Abstract
Brown adipose tissue plays an important role in obesity, insulin resistance, and diabetes. We have previously shown that the transition from brown preadipocytes to mature adipocytes is mediated in part by insulin receptor substrate (IRS)-1 and the cell cycle regulator protein necdin. In this study, we used pharmacological inhibitors and adenoviral dominant negative constructs to demonstrate that this transition involves IRS-1 activation of Ras and ERK1/2, resulting in phosphorylation of cAMP response element-binding protein (CREB) and suppression of necdin expression. This signaling did not include an elevation of intracellular calcium. A constitutively active form of CREB expressed in IRS-1 knockout cells decreased necdin promoter activity, necdin mRNA, and necdin protein levels, leading to a partial restoration of differentiation. By contrast, forkhead box protein (Fox)O1, which is regulated by the phosphoinositide 3 kinase-Akt pathway, increased necdin promoter activity. Based on reporter gene assays using truncations of the necdin promoter and chromatin immunoprecipitation studies, we demonstrated that CREB and FoxO1 are recruited to the necdin promoter, likely interacting with specific consensus sequences in the proximal region. Based on these results, we propose that insulin/IGF-I act through IRS-1 phosphorylation to stimulate differentiation of brown preadipocytes via two complementary pathways: 1) the Ras-ERK1/2 pathway to activate CREB and 2) the phosphoinositide 3 kinase-Akt pathway to deactivate FoxO1. These two pathways combine to decrease necdin levels and permit the clonal expansion and coordinated gene expression necessary to complete brown adipocyte differentiation.
The adipose tissue pool in mammals is composed of at least two functionally different types of fat: white and brown. White adipose tissue is the primary site of energy storage, which in excess leads to obesity, type 2 diabetes, and related morbidities, such as coronary heart disease and cancer (1, 2). Brown adipose tissue is specialized for energy expenditure and thermogenesis through numerous mitochondria and the expression of uncoupling protein 1 (UCP-1). Brown fat affects whole-body metabolism, capable of altering insulin sensitivity (3) and modifying pancreatic β-cell function (4). In humans, brown fat is present at birth and previously had been thought to be metabolically irrelevant in the adult (5). However, recent evidence has shown that brown adipose tissue is prevalent in adult humans at a significant rate and may play a role in protecting against obesity (6, 7). Understanding the regulation of brown adipose tissue differentiation is of great metabolic significance and clinical importance (8).
Brown adipogenesis is a complex process that is tightly regulated. As with the white adipocyte, the differentiation of brown preadipocytes into brown fat cells can be divided into four distinct stages: 1) preconfluent proliferation, 2) confluent growth arrest, 3) hormonal induction with clonal expansion, and 4) permanent growth arrest with terminal differentiation (9). The development of the characteristic mature phenotype beginning at stage 3 is initiated by the activation of the insulin and IGF-I receptors (10), functionally similar heterotetrameric transmembrane tyrosine kinases that autophosphorylate after ligand binding (11). The receptors then tyrosine phosphorylate adapter proteins known as insulin receptor substrates (IRS) that serve as docking proteins for src homology domain 2-domain-containing proteins. Two canonical serine/threonine kinase signaling pathways are then activated: phosphoinositide 3 kinase (PI3K)-Akt and Ras-ERK1/2 MAPK, leading to a myriad of intracellular events, including gene expression, increased glucose transport, cell growth, and differentiation (12, 13).
We previously showed that brown preadipocytes in which IRS-1 is genetically inactivated fail to differentiate, indicating that IRS-1 was critical for transducing insulin/IGF-I signaling to trigger brown adipocyte differentiation (14, 15). Through a combination of microarray and cell biological approaches, we then defined the highly coordinated pattern of gene expression that predicts the potential of brown preadipocytes to become adipocytes (16). Among the new molecules that appeared to be involved in this blockage of adipogenesis, the protein necdin was of particular interest. Necdin comprises 325 amino acids and is a member of the type 2 melanoma-associated antigen family of proteins. Necdin was originally found in neurally differentiated mouse embryonal carcinoma cells, but a wider role has been postulated via its ectopic expression, which suppresses the proliferation of several cell lines (17). In preadipocytes that have defects in differentiation, such as IRS-1 knockout (KO) cells, necdin levels are markedly increased. Reducing necdin levels in IRS-1 KO preadipocytes using small interfering RNA reverses the blockade in differentiation and restores both the phenotype and gene expression profile of wild-type (WT) cells, including the reduction of overexpression of known suppressors of preadipocyte-adipocyte transition, preadipocyte factor-1 and Wnt10a (16). These features indicate that necdin is not only an important negative regulator of brown adipogenesis but that it also functions very early in the process.
What remains unknown is how insulin/IGF-I receptor signal transduction cascades link to necdin. Necdin has not been shown to undergo posttranslational modifications, such as phosphorylation or acetylation, nor has it been demonstrated that it interacts directly with the PI3K-Akt or Ras-ERK1/2 MAPK pathways. Rather, there is evidence that the link between the insulin/IGF-I receptors and necdin likely is mediated in part by two transcription factors, cAMP response element-binding protein (CREB) and forkhead box protein (Fox)O1. In this study, we delineated the signal transduction pathways linking the insulin/IGF-I receptors through PI3K-Akt and Ras-ERK1/2 MAPK to CREB and FoxO1 to regulate necdin gene expression. We found that these two pathways, mediated by complementary arms of the insulin/IGF-I signaling network, initiate the final two phases of brown preadipocyte differentiation.
Materials and Methods
Materials
Human recombinant IGF-I was obtained from PeproTech, Inc. (Rocky Hill, NJ). Human recombinant insulin was purchased from Roche Molecular Biochemicals (Indianapolis, IN). LY294002 and U0126 were from Calbiochem, EMD Biosciences (San Diego, CA). Other chemicals were obtained from Sigma Chemical Co. (St. Louis, MO) unless otherwise specified. Antiphospho-CREB (Ser-133), anti-CREB, antiphospho-FoxO1 (Ser-256), anti-FoxO1 (Forkhead transcription factor), antiphospho-Akt (Ser-473), and antiphospho-p44/42 MAPK (Tyr-204) antibodies were purchased from Cell Signaling Technology (Beverly, MA). Plasmids encoding constitutively active (CA) and dominant negative (DN)-CREB were gifts from M. Montminy (Salk Institute, La Jolla, CA) (18), and plasmids encoding the necdin promoter-luciferase constructs were gifts from T. Braun (University of Halle-Wittenberg, Halle, Germany) (19).
Cell culture, plasmids, transfection, and retroviral and adenoviral infection
Murine brown preadipocytes were isolated and immortalized as previously described (20). Cells were maintained in DMEM supplemented with 10% fetal bovine serum at 37 C in a 5% CO2 environment. For luciferase-based transient transfection, cells were seeded onto 12-well plates and grown overnight at 37 C. At 90–100% confluence, transfection of 1 μg of necdin promoter-luciferase plasmids along with 1 μg of the different CREB and FoxO1 constructs and control pcDNA3.1 (Invitrogen, Carlsbad, CA) was done such that an equal amount of DNA was transfected into each well. As a transfection control, 0.1 μg of a promoterless Renilla luciferase construct (pRL-0) (Promega, Fitchburg, WI) was added to each well. Cell lysis and quantification of luciferase activity was then done as described (16). Retroviral introduction of IRS-1 into IRS-1 KO cells was done by cloning full-length human IRS-1 into the pBabe-bleo vector, transfecting the construct into IRS-1 KO cells, and selecting with Zeocin as described (16, 20). Recombinant adenoviruses of DN-Akt were constructed by substituting Thr-308 to Ala and Ser-473 to Ala as described previously (21). cDNA of DN K-Ras (DN-Ras; substituted Ser-17 to Asn) was kindly provided by Y. Takai (Osaka University, Osaka, Japan) (22). The recombinant adenoviruses were constructed by homologous recombination between the parental virus genome and the expression cosmid cassette or shuttle vector as described previously (23, 24). Infection and expression of the adenoviruses was assessed as described (25).
Oil red O staining
Confluent six-well plates were washed twice with PBS and fixed with 10% buffered formalin for at least 1 h at room temperature. Cells were then stained for 5–6 h at room temperature with Oil Red O solution (0.5% Oil Red O in isopropyl alcohol), washed twice with distilled water, and visualized.
Immunoblotting
To assess CREB and FoxO1 phosphorylation in response to acute IGF-I stimulation, we grew WT and IRS-1 KO cells in 100-mm dishes to 95–100% confluence. The cells were then serum deprived overnight in DMEM containing 0.1% BSA, pretreated for 1 h with pharmacologic inhibitors, and then incubated with IGF-I at a final concentration of 10–100 nm for either 5 or 30 min. After stimulation, cells were washed twice with ice-cold PBS and scraped into 0.5 ml of lysis buffer as previously described (16). Protein concentrations were determined using the Bradford protein assay (Bio-Rad, Hercules, CA). Lysates were subjected to SDS-PAGE followed by immunoblotting using specific antisera and detection with chemiluminescence (ECL; Amersham Biosciences, Piscataway, NJ).
Assay of intracellular calcium concentrations
Murine brown preadipocytes and human embryonic kidney 293T cells growing in DMEM/fetal bovine serum 10%, 5% CO2 at 37 C were serum deprived for more than 6 h in DMEM + 0.1% BSA and then washed with Earle's Balanced Salt Solution with HEPES (EBSSH) buffer [26 mm HEPES (pH 7.4), 125 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgSO4, 1 mm NaH2PO4, 5.6 mm glucose, and 0.1% (wt/vol) BSA] and resuspended in 1 ml EBSSH at a density of 0.5–1.0 × 106 cells/ml. The fluorophore fluo 3-AM (Molecular Probes, Invitrogen) was added to a final concentration of 2 μm, and the cells were incubated for 60–75 min at room temperature. The fluo 3-loaded cells were pelleted and resuspended in 1 ml of EBSSH, then washed again and incubated in EBSSH for 30–45 min. Just before the assay, the cells were pelleted and transferred to a cuvette at a density of 0.25–0.5 × 106 cells/ml. Fluorescence due to intracellular calcium was measured at room temperature in a Hitachi F-2000 fluorescence spectrophotometer (Hitachi, Schaumburg, IL) using excitation and emission wavelengths of 505 and 525 nm, respectively. Intracellular calcium concentrations were measured and calculated as described (26).
Chromatin immunoprecipitation (ChIP)
ChIP were performed as previously described (27). IRS-1KO cells were cotransfected with necdin promoter construct along with CREB or FoxO1 expression vectors (RSV-CREB or pAlter- Forkhead transcription factor-WT). Eighteen hours after transfection, cells were incubated in 1% formaldehyde at room temperature for 10 min for cross-linking. This was followed by addition of 1 m glycine to a final concentration of 0.125 m and incubation at room temperature for 10 min. The cells were washed twice with cold PBS, and harvested in ChIP lysis buffer [0.1% sodium dodecyl sulfate, 1% Triton X-100, 0.15 m NaCl, 1 mm EDTA, and 20 mm Tris (pH 8)]. The samples were then subjected to sonication for 8 min to ensure that DNA was fragmented to the size range 100-1000 bp. We then combined 100 μl of ChIP sample, 20 μg of Protein G-Agarose beads (Amersham), and 1 μg of either anti-CREB (sc-186X; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or anti-FoxO1 (sc-34890X; Santa Cruz Biotechnology, Inc.) antibody to precipitate CREB or FoxO1 proteins. The negative “control” ChIP samples were processed similarly, except that there was an additional 5 μg of CREB or FoxO1 blocking peptide. The positive control “input” for the PCR consisted of adding 10% of the ChIP samples, processed similarly as above, except that no anti-CREB or anti-FoxO1 antibody was added.
The experimental and control ChIP samples were incubated at 4 C overnight, and then pelleted. The precipitated agarose beads were washed in buffers of different salt concentrations and then decross-linked in a buffer containing 1% sodium dodecyl sulfate, 0.1 m NaHCO3, and 0.2 m NaCl at 65 C for 4 h. After phenol-chloroform extraction of proteins, the DNA was precipitated and dissolved in nuclease-free water and then analyzed by PCR using primers specific for the putative CREB or FoxO1 binding elements in necdin promoter (see figure 5). The primers used in the ChIP assay were the following: CREB-forward, 5′-CTC CCT TAG ACC CCA GTG GTT-3′; CREB-reverse, 5′-GGG TGG TAG GGC TGG AAA G-3′; FoxO1-forward, 5′-AGC CCT ACC ACC CTT CTG GC-3′; and FoxO1-reverse, 5′-GCGATATTGCGCATGCG-3′. PCR products were then analyzed in 1.2% agarose gels.
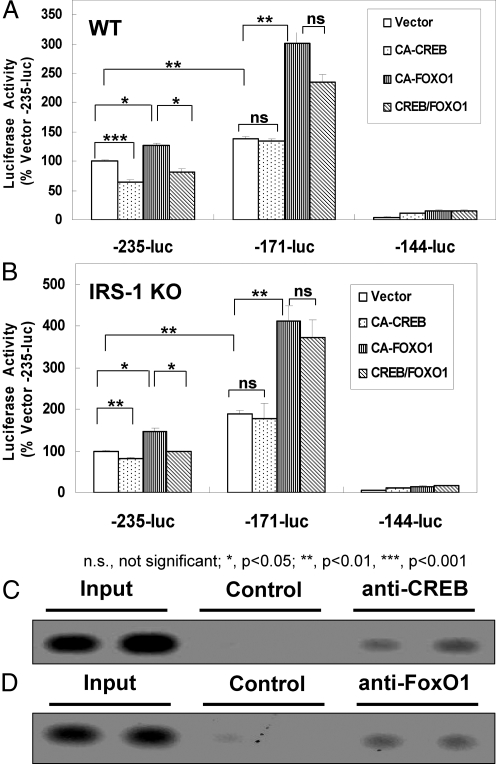
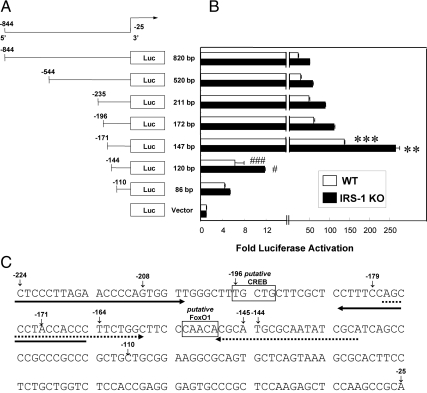
Fig. 5.
Regulation and binding of CREB and FoxO1 to the mouse necdin promoter. A and B, Three deletion constructs were made from the −844/−25 segment of the mouse necdin promoter connected to the luciferase reporter gene vector pTA-LUC-NP. All constructs were transiently transfected into either WT or IRS-1 KO brown preadipocytes together with plasmids expressing vector control, CA-CREB, CA-FoxO1, or CA-CREB + CA-FoxO1, and luciferase activity was measured as described under Materials and Methods. Values were obtained from a minimum of three independent experiments with each treatment group represented by triplicate plates; error bars indicate the sem; *, P < 0.05; **, P < 0.01; ***, P < 0.001. For ChIP analysis of CREB and FoxO1 binding to the necdin promoter, IRS-1KO cells were cotransfected with expression vectors for necdin and either CREB or FoxO1 and then subjected to ChIP followed by analysis of the PCR products using 1.2% agarose gel as described in Materials and Methods. Shown is the binding to CREB (C) or FoxO1 (D), along with the respective positive and negative controls.
Statistical analysis
Data and error bars in the graphs are expressed as mean ± sem. Differences between two groups were evaluated by an unpaired Student's t test. Experiments shown are single experiments with each value measured done in duplicate or triplicate and representative of at least three independent experiments.
Results
IRS-1, CREB, and necdin play significant roles in brown adipogenesis
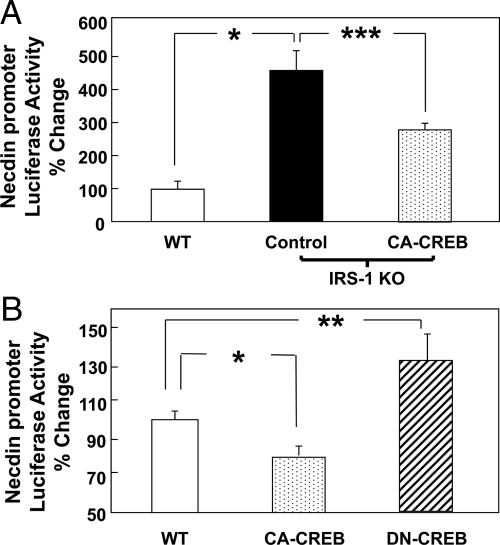
Brown preadipocyte cell lines derived from IRS-1−/− mice demonstrated a profound defect to differentiate when stimulated with a standard differentiation cocktail (28). In the IRS-1-KO cells, necdin levels were elevated more than 50-fold at the mRNA and protein level, and reexpression of IRS-1 restored low-level necdin expression (Supplemental Fig. 1A, left, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org) (16). Stable transfection of IRS-1 KO cells with the CA form of CREB (CA-CREB) profoundly lowered necdin content at the mRNA and protein levels (Supplemental Fig. 1A, right). Furthermore, when IRS-1 was reexpressed in IRS-1 KO cells, the brown preadipocytes regained the ability to differentiate similarly to WT brown preadipocytes, and CA-CREB substantially, although not completely, restored differentiation of IRS-1 KO cells (Supplemental Fig. 1B). These findings demonstrate that CREB lies upstream of necdin, exerts a significant effect on the necdin promoter, transcript, and protein levels, and in doing so, partially restores the WT phenotype.
IGF-I activates Ras-ERK1/2 MAPK, and not PI3K-Akt, to phosphorylate CREB
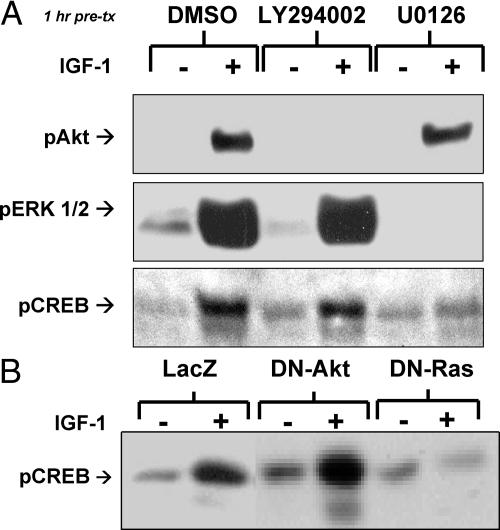
To identify which insulin/IGF-I signaling pathways activate CREB, we used the pharmacologic inhibitors LY294002 and U0126, which block the PI3K-Akt and the Ras-ERK1/2 pathways, respectively. LY294002 (50 μm) effectively blocked IGF-I-mediated phosphorylation of Akt but not of ERK1/2 or CREB. In contrast, U0126 (10 μm) inhibited IGF-I-mediated phosphorylation/activation of ERK1/2 and blocked IGF-I-stimulated phosphorylation of CREB at Ser-133, the site leading to CREB activation (Fig. 1A) (29). Likewise, overexpression of a DN mutant of Ras (DN-Ras) in WT brown preadipocytes blocked IGF-I-mediated activation of CREB phosphorylation, whereas expression of a DN-Akt had no effect (Fig. 1B). To determine whether calcium could be involved in the insulin/IGF-I-mediated activation of CREB phosphorylation, we compared the effects of IGF-I/insulin and other stimulators on intracellular calcium concentration in WT and IRS-1 KO preadipocytes and human embryonic kidney 293 cells that have been previously characterized (26). As shown in Supplemental Fig. 2, in the basal state, the IRS-1 KO cells had a more than 3-fold higher calcium concentration than that in WT cells (P < 0.05). However, IGF-I and insulin were unable to stimulate any detectable change in [Ca2+]i in either WT or IRS-1 KO brown preadipocytes (data not shown), indicating that this pathway does not appear to be involved. Rather, these experiments demonstrate that CREB-mediated regulation of necdin expression is critical to brown adipocyte differentiation. IGF-I activates CREB via the Ras-ERK1/2 MAPK pathway and not PI3K-Akt or calcium-based pathways.
Fig. 1.
Signaling pathways leading from the insulin/IGF-I receptors to CREB. A, WT brown preadipocytes were grown to confluence, serum deprived, pretreated for 1 h with specific inhibitors LY294002 (50 μm) or U0126 (10 μm), incubated with IGF-I (100 nm) for 5 min, and immunoblotted with phosphospecific antibodies to Akt, ERK1/2, or CREB as described under Materials and Methods. B, Brown preadipocytes expressing adenoviruses containing either control vector (LacZ), DN-Akt, or DN-Ras were serum deprived grown to confluence, incubated with IGF-I (10 nm) for 5 min, and immunoblotted with phosphospecific antibodies to CREB as described under Materials and Methods. DMSO, Dimethylsulfoxide; pAKT, phospho-Akt.
CREB down-regulates necdin promoter activity
To see how CREB directly affects necdin gene transcription, we transiently cotransfected a murine necdin-promoter (base pairs −844 to −25)-driven firefly luciferase construct (pTA-LUC-NP) (19, 30) with either the CA-CREB or a DN form of CREB (DN-CREB). Consistent with the increased necdin mRNA and protein levels seen in IRS-1 KO cells (Supplemental Fig. 1A) (16), IRS-1 KO cells demonstrated a more than 4-fold increase in necdin promoter activity compared with WT cells (P < 0.05) (Fig. 2A). This activity in IRS-1 KO cells was reduced by 33% via transfection of CA-CREB (P < 0.001) (Fig. 2A). Likewise, in WT preadipocytes, overexpression of CA-CREB decreased necdin promoter activity by 30% (P < 0.05) (Fig. 2B), whereas overexpression of DN-CREB increased necdin promoter by 30% (P < 0.01) (Fig. 2B). Thus, increased CREB activity resulted in down-regulation of the necdin promoter activity in both WT and IRS-1 KO brown preadipocytes, and with decreased CREB activity the converse was true.
Fig. 2.
Effects of CREB on necdin expression. WT and IRS-1 KO brown preadipocytes were cotransfected with a vector containing the necdin promoter (base pairs −844 to −25)-linked to firefly luciferase, as well as plasmids containing expression vector (control), CA-CREB, or DN-CREB, as described under Materials and Methods. Data are presented as luciferase activity normalized, so that 100% represents the activity seen in the WT cells at basal conditions. Significance was determined by an unpaired, unequal t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Basal necdin promoter activity in WT brown preadipocytes is compared with A, IRS-1 KO cells and IRS-1 KO cells expressing CA-CREB; or B, WT cells expressing CA-CREB or DN-CREB.
FoxO1 up-regulates the necdin promoter
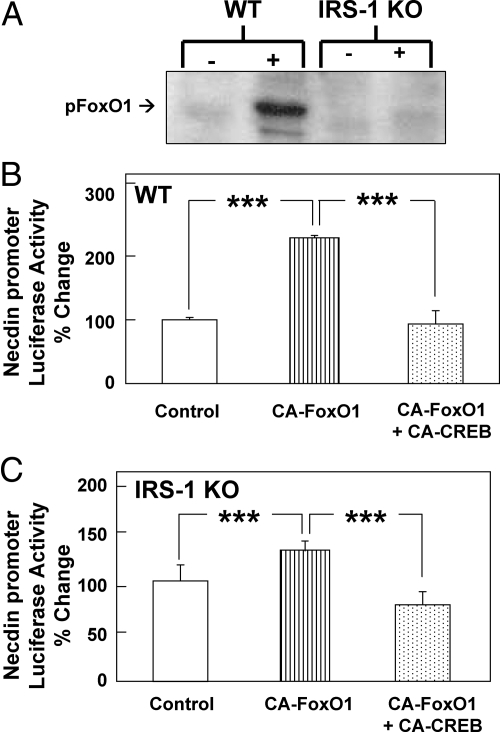
Although CA-CREB substantially restored differentiation of IRS-1 KO brown preadipocytes, it does not completely restore the gene expression profile (16), suggesting that the insulin/IGF-I receptors may regulate necdin through additional pathways in their signal transduction networks. FoxO1 is directly regulated by the insulin/IGF-I-activated PI3K-Akt pathway (31), and in our WT brown preadipocytes, 30 min of IGF-I treatment caused increased phosphorylation of the major regulatory site, Ser-256, in FoxO1. This effect was blunted in IRS-1 KO cells, suggesting that IRS-1 is required for this process (Fig. 3A). To determine whether the FoxO1 pathway interacts with the CREB pathway in the regulation of necdin, we cotransfected the necdin-promoter luciferase construct into either WT or IRS-1 KO brown preadipocytes with either CA-FoxO1 or CA-CREB and measured necdin promoter activity. In both WT and IRS-1 KO cells, CA-FoxO1 increased necdin promoter activity, and this response was reduced by adding CA-CREB (P < 0.001) (Fig. 3, B and C).
Fig. 3.
FoxO1 regulation and its effects on necdin at the promoter and mRNA levels. A, WT and IRS-1 KO brown preadipocytes were serum deprived overnight, treated with IGF-I (100 nm) for 30 min, then immunoblotted with antibody to phosphorylated FoxO1 as described under Materials and Methods. B and C, WT cells (B) or IRS-1 KO cells (C) were cotransfected with a vector containing the necdin promoter (base pairs −844 to −25) linked to firefly luciferase, as well as plasmids containing expression vector (control), CA-FoxO1, or CA-CREB as described under Materials and Methods. Data are presented as luciferase activity normalized, so that 100% represents the activity seen in the WT cells at basal conditions. Significance was determined by an unpaired, unequal t test. ***, P < 0.001. pFoxO1, PhosphoFoxO1.
CREB and FoxO1 binding regions within the necdin promoter
Having demonstrated that CREB and FoxO1 both regulate the necdin promoter, we next sought the regions where they could be involved in binding. Starting with the pTA-LUC-NP described above, we generated a series of truncation mutants from the −844/−25 section of the necdin promoter linked to the luciferase gene (Fig. 4A). We found that, because the promoter was shortened by only 25 bp, from 172 bp (−196/−25) to 147 bp (−171/−25), luciferase activity significantly increased in both the WT (P < 0.001) and IRS-1 KO (P < 0.01) brown preadipocytes, suggesting the presence of a negative regulatory element in this region. However, truncation by another 27 bp (−144/−25) led to a dramatic reduction in promoter activity in both WT (P < 0.001) and IRS-1 KO (P < 0.05) brown preadipocytes, respectively (Fig. 4B), indicating that different positively regulated transcription factor binding sites likely lay in that region. Based on published consensus sequences for CREB (32) and FoxO1 via site-selected amplification studies (33), we identified one putative CREB binding site from −196 to −192 and one putative FoxO1 binding site from −153 to −149 and (Fig. 4C).
Fig. 4.
Localization of putative FoxO1 and CREB responsive elements in the −844/−25 segment of the mouse necdin promoter. A, A series of deletion constructs was made from the −844/−25 segment of the mouse necdin promoter connected to the luciferase reporter gene vector pTA-LUC-NP. B, All constructs were transiently transfected into either WT or IRS-1 KO brown preadipocytes, and luciferase activity was measured as described under Materials and Methods. Fold induction in WT (white bars) or IRS-1 KO (black bars) relative to their respective promoterless control plasmids is shown. Values were obtained from a minimum of three independent experiments with each treatment group represented by triplicate plates; error bars indicate the sem. C, Nucleotide sequence of the −224/−25 region. Putative response elements for CREB (−196/−192) and FoxO1 (−153/−149) are boxed. Horizontal arrows beneath the sequence show the primer sequences used as probes in ChIP, with the ones for CREB in bold and for FoxO1 dashed. ***, P < 0.001 for WT cells 172-bp construct vs. 147-bp construct; **, P < 0.01 for IRS-1 KO cells 172-bp construct vs. 147-bp construct; ###, P < 0.001 for WT cells 147-bp construct vs. 120-bp construct; #, P < 0.05 for IRS-1 KO cells 147-bp construct vs. 120-bp construct.
We confirmed that these putative CREB and FoxO1 binding sites affected necdin promoter activity by transfecting different combinations of CA-CREB and CA-FoxO1 constructs into WT and IRS-KO cells expressing the truncated necdin protomer-luciferase constructs. The −235/−25 construct has both putative bindings sites, and in WT cells (Fig. 5A), CA-CREB significantly suppressed (P < 0.001), and CA-FoxO1 significantly increased (P < 0.05), necdin promoter activity. Cotransfection of CA-CREB on top of CA-FoxO1 reversed the induction of promoter activity by FoxO1 (P < 0.05). This suggests that the CREB and FoxO1 elements present in the −235/−25 necdin-luciferase were able to exert their effect as negative and positive regulatory elements, respectively. Analogous patterns were significantly seen in the IRS-1 KO cells transfected with CA-CREB (P < 0.01), CA-FoxO1 (P < 0.05), and CA-CREB along with CA-FoxO1 (P < 0.05) (Fig. 5B).
In the −171/−25 construct, the CREB element was deleted, but the FoxO1 consensus sequence remained. In both the WT (Fig. 5A) and IRS-1 KO (Fig. 5B) cells, CA-CREB did not affect necdin promoter activity (P > 0.05), whereas CA-FoxO1 increased activity in both cell types (P < 0.01 and P < 0.01, respectively). Cotransfection of CA-CREB on top of CA-FoxO1 also failed to decrease necdin promoter compared with FoxO1 transfection alone (both P > 0.05). The absence of both the CREB and FoxO1 consensus sequences, as seen in the −144/−25 necdin-luciferase promoter construct, resulted in a profound reduction in basal promoter activity, indicating that the principal DNA sequences necessary for activation of the promoter had been eliminated.
Finally, we determined whether CREB and FoxO1 could bind to these putative consensus sequences using a modified ChIP assay. IRS-1KO cells were cotransfected with the necdin promoter construct and expression vectors for CREB or FoxO1. This was followed 18 h later by ChIP via antibodies to CREB or FoxO1. Using specific primers flanking the putative consensus elements, we demonstrated that CREB (Fig. 5C) and FoxO1 (Fig. 5D) interact with the necdin promoter at distinct sites.
Discussion
With the recent identification of human brown adipose tissue as a thermogenic organ that could be used to treat obesity comes the need to determine how to increase its mass and activity. A central challenge is that brown adipogenesis is a complex process that is tightly regulated as it progresses through very different phases of development, with alternating proliferation, growth arrest, and finally, terminal differentiation. This involves coordinated changes in cytoskeletal and matrix modifiers, inhibition of cell cycle stimulators and expression of characteristic transcriptional regulators, such as the CCAAT/enhancer-binding protein (C/EBP)-β, C/EBP-δ, and C/EBP-α, peroxisome proliferator-activated receptor γ (PPARγ) and PPARγ coactivator-1 (PGC-1α), and PR domain containing 16 (PRDM16) (34–36). The fully differentiated phenotype follows with the expression of fatty acid synthase, glucose transporter 4, UCP-1, and other mitochondrial proteins. However, the specific signaling cascades that push the brown preadipocytes past the threshold of growth arrest into clonal expansion and differentiation are less clear. We have previously shown that early in the sequence after hormonal triggering, two important proteins were involved, IRS-1 and CREB, as was the type 2 melanoma-associated antigen protein necdin, a known regulator of cell cycle in neuronal cells (16).
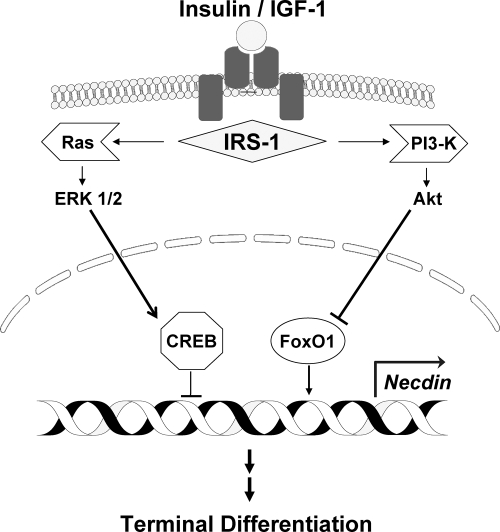
In the current study, we define the earliest events that trigger brown adipogenesis. The insulin and IGF-I receptors initiate this process. These two heterotetramers differ in expression levels, with IGF-I receptor higher earlier, and insulin receptors increased later, in adipocyte differentiation, which is reflected by the different potencies of the two hormones in stimulating differentiation (37). However, at the signal transduction level, the two receptors function identically (38). With the binding of insulin/IGF-I to their heterotetrameric tyrosine kinase cell surface receptors, IRS-1 is activated via phosphorylation to stimulate the Ras-ERK1/2 kinase pathways that then phosphorylate and activate CREB as well as the PI3K-Akt pathway to phosphorylate and inhibit FoxO1. These two effects combine to down-regulate necdin promoter activity, mRNA levels, and protein expression. The lower necdin levels alter the activity of the series of previously described transcription factors that permit the clonal expansion necessary before terminal differentiation into mature brown adipocytes (Fig. 6).
Fig. 6.
Diagram showing the signal transduction pathways leading from the insulin/IGF-I receptors to necdin. In brown preadipocytes, progression past preconfluent growth arrest begins with the binding of insulin/IGF-I to their heterotatrameric tyrosine kinase cell surface receptors, leading to phosphorylation of IRS-1. The activated IRS-1 stimulates the PI3K-Akt pathway to phosphorylate and inhibit FoxO1 and also stimulates the Ras-ERK1/2 MAPK pathways to phosphorylate and activate CREB. These two effects combine to down-regulate necdin promoter activity, mRNA levels, and protein expression. With the regulation of other transcription factors, the reduction in necdin levels permits the clonal expansion necessary before the terminal differentiation into mature brown adipocytes. Arrows indicate activation and T-shaped lines indicate suppression.
Several lines of evidence have suggested CREB as a transcriptional activator in both brown and white adipocyte differentiation in response to insulin or other stimulations. At the molecular level, CREB directly binds to the proximal promoter of C/EBPβ and activates its gene transcription (39). Importantly, CREB activation by a cAMP-dependent pathway also plays a significant role in the expression of PPARγ coactivator-1 (PGC-1α) and UCP-1, two molecular and functional signatures defining the thermogenic program in mature brown adipocytes (40). Our work shows that CREB activation by insulin/IGF-I stimulation is also important even earlier in brown adipogenesis. One of the functions of CREB in the brown preadipocytes is to reduce necdin promoter activity, and truncating a putative consensus site led to an increase in promoter activity. Taken together, our data provide a signaling link from the insulin/IGF-I receptors via Ras-ERK1/2 to CREB activation and further highlight the critical role of CREB in the control of brown fat development.
FoxO1 is a member of the forkhead box family of transcriptional regulators, in which several members play a role in brown and white adipogenesis (41). CA-FoxO1 inhibits adipogenesis through the induction of several cell cycle inhibitors, including the retinoblastoma protein (pRb), blocking the third phase, hormone-triggered clonal expansion, of adipocyte differentiation (31). We postulate that FoxO1 may be involved in a similar process in brown adipogenesis. Previously, we showed that necdin interacts with E2 transcription factor (E2F) to suppress the G1/S transition and limit expression of the master adipogenic transcription factor PPARγ (16). FoxO1 thereby exerts its effects to block adipogenesis in two ways: first, it stimulates expression of necdin, which is a negative regulator of brown fat differentiation, and second, FoxO1 inhibits the adipogenic transcriptional cascade involving PPARγ and ultimately the C/EBP (31, 42). When insulin/IGF-I trigger adipogenesis, the signaling pathway acting through PI3K-Akt leads to phosphorylation of FoxO1, resulting in nuclear exclusion, thus reducing its effect to stimulate the necdin promoter. Simultaneously, activation of the Ras-ERK1/2 pathway stimulates phosphorylation and activation of CREB also directly reducing necdin expression and blocking the effects of FoxO1 to stimulate its expression.
Based on its original identification as a potential regulator of neuronal growth, necdin's pivotal role in adipogenesis would not have been anticipated. Necdin is known to be a maternally imprinted gene mapping to human chromosome 15q11.2-q12, a region deleted in the neurodevelopmental disorder Prader-Willi syndrome that is distinguished by feeding problems, gross obesity, and hypogonadism (43, 44). Subsequent studies have shown that necdin directly interacts with viral oncoproteins Simian virus 40 large T antigen and adenovirus early region 1a, the p75 neurotrophin receptor, as well as transcription factors E2F1, E2F4, p53, and Msx2 (17, 45–47). Although structurally very different, this functional profile highlights the similarities among necdin and the pRb family of pocket proteins that act as principal growth suppressors involved in terminal mitosis and differentiation. Although different from pRb in distinct ways, including expression in preadipocytes (48, 49) and lack of regulation by cyclin-dependent kinase 4-kinase-cyclin D1 (50), necdin behaves like pRb by preventing proliferation, differentiation, or apoptosis (51).
An increasing number of transcriptional regulators have been shown to affect necdin gene activity. The basic helix-loop-helix factors non-small cell lung cancer-1 and non-small cell lung cancer-2 (19) and PPARγ agonists (52) have also been shown to affect necdin gene expression in vivo in hypothalamus and white adipose tissue, respectively. Necdin levels are also markedly elevated in brown preadipocytes that are unable to differentiate due to impaired insulin signaling, and knockdown of necdin via CREB and FoxO1 helps restore brown adipogenesis with subsequent down-regulation of adipogenic suppressors preadipocyte factor-1 and Wnt10a expression (16). Nygard et al. (53) have shown that necdin expression is also negatively regulated by thyroid hormone, a molecule that regulates brown adipogenesis. Together, these data suggest that in brown preadipocytes, suppression of necdin by insulin/IGF-I and IRS-1, and possibly also by thyroid hormone, is required for these precursors to enter the brown adipogenic program. We acknowledge the limitation that these conclusions are based on in vitro studies. Unfortunately, to date, there are no data on brown fat in the necdin KO mouse (44) or in patients with Prader-Willi syndrome (54), who lack necdin. Therefore, future studies are still required to correlate necdin expression with brown adipogenesis in vivo.
As with CREB and FoxO1, necdin's role must be explained in the context of the paradoxical sequence of growth arrest followed by clonal expansion and then terminal differentiation. We posit that in brown fat, necdin's role is similar to what has been seen in neurons (55), osteosarcoma cells (46), and myoblasts (56), where it facilitates terminal differentiation by facilitating cell cycle exit and promoting survival (57). Indeed, in IRS-1 KO brown preadipocytes, down-regulation of necdin expression reversed the defect in mitotic clonal expansion (16). Thus, in the brown preadipocyte, two pathways are linked to progression through differentiation. The normally nuclear FoxO1 stimulates expression of necdin and inhibits the expression of PPARγ. Insulin/IGF-I acting through PI3K-Akt stimulates FoxO1 phosphorylation, preventing its stimulation of necdin expression and the associated repression of PPARγ. Through IRS-1 and then Ras-ERK1/2, insulin/IGF-I also causes CREB phosphorylation and activation leading to decreased necdin levels. Together, these two pathways allow the transition from confluent growth arrest to clonal expansion and the activation of transcription factors and cellular proteins necessary to complete the terminal differentiation into functional brown adipose tissue. With this more detailed description of the pathways leading to thermogenic brown adipose tissue, there can be more focused attempts to develop interventions using brown fat as a treatment for obesity and diabetes.
Acknowledgments
We thank C. Ronald Kahn for his advice in preparing the manuscript, Marcus Krüger and Karen Ruschke for the construction of the necdin-luciferase promoter, Marc Montminy for providing the CREB constructs, M. Fasshauer and J. Klein for preparation of cell lines, Koji Ueki for generating the adenoviruses, and Kazuaki Yoshikawa and Michio Niinobe for necdin antibodies. We also thank Tom Sakmar and his lab members Manija Kazmi, David Kastner, and Pallavi Sachdev, who provided the resources to conduct the calcium experiments.
This work was supported by the Eli Lilly Foundation and National Institutes of Health Grants DK070722, DK077097, P30 DK46200, and P30 DK040561 (to Y.-H.T.), DK33201, DK046200, DK081604, DK087317, and RR025757 (to A.M.C.), and DK41430 (to T.G.U.), the Eleanor and Miles Shore 50th Anniversary Fellowship Program from Harvard Medical School (Y.-H.T.), the Clinical Investigator Training Program, Beth Israel Deaconess Medical Center-Harvard/Massachusetts Institute of Technology Health Sciences and Technology, in collaboration with Pfizer, Inc. and Merck & Co. (A.M.C.), and the Department of Veterans Affairs Merit Review Program (T.G.U.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CA
- Constitutively active
- C/EBP
- CCAAT/enhancer-binding protein
- ChIP
- chromatin immunoprecipitation
- CREB
- cAMP response element-binding protein
- DN
- dominant negative
- EBSSH
- Earle's Balanced Salt Solution with HEPES
- E2F
- E2 transcription factor
- Fox
- forkhead box protein
- IRS
- insulin receptor substrate
- KO
- knockout
- PI3K
- phosphoinositide 3 kinase
- PPARγ
- peroxisome proliferator-activated receptor γ
- pRb
- retinoblastoma protein
- UCP-1
- uncoupling protein 1.
References
- 1. Wolf AM, Colditz GA. 1998. Current estimates of the economic cost of obesity in the United States. Obes Res 6:97–106 [DOI] [PubMed] [Google Scholar]
- 2. Rosen ED, Spiegelman BM. 2006. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lowell BB, S-Susulic V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. 1993. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 366:740–742 [DOI] [PubMed] [Google Scholar]
- 4. Guerra C, Navarro P, Valverde AM, Arribas M, Brüning J, Kozak LP, Kahn CR, Benito M. 2001. Brown adipose tissue-specific insulin receptor knockout shows diabetic phenotype without insulin resistance. J Clin Invest 108:1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cunningham S, Leslie P, Hopwood D, Illingworth P, Jung RT, Nicholls DG, Peden N, Rafael J, Rial E. 1985. The characterization and energetic potential of brown adipose tissue in man. Clin Sci 69:343–348 [DOI] [PubMed] [Google Scholar]
- 6. Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. 2009. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 360:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nedergaard J, Bengtsson T, Cannon B. 2007. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab 293:E444–E452 [DOI] [PubMed] [Google Scholar]
- 8. Tseng YH, Cypess AM, Kahn CR. 2010. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov 9:465–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cowherd RM, Lyle RE, McGehee REJ. 1999. Molecular regulation of adipocyte differentiation. PMID 10:3–10 [DOI] [PubMed] [Google Scholar]
- 10. Rubin CS, Hirsch A, Fung C, Rosen OM. 1978. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J Biol Chem 253:7570–7578 [PubMed] [Google Scholar]
- 11. Cheatham B, Kahn CR. 1995. Insulin action and the insulin signaling network. Endocr Rev 16:117–142 [DOI] [PubMed] [Google Scholar]
- 12. Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, Franklin RA, McCubrey JA. 2003. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia 17:1263–1293 [DOI] [PubMed] [Google Scholar]
- 13. Taniguchi CM, Kondo T, Sajan M, Luo J, Bronson R, Asano T, Farese R, Cantley LC, Kahn CR. 2006. Divergent regulation of hepatic glucose and lipid metabolism by phosphoinositide 3-kinase via Akt and PKCλ/ζ. Cell Metab 3:343–353 [DOI] [PubMed] [Google Scholar]
- 14. Tseng YH, Ueki K, Kriauciunas KM, Kahn CR. 2002. Differential roles of insulin receptor substrates in the anti-apoptotic function of insulin-like growth factor-1 and insulin. J Biol Chem 277:31601–31611 [DOI] [PubMed] [Google Scholar]
- 15. Tseng YH, Kriauciunas KM, Kokkotou E, Kahn CR. 2004. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol 24:1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tseng YH, Butte AJ, Kokkotou E, Yechoor VK, Taniguchi CM, Kriauciunas KM, Cypess AM, Niinobe M, Yoshikawa K, Patti ME, Kahn CR. 2005. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat Cell Biol 7:601–611 [DOI] [PubMed] [Google Scholar]
- 17. Yoshikawa K. 2000. Cell cycle regulators in neural stem cells and postmitotic neurons. Neurosci Res 37:1–14 [DOI] [PubMed] [Google Scholar]
- 18. Asahara H, Santoso B, Guzman E, Du K, Cole PA, Davidson I, Montminy M. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol Cell Biol 21:7892–7900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krüger M, Ruschke K, Braun T. 2004. NSCL-1 and NSCL-2 synergistically determine the fate of GnRH-1 neurons and control necdin gene expression. EMBO J 23:4353–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fasshauer M, Klein J, Kriauciunas KM, Ueki K, Benito M, Kahn CR. 2001. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol Cell Biol 21:319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kitamura T, Ogawa W, Sakaue H, Hino Y, Kuroda S, Takata M, Matsumoto M, Maeda T, Konishi H, Kikkawa U, Kasuga M. 1998. Requirement for activation of the serine-threonine kinase Akt (protein kinase B) in insulin stimulation of protein synthesis but not of glucose transport. Mol Cell Biol 18:3708–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ueki K, Yamamoto-Honda R, Kaburagi Y, Yamauchi T, Tobe K, Burgering BM, Coffer PJ, Komuro I, Akanuma Y, Yazaki Y, Kadowaki T. 1998. Potential role of protein kinase B in insulin-induced glucose transport, glycogen synthesis, and protein synthesis. J Biol Chem 273:5315–5322 [DOI] [PubMed] [Google Scholar]
- 23. He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. 1998. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA 95:2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miyake S, Makimura M, Kanegae Y, Harada S, Sato Y, Takamori K, Tokuda C, Saito I. 1996. Efficient generation of recombinant adenoviruses using adenovirus DNA- terminal protein complex and a cosmid bearing the full-length virus genome. Proc Natl Acad Sci USA 93:1320–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suzuma I, Suzuma K, Ueki K, Hata Y, Feener EP, King GL, Aiello LP. 2002. Stretch-induced retinal vascular endothelial growth factor expression is mediated by phosphatidylinositol 3-kinase and protein kinase C (PKC)-ζ but not by stretch-induced ERK1/2, Akt, Ras, or classical/novel PKC pathways. J Biol Chem 277:1047–1057 [DOI] [PubMed] [Google Scholar]
- 26. Cypess AM, Unson CG, Wu CR, Sakmar TP. 1999. Two cytoplasmic loops of the glucagon receptor are required to elevate cAMP or intracellular calcium. J Biol Chem 274:19455–19464 [DOI] [PubMed] [Google Scholar]
- 27. Zhang H, Schulz TJ, Espinoza DO, Huang TL, Emanuelli B, Kristiansen K, Tseng YH. 2010. Cross talk between insulin and bone morphogenetic protein signaling systems in brown adipogenesis. Mol Cell Biol 30:4224–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fasshauer M, Klein J, Ueki K, Kriauciunas KM, Benito M, White MF, Kahn CR. 2000. Essential role of insulin receptor substrate-2 in insulin stimulation of glut4 translocation and glucose uptake in brown adipocytes. J Biol Chem 275:25494–25501 [DOI] [PubMed] [Google Scholar]
- 29. Shaywitz AJ, Greenberg ME. 1999. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 68:821–861 [DOI] [PubMed] [Google Scholar]
- 30. Uetsuki T, Takagi K, Sugiura H, Yoshikawa K. 1996. Structure and expression of the mouse necdin gene. Identification of a postmitotic neuron-restrictive core promoter. J Biol Chem 271:918–924 [DOI] [PubMed] [Google Scholar]
- 31. Nakae J, Kitamura T, Kitamura Y, Biggs WH, 3rd, Arden KC, Accili D. 2003. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell 4:119–129 [DOI] [PubMed] [Google Scholar]
- 32. Conkright MD, Guzmán E, Flechner L, Su AI, Hogenesch JB, Montminy M. 2003. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell 11:1101–1108 [DOI] [PubMed] [Google Scholar]
- 33. Furuyama T, Nakazawa T, Nakano I, Mori N. 2000. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349:629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rosen ED, MacDougald OA. 2006. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 7:885–896 [DOI] [PubMed] [Google Scholar]
- 35. Hansen JB, Kristiansen K. 2006. Regulatory circuits controlling white versus brown adipocyte differentiation. Biochem J 398:153–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. 2007. Transcriptional control of brown fat determination by PRDM16. Cell Metab 6:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scavo LM, Karas M, Murray M, LeRoith D. 2004. Insulin-like growth factor-I stimulates both cell growth and lipogenesis during differentiation of human mesenchymal stem cells into adipocytes. J Clin Endocrinol Metab 89:3543–3553 [DOI] [PubMed] [Google Scholar]
- 38. Boucher J, Tseng YH, Kahn CR. 2010. Insulin and insulin-like growth factor-1 receptors act as ligand-specific amplitude modulators of a common pathway regulating gene transcription. J Biol Chem 285:17235–17245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang QQ, Zhang JW, Lane DM. 2004. Sequential gene promoter interactions by C/EBPβ, C/EBPα, and PPARγ during adipogenesis. Biochem Biophys Res Commun 318:213–218 [DOI] [PubMed] [Google Scholar]
- 40. Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, Collins S. 2004. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol 24:3057–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gerin I, Bommer GT, Lidell ME, Cederberg A, Enerback S, MacDougald OA. 2009. On the role of FOX transcription factors in adipocyte differentiation and insulin-stimulated glucose uptake. J Biol Chem 284:10755–10763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Farmer SR. 2006. Transcriptional control of adipocyte formation. Cell Metab 4:263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. MacDonald HR, Wevrick R. 1997. The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet 6:1873–1878 [DOI] [PubMed] [Google Scholar]
- 44. Muscatelli F, Abrous DN, Massacrier A, Boccaccio I, Le Moal M, Cau P, Cremer H. 2000. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet 9:3101–3110 [DOI] [PubMed] [Google Scholar]
- 45. Taniura H, Taniguchi N, Hara M, Yoshikawa K. 1998. Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. J Biol Chem 273:720–728 [DOI] [PubMed] [Google Scholar]
- 46. Taniura H, Matsumoto K, Yoshikawa K. 1999. Physical and functional interactions of neuronal growth suppressor necdin with p53. J Biol Chem 274:16242–16248 [DOI] [PubMed] [Google Scholar]
- 47. Kuwajima T, Taniura H, Nishimura I, Yoshikawa K. 2004. Necdin interacts with the Msx2 homeodomain protein via MAGE-D1 to promote myogenic differentiation of C2C12 cells. J Biol Chem 279:40484–40493 [DOI] [PubMed] [Google Scholar]
- 48. Puigserver P, Ribot J, Serra F, Gianotti M, Bonet ML, Nadal-Ginard B, Palou A. 1998. Involvement of the retinoblastoma protein in brown and white adipocyte cell differentiation: functional and physical association with the adipogenic transcription factor C/EBPα. Eur J Cell Biol 77:117–123 [DOI] [PubMed] [Google Scholar]
- 49. Boeuf S, Klingenspor M, Van Hal NL, Schneider T, Keijer J, Klaus S. 2001. Differential gene expression in white and brown preadipocytes. Physiol Genomics 7:15–25 [DOI] [PubMed] [Google Scholar]
- 50. Grafstrom RH, Pan W, Hoess RH. 1999. Defining the substrate specificity of cdk4 kinase-cyclin D1 complex. Carcinogenesis 20:193–198 [DOI] [PubMed] [Google Scholar]
- 51. Hansen JB, Jørgensen C, Petersen RK, Hallenborg P, De Matteis R, Bøye HA, Petrovic N, Enerbäck S, Nedergaard J, Cinti S, te Riele H, Kristiansen K. 2004. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proc Natl Acad Sci USA 101:4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Goldfine AB, Crunkhorn S, Costello M, Gami H, Landaker EJ, Niinobe M, Yoshikawa K, Lo D, Warren A, Jimenez-Chillaron J, Patti ME. 2006. Necdin and E2F4 are modulated by rosiglitazone therapy in diabetic human adipose and muscle tissue. Diabetes 55:640–650 [DOI] [PubMed] [Google Scholar]
- 53. Nygard M, Becker N, Demeneix B, Pettersson K, Bondesson M. 2006. Thyroid hormone-mediated negative transcriptional regulation of Necdin expression. J Mol Endocrinol 36:517–530 [DOI] [PubMed] [Google Scholar]
- 54. Schrander-Stumpel CT, Curfs LM, Sastrowijoto P, Cassidy SB, Schrander JJ, Fryns JP. 2004. Prader-Willi syndrome: causes of death in an international series of 27 cases. Am J Med Genet A 124A:333–338 [DOI] [PubMed] [Google Scholar]
- 55. Takazaki R, Nishimura I, Yoshikawa K. 2002. Necdin is required for terminal differentiation and survival of primary dorsal root ganglion neurons. Exp Cell Res 277:220–232 [DOI] [PubMed] [Google Scholar]
- 56. Deponti D, François S, Baesso S, Sciorati C, Innocenzi A, Broccoli V, Muscatelli F, Meneveri R, Clementi E, Cossu G, Brunelli S. 2007. Necdin mediates skeletal muscle regeneration by promoting myoblast survival and differentiation. J Cell Biol 179:305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Barker PA, Salehi A. 2002. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res 67:705–712 [DOI] [PubMed] [Google Scholar]