SUMMARY
Herpesvirus saimiri (HVS) is a γ-herpesvirus that expresses Sm-class U RNAs (HSURs) in latently-infected marmoset T cells. By deep sequencing, we identified six HVS microRNAs (miRNAs) that are derived from three hairpin structures located immediately downstream of the 3′-end processing signals of three of the HSURs. The viral miRNAs associate with Ago proteins and are biologically active. We confirmed that the expression of the two classes of viral non-coding RNAs is linked by identifying chimeric HSUR-pre-miR transcripts. We show that HVS miRNA biogenesis relies on cis-acting elements specifically required for synthesis and processing of Sm-class RNAs. Knockdown of protein components in vivo and processing assays in vitro demonstrated that HVS does not utilize the Microprocessor complex that generates most host miRNAs. Instead, the Integrator complex cleaves to generate the 3′ end of the HSUR and the pre-miRNA hairpin. Exportin-5 and Dicer are then required to generate mature viral miRNAs.
INTRODUCTION
Non-coding RNAs (ncRNAs) are key players in gene expression in all organisms. It is therefore not surprising that viruses express many kinds of ncRNAs as part of their strategy to redirect host cell metabolism to their own advantage. Many ncRNAs expressed by viruses resemble ncRNAs produced by their hosts. MicroRNAs (miRNAs) are examples of ncRNAs expressed by both host and pathogen.
MiRNAs are short [~22-nucleotide (nt) long] ncRNAs that post-transcriptionally regulate gene expression in metazoan eukaryotes. Most cellular miRNAs are processed from polyadenylated, capped RNA pol II primary (pri-miRNA) transcripts (Cai et al., 2004; Lee et al., 2004). Mature miRNAs are imbedded in ~80-nt hairpin structures that are recognized and excised from pri-miRNAs by the Microprocessor complex in the nucleus (Denli et al., 2004; Gregory et al., 2004; Han et al., 2004; Landthaler et al., 2004). Essential components of the Microprocessor are the RNAse III-like enzyme Drosha and the dsRNA binding protein DGCR8. The released ~60-nt precursor miRNA (pre-miRNA) is exported to the cytoplasm by Exportin-5 (Bohnsack et al., 2004; Lund et al., 2004; Yi et al., 2003), where it is cleaved by another RNAse III-type enzyme called Dicer (Hutvagner et al., 2001). One strand of the resulting ~22-nt duplex is loaded into the RNA-induced silencing complex (RISC) to function in regulatory networks (Yang et al., 2011).
DNA viruses encode almost all viral miRNAs known to date. Most viral miRNAs are believed to be generated by the canonical host miRNA processing pathway and to form similar effector complexes (Grundhoff and Sullivan, 2011). One exception is the murine γ-herpesvirus 68 (MHV68) miRNAs, whose primary transcript also contains tRNAs (Diebel et al., 2010; Pfeffer et al., 2005). The tRNA processing enzyme RNAse Z cleaves at the 3′ end of the tRNA, releasing pre-miRNA hairpins that are subsequently processed by Dicer, thus bypassing the Microprocessor complex (Bogerd et al., 2010).
Another class of ncRNAs shared by host and pathogen is found in Herpesvirus saimiri (HVS), an oncogenic γ-herpesvirus that infects New World monkeys (Ensser and Fleckenstein, 2005). HVS expresses seven small nuclear RNAs (snRNAs) of the Sm-class called HSURs, which are the most abundant viral transcripts in latently-infected marmoset T cells (Albrecht and Fleckenstein, 1992; Lee et al., 1988; Lee and Steitz, 1990; Murthy et al., 1986; Wassarman et al., 1989). Like their cellular counterparts, each HSUR is flanked by a canonical snRNA promoter and a 3′ box, a processing signal that is recognized by the Integrator complex (Baillat et al., 2005). HSURs contain Sm-binding sites and their predicted secondary structures resemble those of cellular Sm-class U RNAs (Albrecht and Fleckenstein, 1992; Lee et al., 1988; Wassarman et al., 1989). During their biogenesis HSURs associate with the SMN (survival of motor neurons) protein complex (Golembe et al., 2005a; Golembe et al., 2005b) to associate stably with Sm proteins and acquire a trimethyl 5′ guanosine cap (Lee et al., 1988; Lee and Steitz, 1990). HSURs 1 and 2, which are the most conserved among HVS strains, upregulate the expression of a handful of host genes by a yet-to-be-described mechanism (Cook et al., 2005). We recently showed that HVS uses HSUR1 to manipulate the host miRNA pathway to regulate host gene expression (Cazalla et al., 2010).
Here, we used deep-sequencing to identify miRNAs in latently-infected T cells and discovered that HVS expresses six miRNAs. These miRNAs reside in three pre-miRNA hairpin structures located directly downstream of the 3′-end processing signals of HSURs 2, 4, and 5. They are co-transcribed with the upstream viral Sm-class RNAs into common primary transcripts. HVS miRNA expression therefore relies on Sm-class transcription and processing signals. In vivo knockdown of processing factors and in vitro processing assays show that the HVS miRNA biogenesis pathway does not require the Microprocessor complex, but instead uses the Integrator complex to generate viral pre-miRNAs. As in the canonical miRNA biogenesis pathway, HVS pre-miRNAs require Exportin-5 for transit to the cytoplasm, where they are cleaved by Dicer into mature functional viral miRNAs.
RESULTS
Identification of H. Saimiri miRNAs
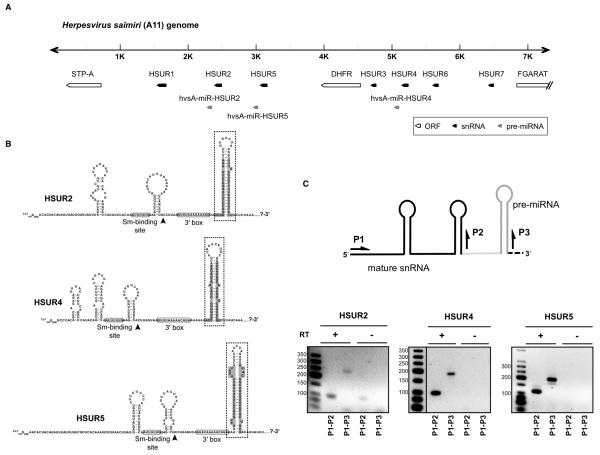
To identify miRNAs encoded by HVS, small RNAs between 18 and 30 nt were isolated from marmoset T cells transformed with HVS (strain A11) (Cook et al., 2004) and cloned. Illumina sequencing yielded 12.2 million high quality reads, representing approximately 593,000 unique sequences. To identify those corresponding to HVS miRNAs, we aligned sequence reads with the viral genome using the following criteria. Reads (i) should map to a unique locus in the HVS A11 genome with two or fewer mismatches, (ii) should be at least 15 nt long, and (iii) should contain no sequencing errors (non-called bases, i.e. “N” bases). Approximately 1.92% of reads perfectly matched the HVS A11 genome, with most corresponding to one of six unique small RNAs (Table S1). These small RNAs derived from both arms (5p and 3p) of the stems of three putative pre-miRNA-like structures (Figure 1B), all located within 7 kbp of the left end of the HVS genome, close to the 3′ ends of the genes for HSURs 2, 4, and 5 (Figure 1A). Following conventional nomenclature (Ambros et al., 2003), we named these putative miRNAs according to the species of origin (hvsA, for Herpesvirus saimiri strain A11), their genomic position downstream of HSUR genes (HSUR2, HSUR4, and HSUR5), and the stem-loop arm of origin (5p or 3p) (Table S1).
Figure 1. HVS miRNAs are located directly downstream of HSUR genes.
(A) Genomic locations of HSURs, protein-coding genes, and miRNAs. The first 7.4 kbp of the HVS A11 genome are shown (Albrecht et al., 1992). (B) Predicted secondary structures of primary transcripts for HSURs and miRNAs. The 3′ ends of mature HSURs are marked by black arrowheads. Mature miRNAs are highlighted in gray. Pre-miRNAs hairpin structures are boxed with dashed lines. (C) RT-PCR identification of transcripts containing both HSURs and miRNAs in marmoset T cells latently infected with HVS. Arrows show the primers used to amplify sequences of mature HSURs (P1+P2) or longer primary transcripts (P1+P3). The mature HSUR is in black, the pre-miRNA hairpin in dark gray, and the intervening sequence (containing the 3′ box, not depicted) in light gray. The lower panel shows fractionation of products obtained in the presence (+) or absence (−) of reverse transcriptase (RT) with size markers.
Some miRNAs expressed by herpesviruses show sequence similarity to host miRNAs (Grundhoff and Sullivan, 2011). We searched the current version (release 16) of miRBase (Griffiths-Jones, 2004; Griffiths-Jones et al., 2006; Griffiths-Jones et al., 2008; Kozomara and Griffiths-Jones, 2011) to see if HVS miRNAs “mimic” known miRNAs. Extensive sequence similarity (including the entire seed region) was found only for hvsA-miR-HSUR-5p and miR-937, a miRNA identified in human cervical cancer cell lines (Lui et al., 2007) that is highly conserved between human, chimpanzee, orangutan, and mouse (Figure S1).
The three HVS pre-miRNA hairpin structures are located immediately downstream of the Sm-class RNA 3′-end formation signals (the 3′ box) for HSURs 2, 4, and 5 (Figure 1B). This suggested that HVS Sm-class RNAs and pre-miRNAs could arise from common primary transcripts. We used reverse transcription polymerase chain reaction (RT-PCR) to detect putative primary transcripts that contain both an Sm-class RNA and miRNAs (Figure 1C). A band corresponding to the region spanning the 5′ end of the Sm-class RNA and the 3′ end of the pre-miRNA hairpin structure was amplified in each case. This suggests that all three HVS pre-miRNAs are co-transcribed with the respective upstream HSUR into pre-Sm-class RNA-pre-miRNA chimeras.
For HVS pre-hvsA-miR-HSUR4, we performed primer extension analysis and RNase protection assays to map the 5′ and the 3′ ends of the predicted ~60-nt pre-miRNA intermediates (Figure S2). These experiments confirmed the structure shown in Figure 1B for pre-hvsA-miR-HSUR4, suggesting that all six HVS miRNAs are processed through ~60-nt hairpin intermediates (boxed in Figure 1B).
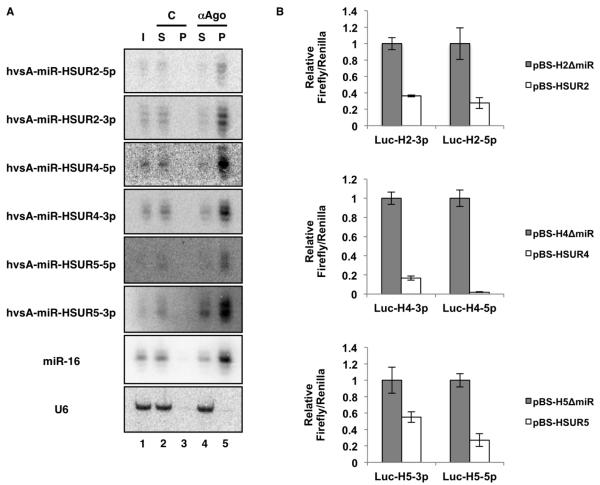
Since functional miRNAs associate with Ago proteins (Hammond et al., 2001; Martinez et al., 2002), we performed co-immunoprecipitation experiments on extracts prepared from virally-transformed marmoset T cells using antibodies that recognize all four mammalian Ago proteins (Nelson et al., 2007) (Figure 2A). All six HVS miRNAs were efficiently co-immunoprecipitated, suggesting they are indeed associated with Ago proteins. Comparable results were obtained when Ago proteins were immunoprecipitated from extracts prepared from 293T cells transiently transfected with a plasmid containing the first 7.4 kbp of the genome of HVS A11 (Figure S3A), arguing that this region contains all the information necessary for proper transcription and processing of HVS miRNAs.
Figure 2. HVS miRNAs are part of active RISC complexes.
(A) Northern blot showing co-immunoprecipitation of HVS miRNAs or host-encoded miR-16 from extracts of virally transformed marmoset T cells with control (C) anti-HA (lane 3) or anti-Ago (αAgo, lane 5) antibody. I: Input (5%); S: Supernatant (5%); P: Pellet (100%). Host U6 serves as a loading control. (B) Luciferase assays confirm the biological activity of HVS miRNAs. 293T cells were co-transfected with Firefly-based reporters containing artificial 3′ UTRs with four perfect target sites for each of the six HVS miRNA, together with vectors expressing HSUR2, 4, or 5 either alone (pBS-HSUR2, pBS-HSUR4, or pBS-HSUR5) or together with their corresponding miRNAs (pBS-H2ΔmiR, pBS-H4ΔmiR, or pBS-H5ΔmiR). Averages of three independent experiments with SD are shown.
Finally, to confirm that the identified HVS miRNAs program functional RISC complexes, we constructed HSUR- and miRNA-expressing vectors containing genomic regions of HVS A11 that included HSUR2, HSUR4, or HSUR5, each with its own promoter and processing signals, with or without its corresponding downstream miRNA. These vectors expressed either a HSUR alone, or a HSUR and its miRNAs (Figure S3B). 293T cells were co-transfected with these expression vectors along with vectors containing the Firefly luciferase reporter gene linked to four artificial target sites perfectly complementary to each HVS miRNA. Significant downregulation of luciferase expression was observed compared to vectors expressing only a HSUR (Figure 2B). We conclude that HVS miRNAs assemble a functional RISC complex and are therefore bona fide miRNAs.
Sm-class RNA Processing Signals Are required to Express H. Saimiri miRNAs
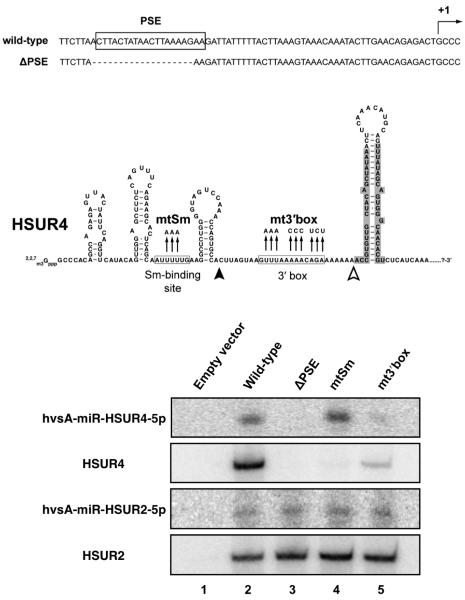
The existence of chimeric transcripts containing both HSUR and miRNA sequences (Figure 1C) strongly suggested that the synthesis and processing of these two classes of ncRNAs are coupled in this virus. To confirm this hypothesis, we mutated the promoter and processing signals of the Sm-class RNAs and analyzed the expression of the linked miRNAs. We focused on HSUR4 and its downstream miRNAs, hsvA-miR-HSUR4-5p and -3p, since these were the most easily detected in transfected 293T cell extracts by Northern blot.
First, we deleted from a vector containing the first 7.4 kbp of the genome of HVS A11 (Figure 1A) the proximal sequence element (PSE) of HSUR4 (Figure 3). The PSE is required for expression of Sm-class RNAs (Hernandez, 2001). This construct generated neither HSUR4 nor hvsA-miR-HSUR4-5p (Figure 3), indicating that HVS miRNAs are indeed transcribed together with HSURs, and not from unknown separate promoters. We next mutagenized the Sm-binding site, an element required for recognition by the SMN complex and binding of Sm proteins in the cytoplasm during snRNP biogenesis (Golembe et al., 2005a). This mutation abolished the appearance of HSUR4, but did not affect the level of hvsA-miR-HSUR4-5p compared to the wild-type construct (mtSm, Figure 3), suggesting that recognition by the SMN and Sm complexes is not required for miRNA expression. Finally, we mutagenized the 3′ box located downstream of HSUR4 that directs the processing of Sm-class RNAs (Egloff et al., 2008). These mutations lowered the levels of both HSUR4 and hvsA-miR-HSUR4-5p (mt3′box, Figure 3). Thus, this signal is required for production of the downstream HVS miRNAs as well as of HSUR4, implicating the Sm-class RNA processing machinery in the production of HVS miRNAs.
Figure 3. Mutational analysis of cis elements required for expression of HSUR4-linked miRNAs.
The HSUR4 proximal sequence element (PSE) and the primary transcript containing HSUR4 and its linked miRNAs (highlighted in gray) are shown. The HSUR4 transcription initiation site (+1), nucleotides deleted (ΔPSE), and mutations disrupting the Sm-binding site or the 3′ box are indicated. Lower panels show Northern blots probed for HSURs 2 and 4, and for hvsA-miR-HSUR2-5p and hvsA-miR-HSUR4-5p. Total RNA was isolated from 293T cells transfected with either empty vector (lane 1) or a plasmid containing the first 7.4 kbp of HVS A11 genome carrying wild-type HSUR4 (lane 2), a promoter deletion (ΔPSE, lane 3), a mutated Sm-binding site (mtSm, lane 4), or a mutated 3′ box (mt3′box, lane 5).
To determine if the sequence or the structure of the HVS miRNA-containing stem-loop is important for the expression of HVS miRNAs, we constructed mutants with alterations in the hairpin predicted downstream of the HSUR4 3′ box (Figure S4). We converted the 3′ arm to the mirror image of the 5′ arm of the stem (construct “3pmt”, Figure S4A), but left the sequence of both hvsA-miR-HSUR4-5p and the loop intact. A second construct derived from “3pmt” altered the 5′ arm of the stem to restore base-pairing (construct “3p5pmt”, Figure S4A). The resulting stem-loop structure is similar to the wild-type but with the 3′ and 5′ arms swapped and their directionality inverted. When these constructs were transiently transfected into 293T cells (Figure S4B), miRNAs were observed only when the stem-loop structure was reconstituted, suggesting that its secondary structure but not its sequence is important for HVS miRNA biogenesis.
Since all three hairpin structures that give rise to HVS miRNAs are located immediately downstream of the 3′ box of a Sm-class RNA (6 or fewer nucleotides away, Figure 1B), we asked whether the distance between the stem-loop structure and the 3′ box is important for generation of miRNAs. We introduced either 6 or 12 extra nucleotides between the 3′ box and the stem-loop structure for HSUR4-miRNAs (“3′box-6nt-insert” and “3′box-12nt-insert”, Figure S4A) and found no effect on the expression of miRNAs (Figure S4B). Together, these data argue that recognition of both the 3′ box (sequence-dependent) and the stem-loop structure (sequence-independent) is important for expression of HVS miRNAs. Furthermore, the 5′ end of the pre-miRNA is determined by a mechanism that does not involve measuring its distance from the 3′ box.
HVS miRNAs Are Processed by a Microprocessor-independent, Dicer-dependent Pathway
Canonical metazoan pri-miRNA transcripts that are Microprocessor substrates consist of a stem of ~33 base pairs (bp), a terminal loop, and flanking ssRNA (Kim et al., 2009). Upon recognition of the ssRNA and the base of the stem by DGCR8, Drosha cleaves the stem ~11bp away from the ssRNA-stem junction (Han et al., 2006), creating both the 5′ end of the 5p arm miRNA and the 3′ end of the 3p arm miRNA. For HVS miRNAs, the 5′ end of the mature 5p miRNA and the 3′ end of the mature 3p miRNA reside in or very close to the flanking ssRNA regions of the putative pri-miRNA (Figure 1B). This hints that they are not substrates for the Microprocessor (Figure 1B).
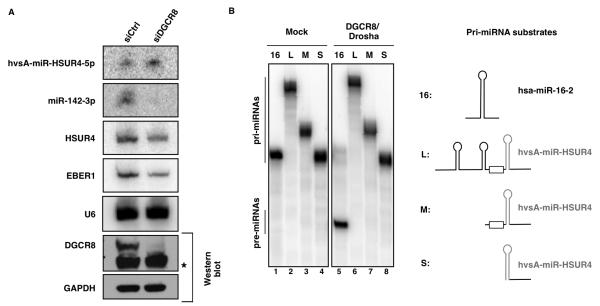
To establish whether the Microprocessor complex is required for expression of HVS miRNAs, we used RNA interference (RNAi)-mediated knockdown of Microprocessor components in 293T cells. We transfected cells with one siRNA against DGCR8 mRNA, followed by a second co-transfection with the same siRNA together with three plasmids: one expressing HSUR4 and its downstream miRNAs; a second expressing the primary transcript of host miR-142, which is not endogenously expressed in these cells (Chen et al., 2004) and is known to be processed by Microprocessor (Wu et al., 2009); and a third expressing EBER1 (Rosa et al., 1981), an RNA pol III transcript, as a transfection control. Efficient depletion of DGCR8 blocked the expression of miR-142-3p (Figure 4A), but had no detectable effect on the level of hvsA-miR-HSUR4-5p, indicating that the Microprocessor complex is not required for in vivo expression of HVS miRNAs. Drosha knockdown in these cells resulted in high cell mortality and very low transfection efficiency (data not shown) and was not further pursued. The lack of Microprocessor involvement was instead confirmed by in vitro Drosha processing assays (Figure 4B). Myc-Drosha together with FLAG-DGCR8 were overexpressed in 293T cells and isolated on anti-Myc or anti-FLAG agarose beads. The purified Microprocessor efficiently processed the canonical pri-miR-16-2 substrate, but no processed pre-miRNA product was observed upon incubation with several versions of the primary transcript for hvsA-miR-HSUR4 under the same experimental conditions (Figure 4B). These in vivo and in vitro data strongly argue that HVS miRNAs are produced through a Microprocessor-independent pathway.
Figure 4. HVS does not utilize the Microprocessor complex.
(A) Northern blot analysis of the expression of hvsA-miR-HSUR4-5p, miR-142-3p, and EBER1 in 293T cells treated with a control siRNA (siCtrl) or a siRNA specific for DGCR8 (siDGCR8) for 48 hr, followed by co-transfection with siDGCR8 and three plasmids expressing HSUR4 and downstream miRNAs, miR-142, or EBER1. U6 provides a loading control. Western blots (lower right two panels) show the knockdown of DGCR8 (a cross-reacting band is marked by an asterisk) with GAPDH as a loading control. (B) 32P-labeled T7-transcribed RNA substrates corresponding to pri-miR-16-2 or to variants of the primary transcript for HSUR4 and its linked miRNAs were incubated with buffer alone (lanes 1-4) or immunoprecipitates containing FLAG-Drosha and FLAG-DGCR8 (lanes 5-8) expressed in 293T cells. Cleavage products were separated on a denaturing polyacrylamide gel. Pri-miRNA substrates are diagramed on the right, with the pre-miRNA hairpin in gray and the box representing the HSUR4-3′ box.
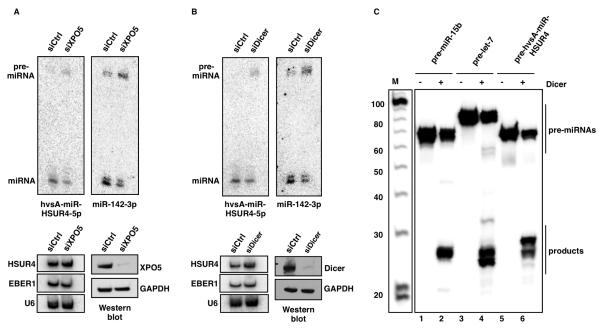
After nuclear processing, canonical pre-miRNAs are exported to the cytoplasm by Exportin-5 (Yi et al., 2003). To test whether Exportin-5 (XPO5) is required for HVS miRNA generation, we performed RNAi-mediated knockdown and observed a modest but reproducible reduction in the levels of both hvsA-miR-HSUR4-5p and host miR-142-3p, accompanied by accumulation of pre-miR-HSUR4 and pre-miR-142 (Figure 5A). These results are consistent with reports showing mild perturbation of host miRNA processing by RNAi-mediated knockdown of Exportin-5 (Lund et al., 2004; Shibata et al., 2006). We conclude that HVS pre-miRNAs likely require Exportin-5 for their export to the cytoplasm.
Figure 5. HVS miRNAs are exported by Exportin-5 and processed by Dicer.
(A) Same as Figure 4A, but cells were treated with an Exportin-5 specific siRNA (siXPO5). Mature miRNAs and pre-miRNAs are shown. (B) Same as in (A), but cells were treated with a Dicer-specific siRNA (siDicer). (C) In vitro assays using purified Dicer (kind gift of J. Doudna). In vitro transcribed 32P-labeled RNAs corresponding to pre-miR-15b (lanes 1 and 2), pre-let-7 (lanes 3 and 4), or the viral pre-hvsA-miR-HSUR4 (lanes 5 and 6) were incubated in the presence of buffer alone (lanes 1, 3, and 5) or recombinant human Dicer (lanes 2, 4, and 6). Cleavage products were separated on a denaturing polyacrylamide gel, with size markers (M).
Once in the cytoplasm, pre-miRNAs are cleaved by Dicer to form mature miRNAs (Kim et al., 2009). Efficient Dicer knockdown in 293T cells produced a modest lowering of the levels of both mature hvsA-miR-HSUR4-5p and miR-142-3p, and a marked accumulation of viral pre-miR-HSUR4 and host pre-miR-142 (Figure 5B). In vitro processing assays performed with recombinant human Dicer confirmed that HSUR4 pre-miRNAs can be processed by Dicer with comparable efficiency to host pre-miR-142 (Figure 5C).
In summary, HVS miRNA biogenesis differs from the canonical host miRNA biogenesis pathway in that it bypasses nuclear processing by the Microprocessor complex. Later, it converges with the canonical pathway that requires Exportin-5 for pre-miRNA export and Dicer for production of mature miRNAs.
The Integrator Complex Is Required for HVS miRNA Biogenesis
Sm-class RNAs are transcribed by RNA pol II, and processing of their 3′ ends occurs co-transcriptionally. Integrator, a complex of 12 proteins that stably associates with the carboxy-terminal domain (CTD) of RNA pol II, is involved in recognition of the 3′ box and cleavage downstream of the mature snRNA 3′ end (Baillat et al., 2005). The pre-Sm-class RNAs are then exported to the cytoplasm where they assemble with the SMN complex that promotes Sm protein binding and undergo 5′-cap trimethylation and 3′-end maturation (Matera et al., 2007).
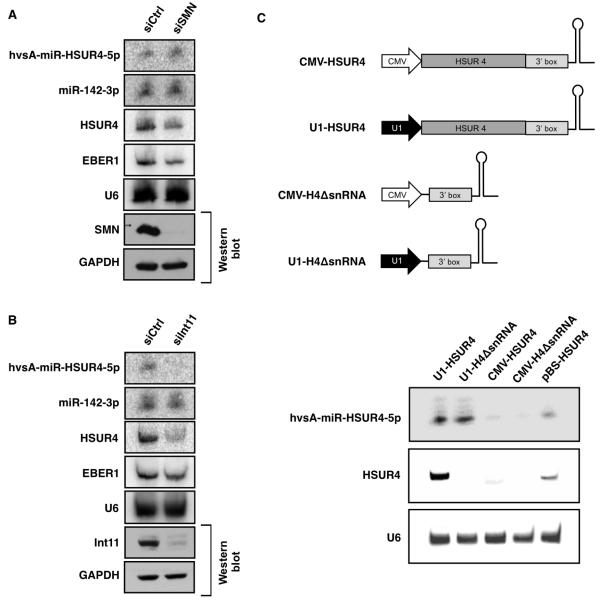
Do the Integrator and SMN complexes also have roles in the biogenesis of HVS miRNAs? RNAi-mediated knockdown of SMN in 293T cells did not affect the expression of either host (miR-142-3p) or viral (hvsA-miR-HSUR4-5p) miRNAs as compared to cells treated with a control siRNA (Figure 6A). In contrast, RNAi-mediated knockdown of Int11 [also called RC-68 (Dominski et al., 2005), a homolog of CPSF-73, the catalytic subunit of the cleavage and polyadenylation specificity complex (CPSF)], resulted in lower levels of both HSUR4 and hvsA-miR-HSUR4-5p compared to the control siRNA; the levels of host miR-142-3p were not affected (Figure 6B). These results indicate that the Integrator complex is important for the biogenesis of HVS miRNAs.
Figure 6. The Integrator complex is required for generation of HVS miRNAs.
(A) Northern blot analysis of the expression of hvsA-miR-HSUR4-5p, miR-142-3p, and EBER1 in 293T cells pre-treated with a control siRNA (siCtrl) or a siRNA specific for SMN (siSMN) and then co-transfected with the same siRNAs and three plasmids expressing HSUR4 and its linked miRNAs, miR-142, or EBER1. The Western blot shows the SMN knockdown, with GAPDH as a loading control. (B) Same as (A), but with Int11-specific siRNA (siInt11). (C) Constructs expressed HSUR4 with its 3′ box and linked miRNAs from a U1 snRNA promoter (U1-HSUR4) or from a cytomegalovirus immediate-early gene (CMV) promoter (CMV-HSUR4), or only the 3′ box of HSUR4 (no snRNA sequence) with its linked miRNAs from the U1 (U1-H4ΔsnRNA) or CMV promoter (CMV-H4ΔsnRNA). The lower panel shows Northern blot analysis of the expression of hvsA-miR-HSUR4-5p and HSUR4 in 293T cells transfected with the above constructs. pBS-HSUR4 (see Figure S3) was a positive control for miRNA expression. U6 snRNA is a loading control.
Coupling between snRNA transcription and 3′-end formation has been suggested based on promoter-swap experiments showing that the 3′ box is efficiently recognized in vivo only in transcripts generated from a Sm-class RNA promoter (Neuman de Vegvar et al., 1986; Hernandez and Weiner, 1986). Since our data indicate that an intact 3′ box (Figure 3) and activity of the Integrator complex (Figure 6B) are both necessary for HVS miRNA expression, we asked whether the nature of the promoter driving transcription and the presence of the snRNA coding sequence are important for the biogenesis of HVS miRNAs. We prepared a series of constructs in which HSUR4, its 3′ box, and its downstream miRNAs would be transcribed from either an snRNA promoter (Figure 6C, U1-HSUR4) or a protein-coding gene promoter (CMV-HSUR4). A second set of constructs contained only a promoter (U1 promoter or CMV promoter), the HSUR4 3′ box, and its downstream miRNAs (Figure 6C, U1-H4ΔsnRNA and CMV-H4ΔsnRNA). When transfected into 293T cells, high expression of hvsA-miR-HSUR4-5p was observed only when transcription was driven by the U1 promoter (Figure 6C). Surprisingly, removal of the entire HSUR4 coding region had no effect on the level of hvsA-miR-HSUR4-5p, suggesting that the 3′ box is necessary and sufficient to direct HVS miRNA processing. Together, these results uncover a Microprocessor-independent, Integrator-dependent miRNA biogenesis pathway used by HVS.
DISCUSSION
HVS Expresses miRNAs
We have identified six miRNAs in HVS transformed marmoset T cells, adding to the roster of miRNAs expressed by herpesviruses (Grundhoff and Sullivan, 2011). A previous report computationally predicted 20 candidate hairpin structures in the HVS genome that could give rise to miRNAs (Walz et al., 2010), but we observed miRNAs from only three of these during latency. The high number of total sequencing reads we obtained (more than 12 million) and the high number of reads for each HVS miRNA (more than 1.8×103 total reads for the least abundant hvsA-miR-HSUR2-5p, Table S1) suggest that it is highly unlikely that other miRNAs have gone undetected, although it is possible that HVS expresses additional miRNAs during lytic infection.
Mutant versions of HVS in which the miRNA-containing regions were deleted are competent for transformation (Ensser et al., 1999; Murthy et al., 1989), indicating that HVS miRNAs are not required for this process. Cells transformed with a mutant version of HVS that lacks HSURs 1 and 2 and its associated miRNAs grow considerably slower than cells transformed with the wild-type virus (Cook et al., 2005; Murthy et al., 1989), suggesting that these two miRNAs could contribute to an enhanced growth rate. Even though herpesviral miRNAs might mediate evolutionarily conserved functions, their sequences are not well conserved (Grundhoff and Sullivan, 2011; and references therein). In the case of HVS, bioinformatics target prediction is further handicapped by the unavailability of the sequence of the genome of the squirrel monkey (Saimiri sciureus), the natural host.
Other herpesviruses express miRNAs that emulate host miRNAs and thus gain access to networks of target transcripts of the host. For instance, host miR-155, an oncogenic miRNA that is upregulated in several different tumors (Croce, 2009), is mimicked by both the Kaposi sarcoma-associated herpesvirus (KSHV)-encoded miR-K11 (Gottwein et al., 2007; Skalsky et al., 2007) and the Marek’s disease virus 1 (MDV1)-encoded miR-M4 (Morgan et al., 2008). One HVS miRNA, hvsA-miR-HSUR5-5p, emulates host miR-937, which is not expressed in HVS-transformed marmoset T cells. This miRNA was identified in human cervical cancer cell lines (Lui et al., 2007) but is not well characterized, making it difficult to discern the advantage for HVS to express a miR-937 mimic. Experimental identification of targets will be necessary to understand the functions of the six HVS miRNAs.
HSURs and miRNAs Are Co-transcribed and Co-processed
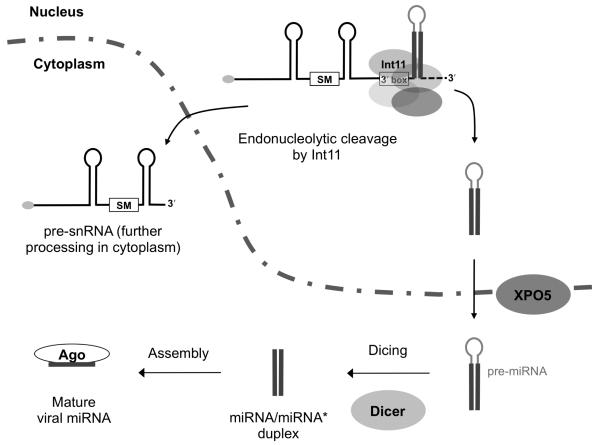
We have shown that HVS miRNAs use the same promoters as their upstream HSURs (Figure 3) to generate primary transcripts (Figures 1B and 1C) that are processed to both Sm-class RNAs and miRNAs (Figure 6B). Recognition of the 3′ box and cleavage of pri-snRNAs by the Integrator complex apparently occur co-transcriptionally as the 3′ box is utilized in vivo only when transcribed from an Sm-class RNA promoter (Neuman de Vegvar et al., 1986; Hernandez and Weiner, 1986). The CTD of RNA pol II is both required for Sm-class RNA 3′-end processing in vivo (Jacobs et al., 2004; Medlin et al., 2003) and directly associates with the Integrator complex (Baillat et al., 2005). Therefore, co-transcriptional cleavage of the 3′ end of HSURs by Integrator releases both the pre-snRNA, which will be exported to the cytoplasm for maturation, and an unstable intermediate that gives rise to a more stable, detectable pre-miRNA (Figures 5A, 5B, and S2). This viral pre-miRNA then enters the canonical miRNA biogenesis pathway, undergoing Exportin-5-facilitated export to the cytoplasm (Figure 5A), where the mature miRNAs produced by Dicer (Figure 5B) are loaded into functional RISC complexes (Figure 2). These results are summarized in a model (Figure 7).
Figure 7. Model of HVS miRNA biogenesis pathway.
The Integrator complex recognizes the 3′ box and Int11 co-transcriptionally cleaves the HSUR-miRNA primary transcript. The released pre-snRNA molecule exits the nucleus for further processing, while the viral pre-miRNA undergoes export directed by Exportin-5 (XPO5) to the cytoplasm, where it is recognized and processed by Dicer to yield two mature Ago-associated HVS miRNAs.
Even though the 3′ box and action of the Integrator complex are required for appearance of the viral pre-miRNAs (Figures 3 and 6), this event is unlikely to generate the 5′ end of the viral pre-miRNA directly. The site of pri-snRNA cleavage after 3′-end processing was reported to reside either within or upstream of the 3′ box (Hernandez, 1985; Kleinschmidt and Pederson, 1987; Neuman de Vegvar and Dahlberg, 1989; Uguen and Murphy, 2003). Since introducing nucleotides between the 3′ box and the pre-miRNA hairpin did not affect the production of HVS miRNAs (Figure S4), cleavage of the primary transcript by the Integrator complex apparently releases an unstable, undetectable pre-miRNA intermediate. One possibility is that the ssRNA ends of this intermediate are trimmed by 5′ and 3′ exonucleases. RNAi-mediated knockdown of Xrn1 or Xrn2 had no effect on the expression of HVS miRNAs (Figure S5). Interestingly, Int11 is a homolog of CPSF-73, which has both endonuclease and 5′ exonuclease activity (Yang et al., 2009). Int11 could therefore potentially cleave the pri-snRNA/miRNA transcript and trim from the 3′ box to the base of the stem, generating the 5′ end of the pre-miRNA. The exosome mediates trimming of 3′-tailed mirtrons (Flynt et al., 2010) in another mode of Microprocessor-independent miRNA biogenesis. We are currently testing whether the same is true for HVS miRNAs.
HVS miRNAs Are Generated in a Microprocessor-independent Manner
HVS miRNAs do not require the Microprocessor for their expression (Figure 4). Previously described examples of Microprocessor-independent miRNA biogenesis include the splicing-dependent mirtrons (Okamura et al., 2007; Ruby et al., 2007), miRNAs derived from snoRNAs (Ender et al., 2008; Saraiya and Wang, 2008), and miRNAs generated from endo-shRNAs (Babiarz et al., 2008). Interestingly, another γ-herpesvirus, MHV68, expresses its miRNAs in the same primary transcripts as a tRNA (Bogerd et al., 2010; Diebel et al., 2010; Pfeffer et al., 2005), which are first processed by the cellular tRNA processing enzyme, RNAse Z (Bogerd et al., 2010), releasing pre-miRNA hairpins that are subsequently matured by Dicer. Why have these two related viruses coordinated the expression of their miRNAs with that of another class of ncRNAs, making their biogenesis pathways independent of the Microprocessor? By linking the expression of two ncRNAs, viruses may economize the use of their genome, and avoid competition between viral and host pri-miRNAs for Microprocessor. Bypassing the Microprocessor allows viral miRNA production to become independent of the many regulatory mechanisms that influence the expression of host miRNAs (Krol et al., 2010).
Viruses and their hosts often share genetic strategies. Indeed, like MHV68, the mouse genome encodes and expresses tRNA/miRNA polycistrons (Reese et al., 2010). Similarly, monkey (and other primate) cells may use the Sm-class RNA processing machinery to generate miRNAs. Detailed inspection of sequences downstream of known cellular snRNAs did not reveal stem-loop structures that could be processed into miRNAs (data not shown), but further investigation is required.
EXPERIMENTAL PROCEDURES
Transient RNAi Knockdown Assays
Specific proteins were depleted in 293T cells by first transfecting 5×104 cells in 6-well plates with either specific or control siRNAs for 48 hr, followed by a second transfection of siRNAs and 0.2 μg of EBER1-expressing plasmid, 0.8 μg of pCDNA3-pri-miR-142, and 1 μg of pBS-HSUR4 using Lipofectamine 2000 (Invitrogen). 48 hr later, total RNA was isolated and analyzed by Northern blot, and total protein was analyzed by Western blot. The following siRNAs were used: for SMN, siSMN (5′-GAAGAAUACUGCAGCUUCCUUA-3′); for Int11, siInt11 (5′-UCGAAGGCCUUGAUGUGCU-3′); for DGCR8, siDGCR8 (5′-AUCACACUCUUGUCCGAUG-3′); for DICER, siDICER (5′-UCCAGAGCUGCUUCAAGCA-3′); for Exportin-5, siXPO5 (5′-UACAAUUCGAGACAGAGCAUC-3′). The unspecific control siRNA (siCtrl) was: 5′-AAGCGAUACCUCGUGUGUGA-3′.
Library Preparation and Deep Sequencing
Small RNA and cDNA library preparation were performed as described (Riley et al., 2010). The Yale Keck Sequencing facility (Yale University, New Haven, CT) used standard 75 bp Illumina/Solexa sequencing with the Genome Analyzer II. Sequencing results were processed using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). A total of 12,225,234 quality-filtered reads were aligned to H. saimiri A11 genome (NC_001350) using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml). Uniquely aligned sequences were visualized using Tablet software (http://bioinf.scri.ac.uk/tablet/). Identities of reads were confirmed by manual alignment with the viral genome and further verified by the basic local alignment search tool (BLAST).
Plasmids
For expression of HSURs and HVS miRNAs in 293T cells, a fragment corresponding to the HVS A11 genome (encompassing positions +22 to approximately +7253), including the genes for HSURs 1 to 7 containing their own enhancers, promoters and processing signals, as well as viral miRNAs, was amplified and inserted between the EcoRI and BamHI sites of pBluescript II SK+ (Stratagene) to generate the plasmid pBS7.4. For expression of individual HSURs with or without viral miRNAs, genomic fragments including positions +2183 to +2750 (containing HSUR2 and hvsA-HSUR2-miRNAs), positions +2344 to +2750 (HSUR2 only), positions +4931 to +5500 (containing both HSUR4 with HSUR4-miRNAs), positions +5072 to +5500 (HSUR4 only), positions +2856 to +3550 (containing both HSUR5 with HSUR5-miRNAs), or positions +3015 to +3550 (HSUR5) were amplified and inserted between the EcoRI and BamHI sites of pBluescript II SK+ to generate plasmids pBS-HSUR2, pBS-HSUR4, pBS-HSUR5, pBS-HSUR2ΔmiR, pBS-HSUR4ΔmiR, or pBS-HSUR5ΔmiR, respectively.
For Firefly luciferase assays, fragments containing 4 sites perfectly complementary to each HVS A11 miRNA were generated by PCR using overlapping oligonucleotides and inserted between the XhoI and SacI sites of pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega). Plasmid pCDN3-pri-miR-142, kindly provided by Jan Pawlicki, was generated by inserting a 258-bp fragment of mouse genomic DNA that directs transcription of the miR-142 primary transcript (Yang et al., 2006) between the HindIII and BglII sites of pcDNA3 (Invitrogen). The plasmid pEBV RIJ expressed EBER 1 (Rosa et al., 1981). A plasmid expressing FLAG-DGCR8 was from Addgene (plasmid #10921). A plasmid expressing FLAG-Drosha (Han et al., 2004) was kindly provided by V. Narry Kim.
In vitro Processing of pre-miRNAs
Reactions were based on the Doudna protocol (Ma et al., 2008). In a 10 μl reaction, 250ng of purified recombinant human Dicer (kindly provided by Jennifer Doudna) were incubated with 100,000cpm of pre-miRNA substrates in the presence of 20mM Tris-HCl (pH 6.5), 1.5mM MgCl2, 25mM NaCl, 1% glycerol and 1mM DTT. Reactions were carried out at 37°C for 1 hr, and products were analyzed by electrophoresis on 8M Urea-15% polyacrylamide gels.
Immunoprecipitation and Antibodies
2A8 anti-Ago monoclonal antibody (kindly provided by Zissimos Mourelatos) (Nelson et al., 2007) or anti-HA antibodies were used for immunoprecipitation (Cazalla et al., 2010) of extracts prepared from either 1×107 HVS(A11)-transformed marmoset T cells or from 5×106 293T cells transfected 48 hr earlier with 10 μg of pBS7.4 plasmid. Co-immunoprecipitated RNA was extracted from beads with Trizol (Invitrogen) and analyzed by Northern blot. Antibodies against GAPDH (Cell Signaling Technology), Dicer (Abcam), DGCR8 (ProteinTech Group), Int11 (Abcam), XPO5 (Abcam), and SMN (Santa Cruz Biotechnology) were used for Western blotting.
Luciferase Assays
293T cells were seeded in 24-well plates and co-transfected the next day with 1 pg Luc reporter specific for each of the viral miRNAs and 800 ng plasmid expressing either HSUR2, HSUR4, HSUR5, HSUR2 and hsvA-HSUR2-miRNAs, HSUR4 and hsvA-HSUR4-miRNAs, or HSUR5 and hsvA-HSUR5-miRNAs. Luciferase activity was assayed 48 hr post-transfection using a TD 20/20n (Turner BioSystems) and the Dual Luciferase Assay System (Promega) per manufacturers’ instructions.
Supplementary Material
Highlights.
Primary transcripts for HVS miRNAs have snRNAs upstream of pre-miRNA hairpins.
HVS miRNA expression relies on snRNA-specific promoter and processing signals.
The Integrator complex is required for generation of HVS pre-miRNAs.
HVS miRNAs are produced by a Microprocessor-independent, Dicer-dependent pathway.
ACKNOWLEDGMENTS
We thank R. C. Desrosiers for plasmids and cell lines; J. Doudna for recombinant human Dicer; T. Yario for technical assistance; Z. Mourelatos for antibodies; M. Hastings for siRNA sequences; J. Pawlicki and V. N. Kim for plasmids; T. Samji for preliminary data; K. Riley for technical guidance; A. Vilborg, D. Cifuentes, and A. Bazzani for critical commentary; and A. Miccinello for editorial assistance. This work was supported by grant CA16038 from the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. M.X. is supported by a Leslie H. Warner Cancer Research Foundation Postdoctoral Research Fellowship. J.A.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Albrecht JC, Fleckenstein B. Nucleotide sequence of HSUR 6 and HSUR 7, two small RNAs of herpesvirus saimiri. Nucleic Acids Res. 1992;20:1810. doi: 10.1093/nar/20.7.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht JC, Nicholas J, Biller D, Cameron KR, Biesinger B, Newman C, Wittmann S, Craxton MA, Coleman H, Fleckenstein B, et al. Primary structure of the herpesvirus saimiri genome. J. Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. A uniform system for microRNA annotation. RNA. 2003;9:277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol. Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla D, Yario T, Steitz J. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Cook HL, Lytle JR, Mischo HE, Li MJ, Rossi JJ, Silva DP, Desrosiers RC, Steitz JA. Small nuclear RNAs encoded by Herpesvirus saimiri upregulate the expression of genes linked to T cell activation in virally transformed T cells. Curr. Biol. 2005;15:974–979. doi: 10.1016/j.cub.2005.04.034. [DOI] [PubMed] [Google Scholar]
- Cook HL, Mischo HE, Steitz JA. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host AU-rich element-containing mRNA levels in virally transformed T cells. Mol. Cell. Biol. 2004;24:4522–4533. doi: 10.1128/MCB.24.10.4522-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nature reviews. Genetics. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Diebel KW, Smith AL, van Dyk LF. Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters. RNA. 2010;16:170–185. doi: 10.1261/rna.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 2005;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O’Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem. Soc. Trans. 2008;36:590–594. doi: 10.1042/BST0360590. [DOI] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Ensser A, Fleckenstein B. T-cell transformation and oncogenesis by gamma2-herpesviruses. Adv. Cancer Res. 2005;93:91–128. doi: 10.1016/S0065-230X(05)93003-0. [DOI] [PubMed] [Google Scholar]
- Ensser A, Pfinder A, Muller-Fleckenstein I, Fleckenstein B. The URNA genes of herpesvirus saimiri (strain C488) are dispensable for transformation of human T cells in vitro. J. Virol. 1999;73:10551–10555. doi: 10.1128/jvi.73.12.10551-10555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Battle DJ, Feng W, Wan L, Dreyfuss G. Lymphotropic Herpesvirus saimiri uses the SMN complex to assemble Sm cores on its small RNAs. Mol. Cell. Biol. 2005a;25:602–611. doi: 10.1128/MCB.25.2.602-611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell. Biol. 2005b;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 1985;4:1827–1837. doi: 10.1002/j.1460-2075.1985.tb03857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 2001;276:26733–26736. doi: 10.1074/jbc.R100032200. [DOI] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986;47:249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jacobs EY, Ogiwara I, Weiner AM. Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol. Cell. Biol. 2004;24:846–855. doi: 10.1128/MCB.24.2.846-855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt AM, Pederson T. Accurate and efficient 3′ processing of U2 small nuclear RNA precursor in a fractionated cytoplasmic extract. Mol. Cell. Biol. 1987;7:3131–3137. doi: 10.1128/mcb.7.9.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews. Genetics. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, Steitz JA. Four novel U RNAs are encoded by a herpesvirus. Cell. 1988;54:599–607. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- Lee SI, Steitz JA. Herpesvirus saimiri U RNAs are expressed and assembled into ribonucleoprotein particles in the absence of other viral genes. J. Virol. 1990;64:3905–3915. doi: 10.1128/jvi.64.8.3905-3915.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Medlin JE, Uguen P, Taylor A, Bentley DL, Murphy S. The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J. 2003;22:925–934. doi: 10.1093/emboj/cdg077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Anderson A, Bernberg E, Kamboj S, Huang E, Lagasse G, Isaacs G, Parcells M, Meyers BC, Green PJ, Burnside J. Sequence conservation and differential expression of Marek’s disease virus microRNAs. J. Virol. 2008;82:12213–12220. doi: 10.1128/JVI.01722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy S, Kamine J, Desrosiers RC. Viral-encoded small RNAs in herpes virus saimiri induced tumors. EMBO J. 1986;5:1625–1632. doi: 10.1002/j.1460-2075.1986.tb04405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy SC, Trimble JJ, Desrosiers RC. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J. Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O’Doherty U, Mourelatos Z. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman de Vegvar HE, Lund E, Dahlberg JE. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- Neuman de Vegvar HE, Dahlberg JE. Initiation and termination of human U1 RNA transcription requires the concerted action of multiple flanking elements. Nucleic Acids Res. 1989;17:9305–9318. doi: 10.1093/nar/17.22.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Reese TA, Xia J, Johnson LS, Zhou X, Zhang W, Virgin HW. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J. Virol. 2010;84:10344–10353. doi: 10.1128/JVI.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley KJ, Rabinowitz GS, Steitz JA. Comprehensive analysis of Rhesus lymphocryptovirus microRNA expression. J. Virol. 2010;84:5148–5157. doi: 10.1128/JVI.00110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa MD, Gottlieb E, Lerner MR, Steitz JA. Striking similarities are exhibited by two small Epstein-Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol. Cell. Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721. doi: 10.1093/nar/gkl663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi’s sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uguen P, Murphy S. The 3′ ends of human pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J. 2003;22:4544–4554. doi: 10.1093/emboj/cdg430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz N, Christalla T, Tessmer U, Grundhoff A. A global analysis of evolutionary conservation among known and predicted gammaherpesvirus microRNAs. J. Virol. 2010;84:716–728. doi: 10.1128/JVI.01302-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Lee SI, Steitz JA. Nucleotide sequence of HSUR 5 RNA from herpesvirus saimiri. Nucleic Acids Res. 1989;17:1258. doi: 10.1093/nar/17.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5′ end variation of mature microRNA. PLoS One. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312–326. doi: 10.1261/rna.2537911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sullivan KD, Marzluff WF, Dominski Z. Studies of the 5′ exonuclease and endonuclease activities of CPSF-73 in histone pre-mRNA processing. Mol. Cell. Biol. 2009;29:31–42. doi: 10.1128/MCB.00776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–301. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.