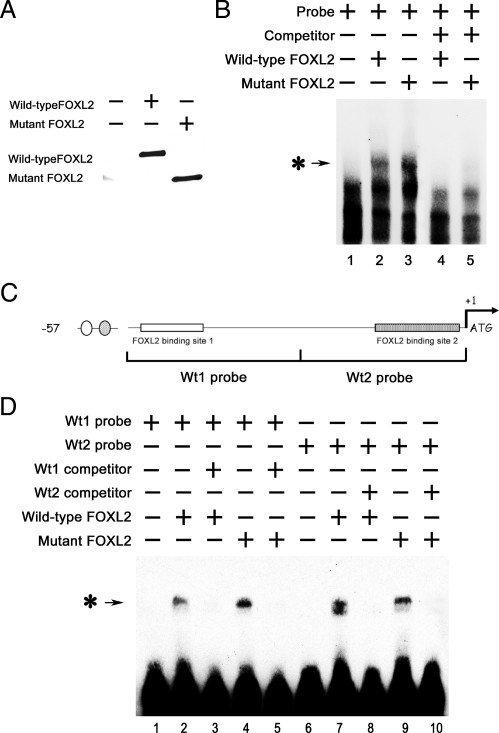
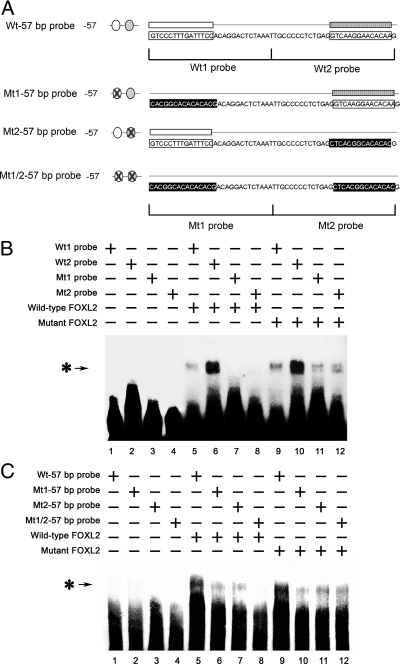
The FOXL2 (aa 1–218) mutant protein lacks the C-terminal alanine/proline-rich repressor domain; however, it retains an intact forkhead DNA-binding domain. Therefore, we used gel shift assays to test whether the inhibitory effects of mutant FOXL2 on wild-type FOXL2's activity as a transcriptional repressor might be due to competition for DNA binding. FLAG-IP products from CHO cells that had been transfected with empty vector backbone (as a control), a FLAG-tagged wild-type FOXL2 expression vector, or a FLAG-tagged mutant FOXL2 expression vector were used as protein substrates (Fig. 4A). An oligonucleotide probe corresponding to the −57-bp region of the CYP19 promoter was labeled with biotin and combined with immunoprecipitates of FLAG-tagged wild-type or mutant FOXL2. The resulting protein-DNA complexes were then analyzed using the LightShift EMSA system. When the biotin-labeled probe was mixed with wild-type FOXL2 or mutant FOXL2, a single protein-DNA complex band was obtained (Fig. 4B, lanes 2 and 3). However, this band was not seen when the biotin-labeled probe was mixed with the IP product from control-transfected CHO cells (Fig. 4B, lane 1). When an excess of competitor (non-biotin-labeled probe) was included in the mixture, the protein-DNA complex bands were eliminated (Fig. 4B, lanes 4 and 5). These results suggest that both wild-type and mutant FOXL2 can bind to this minimal −57-bp CYP19 promoter fragment.
Fig. 4.
Wild-type and mutant FOXL2 bind to the −57-bp region of the CYP19 promoter. CHO cells were transfected with FLAG-tagged wild-type FOXL2 or mutant FOXL2, and after 24 h of transfection, FLAG-tagged proteins were immunoprecipitated. A, Western blot showing the relative amounts of wild-type and mutant FOXL2 protein expressed after transient transfection. B, An oligonucleotide probe corresponding to the −57-bp region of the CYP19 promoter was labeled with biotin and mixed with 3 μl of IP products from CHO cells that had been transfected with empty vector backbone (lane 1), wild-type FOXL2 (lane 2), mutant FOXL2 (lane 3), wild-type FOXL2 and an excess of competitor (non-biotin-labeled probe) (lane 4), or mutant FOXL2 and an excess of competitor (lane 5). These mixtures were then analyzed using the LightShift EMSA system. A protein-DNA complex band was detected when nuclear extracts from CHO cells expressing wild-type or mutant FOXL2 (asterisk-arrow) was combined with the −57-bp CYP19 promoter and was competed out by an excess of unlabeled probe. C, Schematic of the −57-bp CYP19 promoter indicating the two potential FOXL2-binding sites. Corresponding 30-bp biotin-labeled probes (wt1 and wt2 probes) and non-biotin-labeled competitors (wt1 and wt2 competitors) were synthesized. D, Wt1 and wt2 probes (lanes 1–5 and 6–10, respectively) were mixed with wild-type (lanes 2 and 7) or mutant (lanes 4 and 9) FOXL2 or with wild-type (lanes 3 and 8) or mutant FOXL2 (lanes 5 and 10) and excess amounts of competitors. A protein-DNA complex band was detected in mixtures containing wild-type and/or mutant FOXL2 and either the wt1 or wt2 probe (asterisk-arrow), suggesting that both wild-type and mutant FOXL2 can bind to both binding sites.
As described above, the −57-bp CYP19 promoter fragment contains two potential FOXL2-binding regions. The first, more upstream region includes a sequence similar to the nonconsensus FOXL2-binding element identified by Lamba et al. (22) and the reverse complement of the FOXL2-binding site identified by Benayoun et al. (23). The second, downstream, region contains an additional binding sequence identified by Benayoun et al. (23) (Fig. 4C). We then tested whether either of these regions can bind to wild-type and/or mutant FOXL2 by generating corresponding biotin-labeled 30-bp DNA probes (wt1 and wt2), and non-biotin-labeled competitors (wt1 and wt2 competitors) (Fig. 4C). When these 30-bp biotin-labeled probes (Fig. 4D, wt1 probe, lanes 1–5, wt2 probe, lanes 6–10) were mixed with wild-type FOXL2 or mutant FOXL2, a single protein-DNA complex band was obtained (Fig. 4D, wild-type FOXL2, lanes 2 and 7; mutant FOXL2, lanes 4 and 9). However, these bands were not seen when the biotin-labeled probes were mixed with IP products from control-transfected cells (Fig. 4D, lanes 1 and 6). When an excess of either competitor (non-biotin-labeled probe) was included in the mixture, the protein-DNA complex bands were eliminated (Fig. 4D, wild-type FOXL2, lanes 3 and 5; mutant FOXL2, lanes 8 and 10). These results suggest that both wild-type and mutant FOXL2 can bind to either of these two potential FOXL2-binding regions.
Mutation of FOXL2 binding sites in the −57-bp CYP19 promoter region results in loss of FOXL2 transcriptional repressor activity
To determine whether these potential binding regions are involved in transcriptional repression of the CYP19 promoter by FOXL2, we mutated these sequences either individually or together (Fig. 5A). The wild-type and mutant DNA-binding sites 1 and 2 were synthesized as 30-bp biotin-labeled probes (Wt1, Wt2, Mt1, and Mt2). Wild-type and mutant biotin-labeled 57-bp probes were also generated (Wt-57 bp, Mt1-57 bp, Mt2-57 bp, and Mt1/2-57 bp). The binding affinities of each mutant binding region (Mt1 and Mt2) were first determined. As shown in Fig. 5B, when the 30-bp biotin-labeled wild-type and mutant probes, Wt1, Wt2, Mt1, and Mt2, were mixed with IP products from control-transfected cells (Fig. 5B, lanes 1–4), no band was seen. When the 30-bp biotin-labeled wild-type probes (Wt1 and Wt2) were mixed with wild-type FOXL2 (lanes 5 and 6) or mutant FOXL2 (lanes 9 and 10), a single protein-DNA complex band was obtained. These bands were not observed when the biotin-labeled mutant probes (Mt1 and Mt2) were mixed with wild-type FOXL2 (lanes 7 and 8). However, the 30-bp biotin-labeled mutant probes (Mt1 and Mt2) still had some affinity for mutant FOXL2 (lanes 11 and 12). The biotin-labeled 57-bp probes were also tested. Strong binding was observed between the Wt-57 bp probe and wild-type or mutant FOX2 (Fig. 5C, lanes 5 and 9). In contrast, when wild-type and mutant FOXL2 were mixed with the Mt1-57 bp and Mt2-57 bp probes, the intensity of the resulting protein-DNA complex bands was much weaker (Fig. 5C, lanes 6 and 10; lanes 7 and 11). There was no binding between wild-type FOXL2 and the double mutant Mt1/2-57 bp probe (Fig. 5C, lane 8), but mutant FOXL2 was still able to bind to this Mt1/2-57 bp probe (Fig. 5C, lane 12). Taken together, the results shown in Figs. 4 and 5 suggest that both wild-type and mutant FOXL2 can bind to the binding sites identified by Lamba et al. (22) and by Benayoun et al. (23). However, mutant FOXL2 appears to have less specificity of binding than wild-type FOXL2; i.e. the mutant FOXL2 protein appears more able to bind to altered binding sites than wild-type FOXL2.
Fig. 5.
Mutation of FOXL2-binding sites reduces binding ability of FOXL2. A, Schematic of the −57-bp region of the CYP19 promoter illustrating the 30- and 57-bp wild-type (wt) and mutant (mt) oligonucleotide probes used. B, The 30-bp Wt1, Wt2, Mt1, or Mt2 probes were mixed with IP products from CHO cells transfected with the empty vector backbone (lanes 1–4), FLAG-tagged wild-type FOXL2 (lanes 5–8), or FLAG-tagged mutant FOXL2 (lanes 9–12). The protein-DNA complex band was detected in mixtures containing wild-type FOXL2 and either the Wt1 or Wt2 probes (lanes 5 and 6, asterisk-arrow) but not the Mt1 and M2 probes (lanes 7 and 8). This protein-DNA complex band was also strongly detected in mixtures containing mutant FOXL2 and the Wt1 or Wt2 probes (lanes 9 and 10) and more weakly detected in mixtures containing mutant FOXL2 and the Mt1 and Mt2 probes (lanes 11 and 12). C, The 57-bp probes Wt-57 bp, Mt1-57 bp, Mt2-57 bp, and Mt1/2-57 bp were also mixed with IP products from CHO cells transfected with the empty vector backbone (lanes 1–4), FLAG-tagged wild-type FOXL2 (lanes 5–8), or FLAG-tagged mutant FOXL2 (lanes 9–12). A protein-DNA complex band was strongly detected in mixtures containing wild-type FOXL2 and the Wt-57 bp probe (lane 5, asterisk-arrow) and more weakly detected in mixtures containing the Mt1-57 bp and Mt2-57 bp probes (lanes 6 and 7) but not in mixtures containing the Mt1/2-57 bp probe (lane 8). A protein-DNA complex band was strongly detected in mixtures containing mutant FOXL2 and the Wt-57 bp probe (lane 9, asterisk-arrow) and more weakly detected in mixtures containing mutant FOXL2 and the Mt1-57 bp, Mt2-57 bp, and Mt1/2-57 bp probes (lanes 10, 11, and 12, respectively).
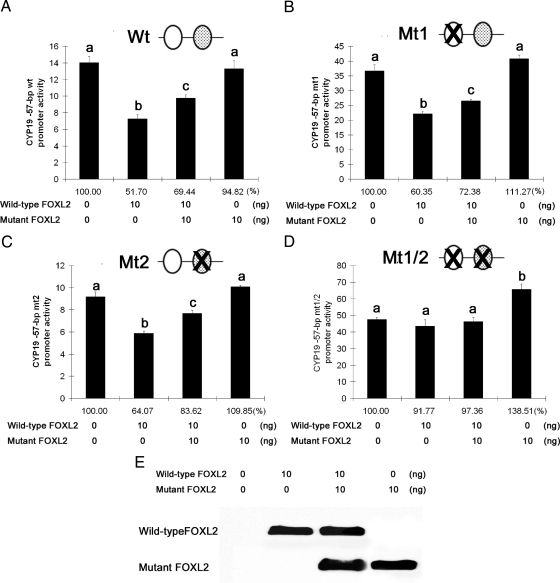
We then tested the effects of the presence of wild-type and mutant DNA-binding sites on the activity of wild-type FOXL2 as a transcriptional repressor of the CYP19 promoter, using luciferase assays. When both binding sites were intact, repression of CYP19 promoter activity by FOXL2 occurred as expected (Wt, Fig 6A). When DNA-binding region 1 or 2 was mutated, this repression still occurred (Mt1 and Mt2, Fig 6, B and C), but when both binding regions were mutated, transcriptional repression was lost (Mt1/2, Fig 6D). Furthermore, wild-type FOXL2's activity as a transcriptional repressor of the Wt, Mt1, and Mt2 promoters diminished when coexpressed with the mutant FOXL2 protein (Fig. 6, A–C), consistent with our previous results. The mutant FOXL2 protein had no repressive effect on any of the constructs (Fig. 6, A–D). These results suggest that the presence of one intact DNA-binding site is sufficient for the repressor activity of wild-type FOXL2 and for the dominant-negative effect of the mutant FOXL2 protein.
Fig. 6.
Mutation of FOXL2-binding sites results in loss of FOXL2 transcriptional repressor activity. A–D, CHO cells were transiently transfected with expression constructs for wild-type FOXL2 and/or mutant FOXL2 and with reporter constructs for the wild-type or mutated −57-bp CYP19 promoter constructs, Wt (A), Mt1 (B), Mt2 (C), and Mt1/2 (D). Twenty-four hours after transfection, luciferase activity was measured. Results were normalized against β-galactosidase activity and reported as fold change relative to the luciferase activity of the empty pGL2 vector backbone under the same experimental conditions. Results were also calculated as the percentage of control (no FOXL2 transfected, 100%) shown below the columns. Each condition was tested in quadruplicate. One-way ANOVA was performed between samples, and different letters (a, b, or c) denote significant differences (P < 0.05) between samples. Transcriptional repression by wild-type FOXL2, and the dominant-negative effect of mutant FOXL2, were not affected when either binding site was mutated individually (B and C), but mutation of both sites resulted in loss of transcriptional repression (D). E, Western blot showing the relative amounts of wild-type and mutant FOXL2 protein expressed after transient transfection.
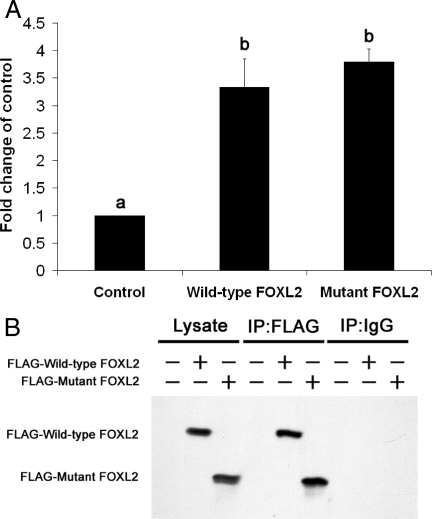
To confirm that wild-type and mutant FOXL2 binds to the aromatase promoter, and to demonstrate binding in vivo, ChIP studies were performed using KGN cells overexpressing wild-type or mutant FOXL2. The KGN cell line (24) has been used as a model of granulosa cells of the postnatal ovary. These adult granulosa cells are derived from granulosa cell tumors. As shown in Fig. 7A, wild-type and mutant FOXL2 bound to more DNA fragments corresponding to the −113- to +8-bp region of human CYP19 promoter (3.3- and 3.8-fold vs. control, respectively) in KGN cells. The expression of wild-type and mutant FOXL2 in KGN cells and the IP activities are shown in Fig. 7B. These results further support that wild-type and mutant FOXL2 bind to the human CYP19 promoter in vivo.
Fig. 7.
ChIP suggests binding of FOXL2 to the human CYP19 promoter. KGN cells were transiently transfected with empty expression construct (control) or expression constructs for FLAG-tagged wild-type or mutant FOXL2. FLAG-tagged wild-type and mutant FOXL2 were then immunoprecipitated using the anti-FLAG M2 antibody. A, The amount of DNA cross-linked with FLAG-tagged wild-type or mutant FOXL2 was then measured by quantitative real-time PCR with primers corresponding to the −113- to +8-bp region of the human ovarian-specific CYP19 promoter. The results shown represent the averages of three independent experiments. The Ct counts for IP with IgG and M2 FLAG antibodies were first normalized against the Ct counts for 1% input, to give the percentage of total input. The percentages of total input from IP using the M2 FLAG antibody were then divided by the percentages of total input from IP using the IgG antibody. The results from ChIP with cells transfected with wild-type FOXL2 and mutant FOXL2 were further normalized against the results from ChIP with cells transfected with empty pFLAG-CMV-2 vector (control). Significant differences (P < 0.05) between samples are denoted by a or b. B, The levels of expression and IP of FLAG-tagged wild-type and mutant FOXL2 were confirmed by Western blotting.
Discussion
FOXL2 is emerging as a central transcription factor in ovarian follicle development. We have previously shown that FOXL2 represses the promoters of several key genes involved in granulosa cell steroidogenesis and proliferation, including StAR, CYP19, and CYP11A, as well as CCND2, which controls cell cycle progression (8, 13). Moreover, we found that these promoters are differentially affected by the truncated FOXL2 (aa 1–218) protein, which results from the heterozygous Q219X mutation in BPES type 1 patients (2). This mutant protein lacks the C-terminal alanine/proline-rich transrepression domain and retains the ability to repress transcription of the CYP11A and CCND2 promoters, but loses the ability to repress transcription of the StAR and CYP19 promoters (13). In this study, we began to determine the mechanisms by which mutant FOXL2 affects the activity of wild-type FOXL2 as a transcriptional repressor of the CYP19 promoter, which was used as a marker of granulosa cell differentiation.
We first tested the effect of the mutant protein on the transcriptional repression of the CYP19 promoter by wild-type FOXL2. We found that, similar to the effects we previously demonstrated for the StAR promoter (8), wild-type FOXL2's activity as a repressor of the CYP19 promoter was lost when coexpressed with the FOXL2 (aa 1–218) mutant (Fig. 1). Furthermore, we confirmed that mutant FOXL2 retains its ability to repress the CCND2 promoter but had no significant effect on the CYP11A promoter, indicating that FOXL2 and mutant FOXL2 may regulate transcription of different genes involved in regulation of granulosa cell steroidogenesis and proliferation via different mechanisms.
The forkhead DNA recognition motif consists of 3 α-helices (H1, H2, and H3) and 3 β-strands (S1, S2, and S3) that form a three-stranded, twisted antiparallel β-sheet (25). Although initial crystallization studies of the HNF-3γ forkhead domain demonstrated that forkhead family members exist as monomers, more recent data demonstrate that some members of this family exist as dimers (21, 22, 26). In fact, the forkhead domain of FOXP2 exists both as monomers and dimers in solution (21) and thus can contribute to gene regulation, flexibility, and complexity (27). We found that wild-type FOXL2 can form homodimers and that the FOXL2 (aa 1–218) mutant, which contains an intact DNA-binding domain, can form both homodimers and heterodimers with wild-type FOXL2 (Fig. 2). This heterodimer formation may underlie the dominant-negative effect of the mutant protein on the transcription of the StAR and CYP19 promoters, both of which are central to granulosa cell steroidogenesis and markers of granulosa cell differentiation. The two α-helices H2 and H3 and the intervening region known as the T loop of the winged helix fold are structurally similar to the helix-turn-helix proteins (15, 16), including other transcription factors such as the POU transcription factor Oct-1, which bind to DNA in various monomer and dimer configurations (17, 18) similar to FOXP2. In addition, dimerization may function as an additional control for regulation of gene expression in conjunction with posttranslational modifications. For example, phosphorylation and sumoylation of FOXL2 (28, 29) may control homo- and heterodimer formation, similar to phosphorylation of the basic helix-loop-helix protein E47, which regulates heterodimer formation with myoblast determination protein 1 and subsequent transcriptional regulation (30).
We also found that the −57-bp region of the CYP19 promoter is sufficient for transcriptional repression by FOXL2 (Fig. 3), indicating that this promoter region contains key FOXL2-binding sites. Interestingly, mutant FOXL2 slightly induced expression of the −633-bp region but fails to repress transcription of the minimal −57-bp promoter region or of the −313- and −114-bp regions (Fig. 3). The latter results were expected because the mutant protein lacks the C-terminal transrepression domain. However, our EMSA results suggest that this is not due to an inability of mutant FOXL2 to bind to the −57-bp region (Fig. 4B). This region contains two potential FOXL2-binding sites (Fig. 4C), which are similar to the nonconsensus FOXL2-binding element identified by Lamba et al., (22) 5′-TGTTTA-3′, and the binding sequence identified by Benayoun et al. (23), 5′-GTCAAGG(T/C)-3′. By testing the binding between FOXL2 and each of these sites (as 30-bp oligonucleotide probes, Fig. 4C), we determined that both sites function as FOXL2-binding sites (Fig. 4D). When either of these two 30-bp binding sites was mutated, wild-type FOXL2 lost its ability to bind to them (Fig. 5B). However, the mutant FOXL2 protein retained some binding affinity to the mutant probes (Fig. 5B). We also tested the effects of these two mutations in the form of 57-bp probes (Fig. 5A). We found that wild-type FOXL2 binds to the wild-type probe and to probes containing one mutation but could not bind to the probe containing two mutations (Fig. 5C). However, the mutant FOXL2 protein not only binds to the wild-type probe and to probes containing one mutation but also binds to the probe containing two mutations (Fig. 5C). These results suggest that the mutant FOXL2 protein has less specificity of binding than the wild-type protein and also suggest that the C-terminal alanine/proline-rich domain of FOXL2 can influence DNA binding, although the basis for this remains to be determined. This reduced specificity of binding of the mutant protein may also explain its ability to slightly induce expression of the −633-bp promoter region, because this might allow it to bind to enhancer elements in these regions that are not bound by wild-type FOXL2. This is currently under investigation in our laboratory.
Coexpression of the wild-type and mutant proteins results in loss of FOXL2 transcriptional repressor activity for all of the CYP19 promoter fragments tested, including the −57-bp region (Fig. 3). When either of the two above-described FOXL2-binding sites in the −57-bp region was mutated, wild-type FOXL2 retained its ability to repress transcription of the promoter fragment, and mutant FOXL2 exerted a dominant-negative effect on this activity (Fig. 6, B and C). However, when both binding sites were mutated, the transcriptional repressor activity of wild-type FOXL2 was lost (Fig. 6D). These data suggest that either of these DNA-binding sites is sufficient for transcriptional repression of the CYP19 promoter by FOXL2, but at least one binding site needs to be present. The results of our ChIP assays using the KGN cell line (Fig. 7) demonstrate that both mutant and wild-type FOXL2 can bind to the CYP19 promoter in vivo, further supporting a role for FOXL2 as a transcriptional repressor of CYP19. Although the KGN cell line is derived from granulosa cell tumors and carries a mutation in FOXL2, C402G (24), similar to other binding studies using this model (31), our studies were conducted with overexpressing wild-type and mutant FOXL2 and demonstrates the ability of these proteins to bind to aromatase in vivo. Furthermore, we have recently demonstrated that mouse Foxl2 represses the activity of mouse Cyp19, and other key genes involved in granulosa cell steroidogenesis and proliferation, in granulosa cells (Kuo, F. T., K. Fan, I. K. Bentsi-Barnes, G. M. Barlow, and M. D. Pisarska, submitted for publication), which further supports our findings in the human KGN cells and CHO cells presented here.
In summary, we have found that the dominant-negative effect of mutant FOXL2 on the wild-type protein may be due to both direct competition for DNA binding and to the formation of heterodimers between the wild-type and mutant FOXL2 proteins. This dominant-negative effect would explain the presence of phenotypic effects in patients with heterozygous FOXL2 Q219X mutations. Thus, homodimer formation may be necessary for cofactor binding, and the conformational change brought about by heterodimer formation may prevent this binding, leading to loss of transcriptional repressor activity. In fact, a number of FOXL2 binding partners in granulosa cells have been identified, including steroidogenic factor-1 (32) and the DEAD box protein DP103 (8), and interactions between FOXL2 and SOX9 have been shown to influence the role of FOXL2 in sex determination as a result of altered cellular differentiation (33). In the pituitary, the intact forkhead box DNA-binding domain of FOXL2 can directly associate with the signal transduction protein Smad3, but not Smad2 or Smad4, and this association enhances the transcriptional activity of FOXL2 (34). We recently demonstrated that all members of the SMAD family of signal transduction proteins are expressed in human granulosa cells (35), although their role with respect to FOXL2 activity in granulosa cells has yet to be determined. Elucidation of FOXL2 interactions with potential binding partners, including SMAD, and the effect of heterodimer formation with the FOXL2 (aa 1–218) mutant protein on these interactions, will be important in determining the functional mechanisms underlying FOXL2's activity as a key transcriptional repressor during granulosa cell steroidogenesis, proliferation, and hence differentiation, and how alterations to these mechanisms may contribute to the POF associated with BPES type 1. Lastly, our results suggest that the CYP19 and StAR genes may also be affected by the presence of the mutant FOXL2 protein in the granulosa cells of BPES type 1 patients, which we hypothesize may contribute to the POF phenotype in patients carrying the FOXL2 Q219X heterozygous mutation.
Acknowledgments
We thank Dr. Masataka Nakamura (Human Gene Sciences Center, Tokyo Medical and Dental University, Tokyo, Japan) for his generous gift of the human cyclin D2 promoter. We also thank Dr. Yoshihiro Nishi and Dr. Toshihiko Yanase (Department of Endocrinology and Diabetes Mellitus, School of Medicine, Fukuoka University, Fukuoka, Japan) for granting permission to use the KGN cell line, which was obtained from the RIKEN BioResource Center Cell Bank, Ibaraki, Japan.
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the Office of Research on Women's Health (R01HD047603 to M.D.P.) and by a grant from the Helping Hands of Los Angeles, Inc. (to M.P.).
Disclosure Summary: F.-T.K., I.K.B.-B., G.M.B., and M.D.P. have nothing to disclose.
Footnotes
- aa
- Amino acids
- BPES
- blepharophimosis-ptosis-epicanthus inversus
- ChIP
- chromatin IP
- Ct
- cycle threshold
- FOXL2
- Forkhead L2
- HA
- hemagglutinin
- IB
- immunoblot
- IP
- immunoprecipitation
- POF
- premature ovarian failure
- SDS
- sodium dodecyl sulfate
- StAR
- steroidogenic acute regulatory protein.
References
- 1. Zlotogora J, Sagi M, Cohen T. 1983. The blepharophimosis, ptosis, and epicanthus inversus syndrome: delineation of two types. Am J Hum Genet 35:1020–1027 [PMC free article] [PubMed] [Google Scholar]
- 2. Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R, Rocchi M, Nicolino M, Lienhardt-Roussie A, Nivelon A, Verloes A, Schlessinger D, Gasparini P, Bonneau D, Cao A, Pilia G. 2001. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nature genetics 27:159–166 [DOI] [PubMed] [Google Scholar]
- 3. Fraser IS, Shearman RP, Smith A, Russell P. 1988. An association among blepharophimosis, resistant ovary syndrome, and true premature menopause. Fertil Steril 50:747–751 [DOI] [PubMed] [Google Scholar]
- 4. Meduri G, Bachelot A, Duflos C, Bstandig B, Poirot C, Genestie C, Veitia R, De Baere E, Touraine P. 2010. FOXL2 mutations lead to different ovarian phenotypes in BPES patients: case report. Hum Reprod 25:235–243 [DOI] [PubMed] [Google Scholar]
- 5. De Baere E, Dixon MJ, Small KW, Jabs EW, Leroy BP, Devriendt K, Gillerot Y, Mortier G, Meire F, Van Maldergem L, Courtens W, Hjalgrim H, Huang S, Liebaers I, Van Regemorter N, Touraine P, Praphanphoj V, Verloes A, Udar N, Yellore V, Chalukya M, Yelchits S, De Paepe A, Kuttenn F, Fellous M, Veitia R, Messiaen L. 2001. Spectrum of FOXL2 gene mutations in blepharophimosis-ptosis-epicanthus inversus (BPES) families demonstrates a genotype-phenotype correlation. Hum Molr Genet 10:1591–1600 [DOI] [PubMed] [Google Scholar]
- 6. Ramírez-Castro JL, Pineda-Trujillo N, Valencia AV, Muñetón CM, Botero O, Trujillo O, Vásquez G, Mora BE, Durango N, Bedoya G, Ruiz-Linares A. 2002. Mutations in FOXL2 underlying BPES (types 1 and 2) in Colombian families. Am J Med Genet 113:47–51 [DOI] [PubMed] [Google Scholar]
- 7. De Baere E, Beysen D, Oley C, Lorenz B, Cocquet J, De Sutter P, Devriendt K, Dixon M, Fellous M, Fryns JP, Garza A, Jonsrud C, Koivisto PA, Krause A, Leroy BP, Meire F, Plomp A, Van Maldergem L, De Paepe A, Veitia R, Messiaen L. 2003. FOXL2 and BPES: mutational hotspots, phenotypic variability, and revision of the genotype-phenotype correlation. Am J Hum Genet 72:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pisarska MD, Bae J, Klein C, Hsueh AJ. 2004. Forkhead l2 is expressed in the ovary and represses the promoter activity of the steroidogenic acute regulatory gene. Endocrinology 145:3424–3433 [DOI] [PubMed] [Google Scholar]
- 9. Treier M, Gleiberman AS, O'Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. 1998. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev 12:1691–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clark BJ, Wells J, King SR, Stocco DM. 1994. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR). J Biol Chem 269:28314–28322 [PubMed] [Google Scholar]
- 11. Lin D, Sugawara T, Strauss JF, 3rd, Clark BJ, Stocco DM, Saenger P, Rogol A, Miller WL. 1995. Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science 267:1828–1831 [DOI] [PubMed] [Google Scholar]
- 12. Stocco DM. 2001. StAR protein and the regulation of steroid hormone biosynthesis. Annu Rev Physiol 63:193–213 [DOI] [PubMed] [Google Scholar]
- 13. Bentsi-Barnes IK, Kuo FT, Barlow GM, Pisarska MD. 2010. Human forkhead L2 represses key genes in granulosa cell differentiation including aromatase, P450scc, and cyclin D2. Fertil Steril 94:353–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Veitia RA. 2002. Exploring the etiology of haploinsufficiency. Bioessays 24:175–184 [DOI] [PubMed] [Google Scholar]
- 15. Pabo CO, Sauer RT. 1992. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem 61:1053–1095 [DOI] [PubMed] [Google Scholar]
- 16. Harrison SC. 1991. A structural taxonomy of DNA-binding domains. Nature 353:715–719 [DOI] [PubMed] [Google Scholar]
- 17. Lins K, Reményi A, Tomilin A, Massa S, Wilmanns M, Matthias P, Schöler HR. 2003. OBF1 enhances transcriptional potential of Oct1. EMBO J 22:2188–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reményi A, Pohl E, Schöler HR, Wilmanns M. 2001. Crystallization of redox-insensitive Oct1 POU domain with different DNA-response elements. Acta Crystallogr D Biol Crystallogr 57:1634–1638 [DOI] [PubMed] [Google Scholar]
- 19. Bilokapic S, Ivic N, Godinic-Mikulcic V, Piantanida I, Ban N, Weygand-Durasevic I. 2009. Idiosyncratic helix-turn-helix motif in Methanosarcina barkeri seryl-tRNA synthetase has a critical architectural role. J Biol Chem 284:10706–10713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lamba P, Wang Y, Tran S, Ouspenskaia T, Libasci V, Hébert TE, Miller GJ, Bernard DJ. 2010. Activin A regulates porcine follicle-stimulating hormone beta-subunit transcription via cooperative actions of SMADs and FOXL2. Endocrinology 151:5456–5467 [DOI] [PubMed] [Google Scholar]
- 21. Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. 2006. Structure of the forkhead domain of FOXP2 bound to DNA. Structure 14:159–166 [DOI] [PubMed] [Google Scholar]
- 22. Lamba P, Fortin J, Tran S, Wang Y, Bernard DJ. 2009. A novel role for the forkhead transcription factor FOXL2 in activin A-regulated follicle-stimulating hormone beta subunit transcription. Mol Endocrinol 23:1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Benayoun BA, Caburet S, Dipietromaria A, Bailly-Bechet M, Batista F, Fellous M, Vaiman D, Veitia RA. 2008. The identification and characterization of a FOXL2 response element provides insights into the pathogenesis of mutant alleles. Hum Mol Genet 17:3118–3127 [DOI] [PubMed] [Google Scholar]
- 24. Nishi Y, Yanase T, Mu Y, Oba K, Ichino I, Saito M, Nomura M, Mukasa C, Okabe T, Goto K, Takayanagi R, Kashimura Y, Haji M, Nawata H. 2001. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 142:437–445 [DOI] [PubMed] [Google Scholar]
- 25. Clark KL, Halay ED, Lai E, Burley SK. 1993. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature 364:412–420 [DOI] [PubMed] [Google Scholar]
- 26. Chae WJ, Henegariu O, Lee SK, Bothwell AL. 2006. The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells. Proc Natl Acad Sci USA 103:9631–9636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG. 2008. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci 33:220–229 [DOI] [PubMed] [Google Scholar]
- 28. Kuo FT, Bentsi-Barnes IK, Barlow GM, Bae J, Pisarska MD. 2009. Sumoylation of forkhead L2 by Ubc9 is required for its activity as a transcriptional repressor of the Steroidogenic Acute Regulatory gene. Cell Signal 21:1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pisarska MD, Kuo FT, Bentsi-Barnes IK, Khan S, Barlow GM. 2010. LATS1 phosphorylates forkhead L2 and regulates its transcriptional activity. Am J Physiol Endocrinol Metab 299:E101–E109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lluís F, Ballestar E, Suelves M, Esteller M, Muñoz-Cánoves P. 2005. E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J 24:974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benayoun BA, Georges AB, L'Hôte D, Andersson N, Dipietromaria A, Todeschini AL, Caburet S, Bazin C, Anttonen M, Veitia RA. 2011. Transcription factor FOXL2 protects granulosa cells from stress and delays cell cycle: role of its regulation by the SIRT1 deacetylase. Hum Mol Genet 20:1673–1686 [DOI] [PubMed] [Google Scholar]
- 32. Park M, Shin E, Won M, Kim JH, Go H, Kim HL, Ko JJ, Lee K, Bae J. 2010. FOXL2 interacts with steroidogenic factor-1 (SF-1) and represses SF-1-induced CYP17 transcription in granulosa cells. Mol Endocrinol 24:1024–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang DS, Kobayashi T, Zhou LY, Paul-Prasanth B, Ijiri S, Sakai F, Okubo K, Morohashi K, Nagahama Y. 2007. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol Endocrinol 21:712–725 [DOI] [PubMed] [Google Scholar]
- 34. Blount AL, Schmidt K, Justice NJ, Vale WW, Fischer WH, Bilezikjian LM. 2009. FoxL2 and Smad3 coordinately regulate follistatin gene transcription. J Biol Chem 284:7631–7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuo FT, Fan K, Ambartsumyan G, Menon P, Ketefian A, Bentsi-Barnes IK, Pisarska MD. 16 July 2011. Relative expression of genes encoding SMAD signal transduction factors in human granulosa cells is correlated with oocyte quality. J Assist Reprod Genet 10.1007/s10815-011-9609-6 [DOI] [PMC free article] [PubMed] [Google Scholar]