Abstract
Hoxgenes play a crucial role during embryonic patterning and organogenesis. Of the 39 Hox genes, Hoxa1 is the first to be expressed during embryogenesis and the only anterior Hox gene linked to a human syndrome. Hoxa1 is necessary for proper development of the brainstem, inner ear and heart in humans and mice; however, almost nothing is known about the molecular downstream targets through which it exerts its function. To gain insight into the transcriptional network regulated by this protein, we performed microarray analysis on tissue microdissected from the prospective rhombomere 3–5 region of Hoxa1 null and wild type embryos. Due to the very early and transient expression of this gene, dissections were performed on early somite stage embryos during an eight-hour time window of development. Our array yielded a list of around 300 genes differentially expressed between the two samples. Many of the identified genes play a role in a specific developmental or cellular process. Some of the validated targets regulate early neural crest induction and specification. Interestingly, three of these genes, Zic1, Hnf1b and Foxd3, were down-regulated in the posterior hindbrain, where cardiac neural crest cells arise, which pattern the outflow tract of the heart. Other targets are necessary for early inner ear development, e.g. Pax8 and Fgfr3 or are expressed in specific hindbrain neurons regulating respiration, e.g. Lhx5. These findings allow us to propose a model where Hoxa1 acts in a genetic cascade upstream of genes controlling specific aspects of embryonic development, thereby providing insight into possible mechanisms underlying the human HoxA1-syndrome.
Keywords: Hoxa1, microarray, neural crest, inner ear, rhombomere
INRODUCTION
Hox proteins constitute a family of transcription factors which control gene expression networks that regulate biological processes such as neurogenesis, patterning, organogenesis and cancer (Alexander et al., 2009; Capecchi, 1997). Mouse knockout studies revealed that Hox genes execute their role in a specific segment or domain of the embryo, often affecting several tissues at a given axial level (Mallo et al., 2010). Although many gain- and loss-of-function experiments have been carried out, little is known about the molecular targets and the developmental pathways regulated by Hox genes (Hueber and Lohmann, 2008). In this study, we set out to identify the downstream targets of a specific Hox gene, Hoxa1. This gene affects the development of a diverse array of tissues in the anterior domain of the embryo including the brainstem, inner ear and heart.
Hoxa1 is strongly expressed in the neuroectoderm and mesoderm at the level of the presumptive hindbrain (precursor of the brainstem) from mouse embryonic day (E) 7.75 to 8.5 (Murphy and Hill, 1991). Hoxa1 knockout mice die at or shortly after birth from breathing defects, which are thought to result from mispatterning of the hindbrain (Chisaka et al., 1992; Lufkin et al., 1991). During development, the hindbrain is subdivided into eight transient territories termed rhombomeres (r) (Lumsden and Krumlauf, 1996) and Hoxa1−/− embryos exhibit abnormalities in r3–r5. Additionally, the otic vesicle (embryonic progenitor of the inner ear) forms but fails to differentiate and cranial ganglia, condensations of sensory neurons in the head, are smaller and do not connect properly with the brain (Mark et al., 1993). Cranial ganglia develop in part from cranial neural crest cells, which migrate from the dorsal hindbrain (Barlow, 2002), where Hoxa1 is expressed. So far it is unclear through which mechanisms Hoxa1 regulates the development of neural crest cells or the inner ear. Hoxa1 lineage analysis suggests that Hoxa1 might play a direct role in early patterning of the otic placode (precursor of the otic vesicle) and specification of neural crest cell precursors, while they reside in the neural tube (Makki and Capecchi, 2010).
More recently, humans with homozygous truncating mutations in HOXA1 have been identified (Athabascan Brainstem Dysgenesis Syndrome and Bosley-Salih-Alorainy Syndrome). These patients suffer from hypoventilation (requiring mechanical ventilation), deafness, facial weakness, vocal cord paralysis and swallowing dysfunction (Holve et al., 2003; Tischfield et al., 2005). In addition, patients display defects in the outflow tract of the heart, which have not been described in mice so far. Notably, development of the cardiac outflow tract depends on the influx of neural crest cells, which originate in the posterior hindbrain at the level of r6–r8 (Brown and Baldwin, 2006), where Hoxa1 is expressed.
Despite of what we know about the importance of Hoxa1 in proper development of several embryonic tissues in humans and mice, almost nothing is known about the transcriptional network that is regulated by this protein. In this study, we carried out genome-wide microarray analysis to identify genes that are differentially expressed between control and Hoxa1 null embryos. For genomic profiling, tissue was microdissected from the prospective rhombomere 3–5 region of Hoxa1Δ/Δ and wild type embryos at the 1–6 somite stage (ss). Our analysis identified novel targets of Hoxa1 that play a role in neural crest specification, otic placode patterning, and reticulospinal neuron development.
MATERIALS AND METHODS
Gene targeting and genotyping
A 7.9 kb genomic DNA fragment containing the Hoxa1 locus was subcloned into a conventional plasmid and an artificial AscI site was placed 36 bp downstream of the stop codon as described previously (Tvrdik and Capecchi, 2006). To generate the Hoxa1 conditional allele (Hoxa1c), one loxP site together with an EcoRI site were inserted 200 bp upstream of the Hoxa1 transcription initiation site into a SwaI site. The downstream loxP site along with an EcoRI site and a PolII-frt-Neo-frt selection cassette were inserted into the artificial AscI site 3′ of the Hoxa1 stop codon. Positive clones were identified by digesting genomic DNA with EcoRI, Southern blotting and hybridization with a 5′ external probe. Selected clones were further analyzed by digestion with KpnI and hybridization with an exon1 and a Neo probe. Positive ES cell clones were injected into C57BL/6 blastocysts and chimeric males were crossed to C57BL/6 females. The neomycin resistance gene was removed by crossing the mice to a Flpe deleter line (Rodriguez et al., 2000). The Hoxa1-deletion allele (Hoxa1Δ) was generated by crossing Hoxa1 conditional mice to an Hprt-Cre deleter mouse (Tang et al., 2002). Recombination was verified by Southern analysis and PCR. Genotyping was performed using multiplex PCR. The following primers were used: wild type forward NM228 5′-TGAGGCTACTCCAGCCCAACTC-3′, deletion forward NM230 5′-CTCTCACCTCTTGCCAGTTCAGC-3′, reverse NM229 5′-CAATTGATGTGGACACCCGATG-3′, generating a 220bp wild type, a 326bp conditional and a 520bp deletion band.
Mouse breeding and tissue dissection
Hoxa1Δ/+ mice were maintained on a C57BL/6 background. Timed matings were set up between Hoxa1Δ/+ mice and embryos were harvested at E8.25. Deciduas were isolated in cold PBS and transferred into HEPES-buffered DMEM with 5% FBS on ice. Each embryo was isolated in a separate dish in PBS, extraembryonic tissues were removed and the number of somites counted. Using fine tungsten knives, the bulge region (rhombomere 3–5), including neuroectoderm, mesoderm and otic ectoderm, was isolated and the tissue trimmed by a horizontal cut at the level of the floorplate. The tissue was then transferred into 40 ul of RLT buffer (Qiagen Micro-RNA Easy kit), vortexed immediately for 1 minute and stored on ice until all embryos were processed. The yolk sac was collected for DNA isolation and genotyping. Finally, the tissue was homogenized by vortexing for 5 minutes followed by snap freezing in liquid nitrogen and storage at −80° C. A total of 221 embryos were collected and sorted according to genotype (verified at least twice) and somite stage. Twenty-four wild type and 24 Hoxa1Δ/Δ embryos at the 1–6 somite stage were chosen for analysis and pooled into four wild type and four mutant samples, containing one embryo of each somite stage.
RNA isolation, array hybridization and statistical analysis
RNA was isolated from the eight samples using the RNAqueous-Micro Kit (Ambion) with an on-column DNase treatment (Qiagen). The concentration and quality of the RNA was determined at the University of Utah Microarray Core Facility using a Nanodrop and Bioanalyzer (Agilent). The RNA Integrity Number (RIN) deduced from this analysis was 9.9–10 for all samples, which denotes an excellent RNA quality with no degradation (Schroeder et al., 2006). The final concentrations of total RNA varied from 15 to 20 ng/μl and 150 ng of RNA from each pool was subjected to a single linear amplification labeling reaction with Cy3. RNA was hybridized to Agilent mouse whole genome 44K microarray slides (Agilent), using the Agilent one-color gene expression hybridization protocol. Slides were scanned (Agilent G2505B) at 5 μm resolution using an extended dynamic range protocol, and images were processed with Agilent Feature Extraction software 10.5.1.1. Within-array normalization was performed using the “Background detrending” software (Agilent). The nonuniform outlier features (spots) were removed and the intensity values were transformed to a log base 2 scale. Signal density blots showed uniform ranges and distributions of intensity values from each array and no between-array normalization was necessary. All eight array files were then compiled into a working directory and imported into the statistical analysis program “R” (Dudoit et al., 2003). Genes significantly differentially expressed were identified using the Rank Products algorithm with the default setting of 100 permutations (Breitling et al., 2004). Rank Products analysis was chosen because of its biologically meaningful emphasis on the fold change of gene expression and the reproducibility in samples with small numbers of replicates. GO analysis was performed using DAVID (Dennis et al., 2003; Huang da et al., 2009) on significantly differentially expressed genes. In case of overlapping and similar GO terms, one representative is listed, and terms that are too general were not included. Data was hierarchically clustered with Spotfire (TIBCO) and heat maps for selected genes were generated. The microarray data can be found in the Gene Expression Omnibus (GEO) of NCBI through accession number GSE25868.
Quantitative real-time PCR (qPCR)
60 ng of total RNA was linearly amplified using the qScript cDNA SuperMix (Quanta Biosciences). Reverse transcription and PCR conditions were essentially as described (Schmittgen and Livak, 2008) using the SYBR Green detection method. Primer pairs (Table S2) were obtained from the PrimerBank database (Wang and Seed, 2003). Reactions were run on a 7900HT thermal cycler (Applied Biosystems) in the Genomics Core Facility at the University of Utah. For the final experiment, three wild type and three Hoxa1Δ/Δ cDNA samples (biological replicates) were analyzed individually in three replicates of each reaction (technical replicates) and the mean threshold cycle (CT) for each gene was derived. Relative expression levels were calculated by the ΔCT method (Schmittgen and Livak, 2008), normalizing to the housekeeping gene β-actin, and data expressed as mean fold-change relative to wild type. Unpaired, two-tailed Student’s t-test was used to calculate P-values between the Hoxa1 null and control samples.
Inner ear paint-filling and RNA in situ hybridization
For inner ear paint-filling, E15.5 embryos were harvested and fixed overnight in Bodian’s fixative. Embryos were washed in PBS, dehydrated in ethanol and cleared in methyl salicylate. Heads were hemisected and inner ears injected with 2% white latex paint in methyl salicylate using a micropipette (Morsli et al., 1998). For RNA in situ hybridization, Digoxigenin-labeled antisense cRNA probes were generated from plasmids carrying cDNA fragments. The following cloned mouse cDNAs were obtained, sequenced and used to prepare riboprobes: Foxd3 (from T. Labosky) (Labosky and Kaestner, 1998), Hnf1b and Lhx5 (from Q. Ma) (Gray et al., 2004), Spry4 (from K. Shim/G. Martin) (Minowada et al., 1999), Pax8 (from A. Groves) (Ohyama and Groves, 2004), Zic1 (from R. Arkell) (Elms et al., 2004), Lefty2 (from Y. Saijoh/H. Hamada) (Meno et al., 1996), Hnf4a (from Y. Saijoh). Probes for Fzd8 and Fgfr3 (from L. Urness) were generated following direct PCR amplification of the 3′ UTR from genomic DNA. A 28-base T7 RNA polymerase promoter (5′-GGATCCTAATACGACTCACTATAGGGAG-3′) was incorporated at the 5′ end of the reverse primer. Whole-mount in situ hybridization was performed on embryos isolated from timed pregnancies essentially as described (Henrique et al., 1995).
RESULTS
Hoxa1Δ/Δ mice exhibit the same phenotypes as previously described Hoxa1 null lines
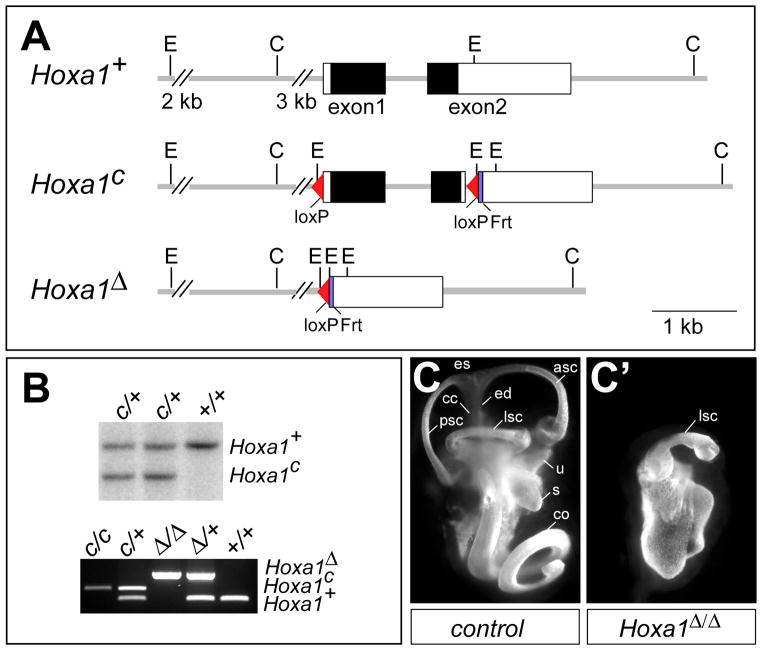
A new Hoxa1 null allele (Hoxa1Δ) was created by flanking the Hoxa1 coding region with loxP sites to generate a conditional allele (Fig. 1A) and then deleting the intervening sequence using Cre recombinase. Hoxa1 conditional (Hoxa1c) mice were generated from targeted ES cells (Fig. 1B) and then crossed to a Flpe-deleter line (Rodriguez et al., 2000) to excise the neomycin resistance gene. Mice homozygous for the Hoxa1 conditional allele are phenotypically wild type. To generate a Hoxa1-deletion allele (Hoxa1Δ), Hoxa1 conditional mice were crossed to an Hprt-Cre deleter line (Tang et al., 2002). As expected, mice with a homozygous deletion of Hoxa1 (Hoxa1Δ/Δ) resemble previously reported Hoxa1 null mice (Chisaka et al., 1992; Mark et al., 1993). Hoxa1Δ/Δ mice are born at normal Mendelian ratios but die shortly after birth at perinatal day P0–P1 (n=34). We also examined Hoxa1Δ/Δ embryos for inner ear defects using the inner ear paint-fill technique (Morsli et al., 1998) and found that the otic vesicle forms but does not differentiate (Fig. 1C, C′) (n=7), as was reported in previous studies (Pasqualetti et al., 2001). Therefore, the Hoxa1Δ allele represents a new Hoxa1 null allele, which was used in all subsequent experiments.
Fig. 1. Hoxa1 targeting and phenotype analysis.
(A) Depiction of Hoxa1 wild-type (Hoxa1+), conditional (Hoxa1c) and deletion (Hoxa1Δ) alleles. The Hoxa1c allele was generated by inserting a 5′ loxP site 200 bp upstream of the Hoxa1 transcription initiation site and a loxP-frt-PolII-Neo-frt cassette 36 bp downstream of the Hoxa1 stop codon. The Neo cassette was removed by recombination, leaving one frt site behind. In the Hoxa1Δ allele, the entire Hoxa1 promoter and coding region are deleted. Black boxes, Hoxa1 coding region; white boxes, UTRs. C, ClaI; E, EcoRI. (B) Upper panel: Southern blot analysis to identify positive Hoxa1c clones. DNA was digested with EcoRI and hybridized with a 5′ external probe to generate an 8.3 kb wt and a 6 kb Hoxa1c band. Lower panel: PCR genotyping to identify the different Hoxa1 alleles. (C, C′) Abnormal inner ear morphology of Hoxa1 mutants. Lateral view of paint-filled inner ears from E15.5 control (C) and Hoxa1Δ/Δ mice (C′). asc, anterior semicircular canal; cc, common crus; co, cochlea; ed, endolymphatic duct; es, endolymphatic sac; lsc, semicircular canal; s, saccule; u, utricle.
Hoxa1 is expressed very transiently in its most anterior domain
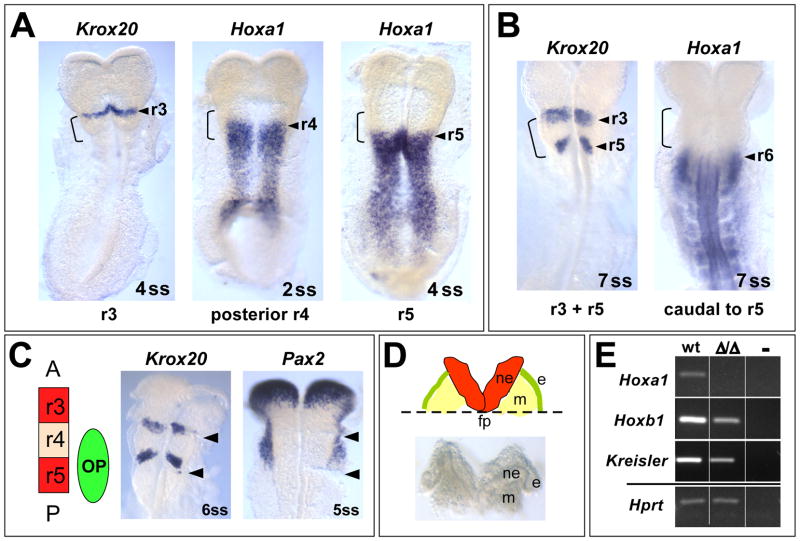
Previous studies showed that Hoxa1 is most strongly expressed in the anterior hindbrain (prospective r3–r5) and neighboring mesoderm (Makki and Capecchi, 2010; Murphy and Hill, 1991) and that all phenotypes resulting from loss of Hoxa1 function are associated with its most anterior expression domain (Chisaka et al., 1992; Lufkin et al., 1991). Therefore, we wanted to identify the exact embryonic time window, during which Hoxa1 is expressed in the prospective r3–r5 region by carrying out RNA in situ hybridization at specific somite stages. As reference we visualized Krox20 expression, which can be detected in r3 from the 4ss and in both r3 and r5 from the 7ss (Fig. 2A, B) (Mechta-Grigoriou et al., 2000). Our analysis revealed that Hoxa1 is only expressed in its most anterior domain (the prospective r3/r4 boundary) from E7.75-2ss (data not shown). From the 2ss (~E8.0), Hoxa1 expression starts retracting to posterior r4. At the 4ss (~E8.25), when Krox20 is first expressed as a single stripe in r3, Hoxa1 has retracted to r5 (Fig. 2A). When the second stripe of Krox20 appears in r5 at the 7ss (~E8.5), Hoxa1 is no longer expressed in this rhombomere (Fig. 2B). Thus, Hoxa1 is expressed for only around twelve hours (E7.75-6ss) in its most anterior domain, which is much more transient than previously believed (Murphy and Hill, 1991).
Fig. 2. Temporal window of Hoxa1 expression and tissue dissection for array analysis.
(A, B)Hoxa1 is expressed very transiently in its most anterior domain as visualized by RNA in situ hybridization in comparison to Krox20. The hindbrain “bulge” region (r3–r5) is marked by an open bracket. (A) At the 4ss, Krox20 labels r3 (the anterior border of the bulge). Hoxa1 is still expressed in posterior r4 at the 2ss but retracts to r5 at the 4ss. (B) At the 7ss, Krox20 also labels r5. At this stage, Hoxa1 is no longer expressed in this rhombomere. (C) In situ hybridization was performed after cutting the hindbrain bulge region (arrowheads indicate cutting sites). Krox20 staining verified that the bulge corresponds to prospective r3–r5 and in situ for Pax2 demonstrates that the dissected region includes the entire otic ectoderm. (D) Schematic depiction (top) and brightfield image (bottom) of the dissected tissue used for RNA isolation. Embryos were flattened out and tissue was cut along the edges of the hindbrain bulge region. The tissue was then trimmed by a horizontal cut along the floorplate of the neural tube (fp), generating a piece of tissue that contains neuroectoderm (ne), otic ectoderm (e) and mesoderm (m). (E) RT-PCR demonstrates that changes in the expression of known downstream effectors of Hoxa1 can be detected in the dissected tissue of wild type and Hoxa1Δ/Δ embryos.
Identifying and isolating the relevant tissue for microarray analysis
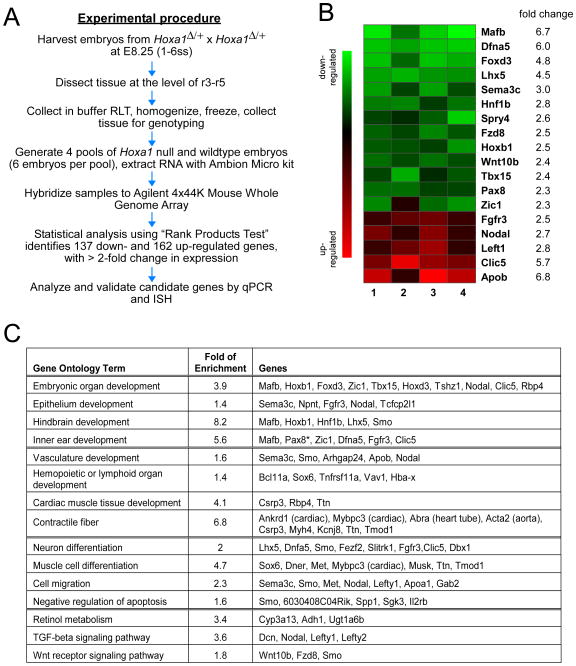
In order to identify genes regulated by Hoxa1, we set out to collect tissue from the prospective r3–r5 region of Hoxa1Δ/Δ and wild type embryos for microarray analysis. Since our in situ experiments revealed that Hoxa1 is expressed in this region from around E7.75 to the 6ss, we chose to collect embryos at the 1–6ss. This is an approximately eight-hour time window (Tam, 1981), around and slightly after the peak of Hoxa1 expression, and before phenotypic manifestations are apparent in Hoxa1 null embryos. Therefore, we believe that our experimental setup would allow identification of both direct and indirect targets of Hoxa1. Conveniently, at this time the prospective r3–r5 region is morphologically visible as a “bulge” that forms in the future hindbrain (Fig. 2A, B open brackets). The bulge region was microdissected by performing two cuts along the edges of the bulge (Fig. 2C) and then trimming the tissue at the level of the floorplate to include neuroectoderm, mesoderm and otic ectoderm at the level of r3–r5 (Fig. 2D). To confirm that the bulge region included the entire r3–r5 region, we performed in situ hybridization for Krox20 after cutting the tissue (Fig. 2C). Since Hoxa1 null mice also exhibit severe inner ear defects, we wanted to include the otic ectoderm, the precursor of the inner ear, which develops at the level of r4–r5 (Ohyama and Groves, 2004). In situ staining for the otic marker Pax2 on cut tissue confirmed that this region was included in our dissection (Fig. 2C). Finally, we wanted to verify that the tissue chosen for dissection would allow us to detect expression changes in known Hoxa1 downstream targets between wild type and Hoxa1 null embryos. Therefore, we isolated RNA from a small number of dissected embryos and performed RT-PCR on two of the few known Hoxa1 targets, Hoxb1 and Kreisler (Mafb) (Pasqualetti et al., 2001). We saw clear changes in RNA levels of these two genes between the two genotypes (Fig. 2E). This gave us confidence to carry out a large scale analysis using this technique (Fig. 3A). A total of 221 embryos from 52 Hoxa1Δ/+ females were dissected and genotyped. The Mendelian ratio was as follows: 21% homozygous, 51% heterozygous and 28% wild type. For microarray analysis, embryos at the 1–6 somite stage were pooled into four wild type and four mutant samples, each containing one embryo per somite stage.
Fig. 3. Microarray analysis identifies novel Hoxa1 targets involved in various developmental processes.
(A) Flowchart showing the experimental procedure from embryo harvesting to validation of microarray targets. (B) Expression heat maps for relative expression of genes of interest obtained from four Agilent microarrays comparing Hoxa1Δ/Δ to control embryos. Green indicates decreased and red increased expression in mutants. Note the reproducible direction and magnitude of the changes. Fold changes are log base 2; P<0.0005. (C) Gene ontology (GO) analysis was performed on significantly differentially expressed genes using DAVID. Enriched GO terms for genes significantly down-regulated (green) or up-regulated (red), as well as fold of enrichment (compared to genome-wide background level) are listed. Asterisk indicates that the gene is involved in corresponding GO function but failed to be recognized by DAVID.
Microarray analysis reveals Hoxa1 candidate targets involved in different developmental processes
To enable a comprehensive assessment of Hoxa1-regulated genes we compared gene expression profiles of four Hoxa1Δ/Δ and four wild type samples using genome-wide microarray analysis. Rank Products analysis (Breitling et al., 2004) yielded a list of 299 differentially expressed genes (137 down-regulated and 162 up-regulated in the mutant) with a ≥ 2-fold change in expression at a false discovery rate of 0.05 and with P-values ≤ 0.0002 (Table S1). As expected, the most highly down-regulated gene in this list is Hoxa1, with a fold change of 70. The two known downstream targets of Hoxa1 were also among the down-regulated genes in the list: Mafb (Kreisler) (Pasqualetti et al., 2001) and Hoxb1 (Barrow et al., 2000), with a 6.7-fold (third most highly down-regulated gene) and a 2.5-fold down-regulation, respectively. In order to identify other “genes of interest”, we scanned the whole list of 299 potential targets for genes that fulfill one of two criteria: (i) known to play a role in a developmental process or (ii) expressed during early embryogenesis. Twelve of the 137 down-regulated and seven of the 162 up- regulated genes were selected as potentially interesting candidates (Table 1). The magnitude of expression changes of the selected genes in each of the four samples is illustrated in the intensity map representations (Fig. 3B). In order to identify biological processes that might be regulated by Hoxa1, we carried out gene ontology (GO) analysis of significantly up- and down-regulated genes using the DAVID software (Database for Annotation, Visualization and Integrated Discovery) (Dennis et al., 2003; Huang da et al., 2009). This analysis identified categories such as hindbrain development, inner ear development, vascular development, neuron differentiation and cell migration (Fig. 3C). Several of the genes listed under one of these categories were also selected as “genes of interest”.
Table 1. Differentially expressed genes of interest from Hoxa1 microarray.
Twelve candidate genes were selected from the list of down-regulated genes and seven from the list of up-regulated genes, based on their published expression pattern and proposed function during development. Candidates were analyzed further by quantitative PCR and/or in situ hybridization. Genes highlighted in green and red were confirmed to be down- or up-regulated, respectively. FC, fold change.
| Gene | FC | Gene name | Proposed function | Expression |
|---|---|---|---|---|
| Dfna5 | 6.0 | Deafness, autosomal dominant 5 | inner ear receptor cell differentiation | E10.5 (northern) |
| Foxd3 | 4.8 | Forkhead box D3 | maintenance/induction of NCC | E9.5, pre-migratory NCC |
| Lhx5 | 4.5 | LIM homeobox protein 5 | respiration (reticulospinal), cardiac dev. | E10.5 |
| Sema3c | 3.0 | Semaphorin 3c | NCC, nervous system, heart dev. | E10.5, cardiac OT |
| Hnf1b | 2.8 | HNF1 homeobox b | hindbrain r5 dev., NCC | E8.0, hindbrain, NCC, foregut |
| Spry4 | 2.6 | Sprouty 4 | Fgf signaling, craniofacial dev. | E8.5, lateral to hindbrain |
| Fzd8 | 2.5 | Frizzled 8 | Wnt receptor | E8.5, head, otic placode |
| Wnt10b | 2.4 | Wingless related 10b | Wnt signaling | E11, 1st arch |
| Tbx15 | 2.4 | T-box 15 | craniofacial dev. | E11, CNS |
| Pax8 | 2.3 | Paired box gene 8 | inner ear (otic placode specification) | ≥ 0ss, otic placode |
| Zic1 | 2.3 | Zinc finger protein of the cerebellum 1 | neural plate patterning, NC specification | ≥ 6ss, hindbrain, neural tube |
| Hoxd3 | 2.1 | Homeobox d3 | cervical vertebrae dev., postnatal death | E9, hindbrain r4/r5 border |
|
| ||||
| Apob | 6.8 | Apolipoprotein B | artery morphogenesis | E7.5 (RT-PCR) |
| Clic5 | 5.7 | Chloride intracellular channel 5 | auditory receptor cell organization | E16.5, cochlea |
| Lefty1 | 2.8 | Left right determination factor 1 | left-right patterning | 3–6ss, floor plate |
| Nodal | 2.7 | Nodal | left-right patterning | 3–5ss, node, mesoderm |
| Hnf4a | 2.7 | Hepatic nuclear factor 4a | endodermal organ development | E8.5, fore-midgut endoderm |
| Fgfr3 | 2.5 | Fibroblast growth factor receptor 3 | inner ear dev. (hearing loss in humans) | E9.0, otic placode |
| Lefty2 | 2.3 | Left right determination factor 2 | left-right patterning | 3–6ss, mesoderm |
Nineteen genes were selected from the total list of 299 differentially expressed genes for further analysis by RT- and qPCR, based on their expression pattern and/or proposed function during development as deduced from the Mouse Genome Informatics webpage (http://www.informatics.jax.org/). Genes highlighted in green or red were confirmed to be down- or up-regulated. FC, fold change.
Validation of microarray targets by quantitative PCR
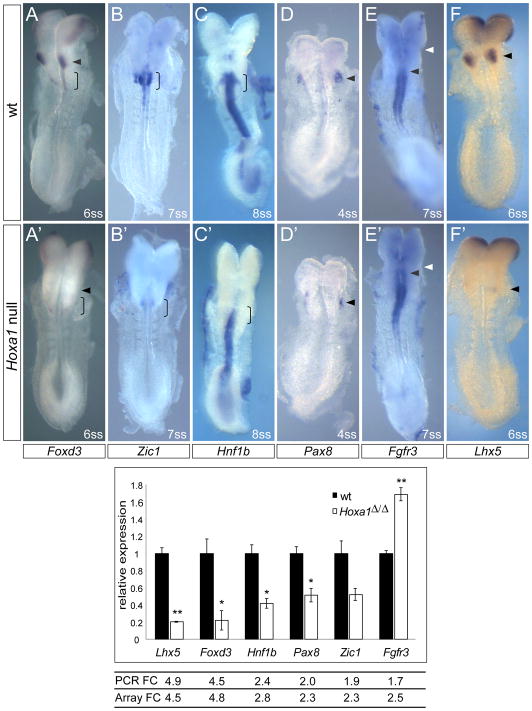
We carried out two qPCR experiments to identify and validate novel downstream targets of Hoxa1. First, we performed an initial qPCR screening of the 19 “genes of interest” (Table 1). For this, RNA from the pool of dissected tissue of three 4–5 somite and two 7–8 somite wild type and Hoxa1Δ/Δ embryos was used. In this screening six genes (Foxd3, Lhx5, Hnf1b, Zic1, Pax8 and Fgfr3) were found to be differentially regulated between Hoxa1 null and wild type embryos in accordance with the microarray data. A second qPCR experiment was performed to further validate these six targets. qPCR validation was based on three biological replicates, each containing dissected tissue from six 1–6 somite embryos (the same samples used for microarray analysis). Unpaired, two-tailed Student’s t-test was used to calculate P-values between Hoxa1 null and control samples. In agreement with the microarray results, Lhx5 and Foxd3 were ~5-fold down-regulated; Hnf1b, Pax8 and Zic1 were ~2-fold down-regulated and Fgfr3 was ~2-fold up-regulated compared to wild type (Fig. 4).
Fig. 4. Validation of novel Hoxa1 targets by RNA in situ analysis and quantitative PCR.
Identification of six novel downstream targets of Hoxa1. Top panel: validation of Hoxa1 microarray candidate genes by in situ hybridization in somite-matched control (A–F) and Hoxa1 null (A′–F′) embryos. (A, A′) Foxd3 expression in premigratory neural crest in the hindbrain is absent in future r4 (arrowhead) and strongly reduced in the posterior hindbrain (future r6–r8; open bracket) of Hoxa1 mutants. (B, B′) Zic1 expression in the posterior hindbrain is absent or reduced in Hoxa1 null embryos. (C, C′) Hnf1b expression is reduced in the posterior hindbrain. (D, D′) Pax8 expression in the otic placode (arrowhead) is reduced. (E, E′) Fgfr3 expression in the hindbrain is expanded anteriorly (black arrowhead: anterior border of Fgfr3 expression, white arrowhead: r2/r3 boundary). (F, F′) Lhx5 expression is absent in the hindbrain bulge region (prospective r4; arrowhead) but is unaffected in the forebrain. 2–4 embryos per genotype were analyzed. Bottom panel: validation of candidate genes by quantitative PCR. Relative changes in gene expression levels were analyzed in three wt and three Hoxa1Δ/Δ samples (biological replicates). The mean threshold cycle (CT) for each gene was derived from triplicate reactions for each sample. Relative expression levels were calculated by the ΔCT method, normalizing to the housekeeping gene β-actin. Expression changes in Hoxa1Δ/Δ samples (white) are plotted as mean fold-change relative to wt samples (black). Fold changes (FC) detected by qPCR are very similar to the fold changes found by microarray analysis. Data are represented as mean +/− SEM. * P<0.02, ** P<0.002 by Student’s two-tailed t-test.
Validation of microarray targets by in situ hybridization
To further validate candidates from our “gene of interest” list, we compared gene expression in somite matched Hoxa1 null and control embryos by whole mount in situ hybridization. Expression patterns of the following genes were examined: Fgfr3, Foxd3, Fzd8, Hnf1b, Hnf4a, Lefty2, Lhx5, Pax8, Spry4, Zic1. No obvious differences in expression of Spy4, Fzd8 or Lefty2 were seen between Hoxa1Δ/Δ and control embryos at the 3–10ss (data not shown) and Hnf4a was not detected in embryonic tissue prior to E8.5. Interesting differences were found in the expression patterns of Foxd3, Zic1 and Hnf1b, all genes known to be expressed in neural crest precursors in the hindbrain. Foxd3 expression in the hindbrain bulge region (prospective r4) was absent in Hoxa1 mutants and expression in the posterior hindbrain (prospective r6–r8) was reduced (Fig. 4A, A′). Similarly, expression of Zic1 and Hnf1b in the posterior hindbrain (future r5–r8) of Hoxa1 mutants was severely reduced (Fig. 4B, C, B′, C′). Moreover, expression of Pax8, a gene required for otic placode specification, was reduced in the placode of Hoxa1 null embryos as early as the 4ss (Fig. 4D, D′). Consistent with upregulation of Fgfr3 expression in the microarray and by qPCR, in situ analysis revealed an anterior expansion of Fgfr3 expression from the r4/r5 boundary in wild type embryos to the r3/r4 boundary in Hoxa1 null embryos (Fig. 4E, E′). Finally, we detected Lhx5 expression in the hindbrain bulge region (prospective r4) as early as E8.25 (6 somite stage) (Fig. 4F). This expression domain was absent in Hoxa1 mutants (Fig. 4F′). Interestingly, three of the validated genes (Foxd3, Zic1, Hnf1b) are known to play a role in neural crest development (Aruga, 2004; Barbacci et al., 1999; Dottori et al., 2001), two of the genes (Pax8, Fgfr3) are important for inner ear development (Mackereth et al., 2005; Pannier et al., 2009) and one gene (Lhx5) is expressed in hindbrain reticulospinal neuron precursors (Cepeda-Nieto et al., 2005; Gray et al., 2004) (Table 1).
DISCUSSION
Although Hoxa1 is crucial for proper development of the hindbrain, inner ear and neural crest in humans and mice, little is known about the downstream genes that are controlled by this transcription factor. Here, we carried out microarray analysis of this early expressed Hox gene and compiled a list of 299 candidate targets. Through systematic analysis of this list, we validated an interesting set of Hoxa1 effector genes. These genes are known to control specific developmental processes such as neural crest induction, inner ear patterning and hindbrain neuron specification and can now be placed in a gene cascade downstream of Hoxa1. This allows us to suggest a new model for how Hoxa1 might regulate the development of the above tissues (Fig. 5) and opens up many new avenues for further investigation. To our knowledge, this is the first microarray analysis performed as early as E8.25 to identify gene expression patterns in the developing mammalian hindbrain and adjacent tissues.
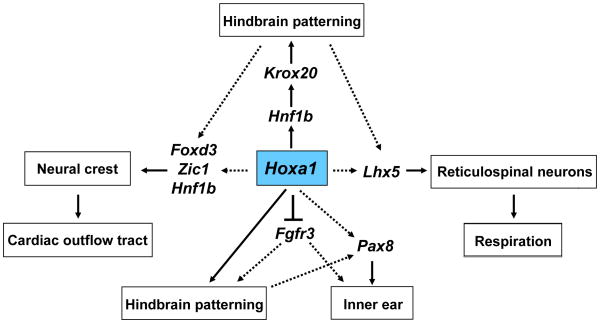
Fig. 5. Proposed model for regulation of hindbrain, inner ear and neural crest development by Hoxa1.
Our data suggests that Hoxa1 influences hindbrain patterning through Hnf1b, which in turn activates Krox20. It also suggests that Hoxa1 might regulate neural crest development, through Foxd3, Zic1 and Hnf1b, which could be the reason for the outflow tract defects in humans. In inner ear development, Hoxa1 acts upstream of Pax8 and Fgfr3. In addition, Hoxa1 might regulate Lhx5 expression in reticulospinal neuron precursors, which could contribute to the respiratory defects in Hoxa1 knockout mice. Whether the above effects are direct or through Hoxa1’s influence on hindbrain patterning remains to be shown (as highlighted by the dotted arrows) and will be the ground for future investigations.
Identification and validation of six novel downstream targets of Hoxa1 involved in development of the neural crest, inner ear and hindbrain neurons
From the list of 299 putative Hoxa1 targets, we selected 19 genes for further analysis. These genes were chosen based on their expression during early embryogenesis and/or a proposed function in a developmental process or signaling pathway. Of the 19 genes, six validated by qPCR and in situ hybridization. Three of the validated Hoxa1 targets, Foxd3, Zic1 and Hnf1b are involved in early neural crest development. Foxd3 is expressed in premigratory neural crest cells in the hindbrain at around E8.5 (Labosky and Kaestner, 1998) and has been shown to promote the development of neural crest from neural tube progenitors (Dottori et al., 2001). Deletion of Foxd3 in neural crest cells using the Wnt1-Cre driver results in loss of neural crest-derived structures (Teng et al., 2008). In Foxd3c/−; Wnt1-Cre embryos cranial neural crest derived ganglia and nerves are smaller. The same phenotype is seen in Hoxa1 null embryos, where cranial ganglia and their associated nerves are reduced in size (Mark et al., 1993), suggesting that Hoxa1 acts upstream of Foxd3 in neural crest development. The second gene, Zic1, is expressed in the neural tube, including the dorsal hindbrain from which neural crest cells arise (Elms et al., 2004; Gaston-Massuet et al., 2005; Nagai et al., 1997). Zic1 was shown to play a role in early neural plate patterning, neural fate acquisition and neural crest specification in Xenopus (Aruga, 2004; Merzdorf, 2007), where it acts upstream of Pax3 and interacts with Gbx2, the earliest factor in neural crest induction (Li et al., 2009). Zic1−/− mice exhibit cerebellar abnormalities but neural crest defects have not been studied in these mice. Besides Foxd3 and Zic1, which play a role in neural crest specification, we identified Hnf1b as a downstream target of Hoxa1. This gene is expressed in the hindbrain, neural crest cells and the foregut at E8.0 and is required for visceral endoderm specification and differentiation (Barbacci et al., 1999; Coffinier et al., 1999; Haumaitre et al., 2005). Because of the role in visceral endoderm development, Hnf1b null mice die at E7.5 (Coffinier et al., 1999) and the role of Hnf1b in mammalian hindbrain and neural crest development has not been studied. Hnf1b is, however, known to play a role in hindbrain development in zebrafish (Choe et al., 2008), where loss of Hnf1b function results in complete absence of Krox20 expression in r5. This is reminiscent of Hoxa1 knockout mice, where r5 is absent and the second stripe of Krox20 expression, which normally marks this rhombomere, is missing (Lufkin et al., 1991). Analysis of cis-regulatory sequences governing Krox20 expression identified a conserved enhancer containing a binding site for the Hnf1b transcription factor, which is necessary for the initiation of Krox20 expression (Chomette et al., 2006). Therefore, our findings suggest that Hoxa1 acts upstream of Hnf1b in the initiation of Krox20 expression in r5 (Fig. 5). Interestingly, our in situ analysis revealed that Foxd3, Hnf1b and Zic1 are strongly reduced in the posterior hindbrain (r6–r8) of Hoxa1 null embryos. This region of the hindbrain is not mispatterned in Hoxa1 mutants and was thought to be unaffected by loss of Hoxa1 function. The posterior hindbrain gives rise to cardiac neural crest cells, which are important for remodeling of the cardiac outflow tract which is affected in humans with mutations in HoxA1. Therefore, reduction of Foxd3, Zic1 and Hnf1b, three neural crest markers, in the posterior hindbrain of Hoxa1 null mice suggests that Hoxa1 might play a direct role in cardiac neural crest development and that this could be the reason for the outflow tract defects in HoxA1-syndrome patients.
Two other confirmed genes, Pax8 and Fgfr3, are known to be important for inner ear development. Pax8 is expressed in the otic placode starting at the pre-somite stage (Ohyama and Groves, 2004) and plays a role in otic placode induction and specification (Mackereth et al., 2005). It is interesting to find changes in Pax8 expression as early as the 4 somite-stage, since gene expression profiles have not been analyzed in the otic placode of Hoxa1 mutants prior to E9.25 (~20 somite stage), when morphological changes have already occurred (Pasqualetti et al., 2001). This suggests that Hoxa1 affects inner ear development at a very early stage, presumably during otic placode specification and might, therefore, play a direct role in inner ear development. The only validated Hoxa1 downstream target that was up-regulated in Hoxa1 mutants was Fgfr3. Expression of Fgfr3 was found to be expanded in the hindbrain of Hoxa1 null embryos, extending from its normal border at the r5/r6 boundary anteriorly into r4. Fgf signaling in several tissues, including the hindbrain, is known to influence inner ear development (Zelarayan et al., 2007) and it was shown that activating Fgfr3 mutations can cause hearing loss and inner ear defects in humans and mice (Pannier et al., 2009). Since Hoxa1 is strongly expressed in r4, it is possible that it acts as an inhibitor of Fgfr3 in the hindbrain and that release of this inhibition leads to ectopic activation of Fgfr3, which might contribute to the inner ear defects in Hoxa1 null mice.
Finally, Lhx5 was identified as a novel downstream target of Hoxa1. Lhx5 expression in the hindbrain has previously been reported at E10.5 (Gray et al., 2004). Our in situ and qPCR data now show that Lhx5 is already expressed as early as E8.25 (6 somite stage). Interestingly, Lhx5 has been implicated in the determination of reticulospinal neuron identity at E12.5 (Cepeda-Nieto et al., 2005). These neurons are involved in modulation of respiration and cardiovascular function both of which are affected by loss of Hoxa1. It is, therefore, possible that Hoxa1 acts upstream of Lhx5 in the development of hindbrain reticulospinal neuron precursors.
In conclusion, we identified Hnf1b, Foxd3 and Zic1 as Hoxa1 downstream targets which are involved in hindbrain and early neural crest development. Interestingly, these markers were reduced in the posterior hindbrain, where cardiac neural crest cells originate suggesting that Hoxa1 might play a role in the development of these cells. Additionally, we identified changes in the expression patterns of Pax8 and Fgfr3, two genes important for inner ear development, which indicates that Hoxa1 affects otic placode specification. Whether it does so directly or through signaling from the hindbrain remains to be shown. Finally, Lhx5, a gene expressed in hindbrain reticulospinal neuron precursors, was down-regulated in Hoxa1 mutants raising the possibility that Hoxa1 acts upstream of Lhx5 in the development of these neurons (Fig. 5). Although our experiments do not allow us to conclude if the identified six genes are direct or indirect targets of Hoxa1, they are likely to play important regulatory roles in the development of the tissues affected by loss of Hoxa1.
In addition to identifying effectors of Hoxa1 in neural crest, inner ear and hindbrain development, our array provides a long list of novel potential targets involved in other developmental and cellular processes such as cardiac and vascular development or neuron and muscle cell differentiation (Fig 3C), which will be the ground for future investigations.
Comparison of Hoxa1 microarray results to other published microarray experiments
Ten microarrays have been published which identified Hox downstream targets in the mouse (reviewed by Hueber and Lohmann, 2008). Of these, six have been carried out on mouse tissue that expresses the gene of interest, whereas the other four, including two Hoxa1 microarrays (Martinez-Ceballos et al., 2005; Shen et al., 2000), have been performed on cultured cell lines. We compared the list of genes identified in our microarray with the lists of the two published Hoxa1 microarrays performed on cultured cells. None of the 28 putative downstream effectors identified in the differential hybridization screening of teratocarcinoma cells overexpressing Hoxa1 (Shen et al., 2000) were found in our microarray. In the second Hoxa1 microarray study, which compared gene expression profiles of wild type and Hoxa1−/− embryonic stem cells treated with retinoic acid, 145 targets were identified (Martinez-Ceballos et al., 2005). Only 45 of these targets are available online and again none of them were identified in our experiment. This is not surprising, since our microarray and the previously published ones constitute very different experiments. The two previous microarrays identified Hoxa1 targets in embryonic stem or cancer cells. Our study now adds a valuable new list of downstream targets, which are controlled by Hoxa1 in the developing embryo.
Microarrays were also performed on Hoxb1 (Tvrdik and Capecchi, 2006), the paralog of Hoxa1 in mice and its ortholog hoxb1a in zebrafish (Rohrschneider et al., 2007). Since Hoxa1 and Hoxb1 are paralogous members and may share some downstream targets, we compared our Hoxa1 dataset with the datasets from the above Hoxb1 studies. The following genes were differentially expressed in both our Hoxa1 microarray as well as either the mouse Hoxb1 or zebrafish hoxb1a microarray and might represent common targets of the two genes: Zinc finger protein of the cerebellum 1 (Zic1), delta/notch-like EGF-related receptor (Dner), nephronectin (Npnt), transthyretin (Ttr), Sjogren syndrome antigen B (Ssb), Nik related kinase (Nrk), DEAD box polypeptide 3 (Ddx3y), leucine rich repeat containing 4 (Lrrc4).
Since Hoxa1 is of profound importance to the development of a variety of tissues, analysis of some of the targets on our list allowed us to propose a model for how Hoxa1 might regulate specific aspects of hindbrain, inner ear and neural crest development. Further investigation into the molecular mechanisms through which Hoxa1 orchestrates the development of these tissues will be necessary to better understand the origin of the defects in HoxA1-syndrome patients. We believe that this study might provide a first stepping stone in this direction.
Supplementary Material
Rank Products analysis revealed 137 down-regulated (green) and 162 up-regulated genes (red) in Hoxa1Δ/Δ samples. Column names are explained in the table of contents tab.
All primer pairs, except for β-actin, were obtained from the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/).
Highlights.
Microarray analysis on the r3–r5 region of 1–6 somite stage Hoxa1 null embryos yields 299 differentially expressed genes.
Some of the validated targets play a role in neural crest specification (Zic1, Hnf1b and Foxd3), inner ear patterning (Pax8 and Fgfr3) and reticulospinal neuron development (Lhx5).
We identified targets (Zic1 and Hnf1b) that are down-regulated in posterior rhombomere-derived cardiac neural crest cells.
Hoxa1 acts in a genetic cascade upstream of genes regulating neural crest, inner ear and hindbrain development.
Acknowledgments
We thank Petr Tvrdik for generating the Hoxa1c targeting vector and members of our tissue culture and mouse facility, in particular Sheila Barnett, Carol Lenz and Karl Lustig for ES cell culture, injection and mouse care; Lisa Urness and Yukio Saijoh for their experimental input and advice, as well as sharing protocols and reagents; Ruth Arkell, Andy Groves, Patricia Labosky, Xiaojing Ma and Katherine Shim for plasmids used to generate riboprobes and Brett Milash and Brian Dalley from the University of Utah Microarray Core for assistance in analyzing the microarray experiment. This manuscript was improved by helpful comments from Anne Boulet and Daniel Kopinke. This work was supported by grants from the NIH (NIH5R01GM021168-34) and Howard Hughes Medical Institute to M.R.C. and the Boehringer Ingelheim Fonds PhD fellowship and the University of Utah Graduate Research Fellowship to N.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander T, Nolte C, Krumlauf R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu Rev Cell Dev Biol. 2009;25:431–56. doi: 10.1146/annurev.cellbio.042308.113423. [DOI] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–21. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Barbacci E, Reber M, Ott MO, Breillat C, Huetz F, Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–805. doi: 10.1242/dev.126.21.4795. [DOI] [PubMed] [Google Scholar]
- Barlow LA. Cranial nerve development: placodal neurons ride the crest. Curr Biol. 2002;12:R171–3. doi: 10.1016/s0960-9822(02)00734-0. [DOI] [PubMed] [Google Scholar]
- Barrow JR, Stadler HS, Capecchi MR. Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development. 2000;127:933–44. doi: 10.1242/dev.127.5.933. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Brown CB, Baldwin HS. Neural crest contribution to the cardiovascular system. Adv Exp Med Biol. 2006;589:134–54. doi: 10.1007/978-0-387-46954-6_8. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Hox genes and mammalian development. Cold Spring Harb Symp Quant Biol. 1997;62:273–81. [PubMed] [Google Scholar]
- Cepeda-Nieto AC, Pfaff SL, Varela-Echavarria A. Homeodomain transcription factors in the development of subsets of hindbrain reticulospinal neurons. Mol Cell Neurosci. 2005;28:30–41. doi: 10.1016/j.mcn.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Chisaka O, Musci TS, Capecchi MR. Developmental defects of the ear, cranial nerves and hindbrain resulting from targeted disruption of the mouse homeobox gene Hox-1.6. Nature. 1992;355:516–20. doi: 10.1038/355516a0. [DOI] [PubMed] [Google Scholar]
- Choe SK, Hirsch N, Zhang X, Sagerstrom CG. hnf1b genes in zebrafish hindbrain development. Zebrafish. 2008;5:179–87. doi: 10.1089/zeb.2008.0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomette D, Frain M, Cereghini S, Charnay P, Ghislain J. Krox20 hindbrain cis-regulatory landscape: interplay between multiple long-range initiation and autoregulatory elements. Development. 2006;133:1253–62. doi: 10.1242/dev.02289. [DOI] [PubMed] [Google Scholar]
- Coffinier C, Thepot D, Babinet C, Yaniv M, Barra J. Essential role for the homeoprotein vHNF1/HNF1beta in visceral endoderm differentiation. Development. 1999;126:4785–94. doi: 10.1242/dev.126.21.4785. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–38. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Dudoit S, Gentleman RC, Quackenbush J. Open source software for the analysis of microarray data. Biotechniques. 2003;(Suppl):45–51. [PubMed] [Google Scholar]
- Elms P, Scurry A, Davies J, Willoughby C, Hacker T, Bogani D, Arkell R. Overlapping and distinct expression domains of Zic2 and Zic3 during mouse gastrulation. Gene Expr Patterns. 2004;4:505–11. doi: 10.1016/j.modgep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Gaston-Massuet C, Henderson DJ, Greene ND, Copp AJ. Zic4, a zinc-finger transcription factor, is expressed in the developing mouse nervous system. Dev Dyn. 2005;233:1110–5. doi: 10.1002/dvdy.20417. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–7. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Haumaitre C, Barbacci E, Jenny M, Ott MO, Gradwohl G, Cereghini S. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc Natl Acad Sci U S A. 2005;102:1490–5. doi: 10.1073/pnas.0405776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–90. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Holve S, Friedman B, Hoyme HE, Tarby TJ, Johnstone SJ, Erickson RP, Clericuzio CL, Cunniff C. Athabascan brainstem dysgenesis syndrome. Am J Med Genet A. 2003;120:169–73. doi: 10.1002/ajmg.a.20087. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hueber SD, Lohmann I. Shaping segments: Hox gene function in the genomic age. Bioessays. 2008;30:965–79. doi: 10.1002/bies.20823. [DOI] [PubMed] [Google Scholar]
- Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185–90. doi: 10.1016/s0925-4773(98)00105-1. [DOI] [PubMed] [Google Scholar]
- Li B, Kuriyama S, Moreno M, Mayor R. The posteriorizing gene Gbx2 is a direct target of Wnt signalling and the earliest factor in neural crest induction. Development. 2009;136:3267–78. doi: 10.1242/dev.036954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufkin T, Dierich A, LeMeur M, Mark M, Chambon P. Disruption of the Hox-1.6 homeobox gene results in defects in a region corresponding to its rostral domain of expression. Cell. 1991;66:1105–19. doi: 10.1016/0092-8674(91)90034-v. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–15. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Mackereth MD, Kwak SJ, Fritz A, Riley BB. Zebrafish pax8 is required for otic placode induction and plays a redundant role with Pax2 genes in the maintenance of the otic placode. Development. 2005;132:371–82. doi: 10.1242/dev.01587. [DOI] [PubMed] [Google Scholar]
- Makki N, Capecchi MR. Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev Biol. 2010;341:499–509. doi: 10.1016/j.ydbio.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M, Wellik DM, Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Dev Biol. doi: 10.1016/j.ydbio.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M, Lufkin T, Vonesch JL, Ruberte E, Olivo JC, Dolle P, Gorry P, Lumsden A, Chambon P. Two rhombomeres are altered in Hoxa-1 mutant mice. Development. 1993;119:319–38. doi: 10.1242/dev.119.2.319. [DOI] [PubMed] [Google Scholar]
- Martinez-Ceballos E, Chambon P, Gudas LJ. Differences in gene expression between wild type and Hoxa1 knockout embryonic stem cells after retinoic acid treatment or leukemia inhibitory factor (LIF) removal. J Biol Chem. 2005;280:16484–98. doi: 10.1074/jbc.M414397200. [DOI] [PubMed] [Google Scholar]
- Mechta-Grigoriou F, Garel S, Charnay P. Nab proteins mediate a negative feedback loop controlling Krox-20 activity in the developing hindbrain. Development. 2000;127:119–28. doi: 10.1242/dev.127.1.119. [DOI] [PubMed] [Google Scholar]
- Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature. 1996;381:151–5. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236:922–40. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–75. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Morsli H, Choo D, Ryan A, Johnson R, Wu DK. Development of the mouse inner ear and origin of its sensory organs. J Neurosci. 1998;18:3327–35. doi: 10.1523/JNEUROSCI.18-09-03327.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P, Hill RE. Expression of the mouse labial-like homeobox-containing genes, Hox 2.9 and Hox 1.6, during segmentation of the hindbrain. Development. 1991;111:61–74. doi: 10.1242/dev.111.1.61. [DOI] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Takada S, Gunther T, Sporle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–6. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–9. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- Pannier S, Couloigner V, Messaddeq N, Elmaleh-Berges M, Munnich A, Romand R, Legeai-Mallet L. Activating Fgfr3 Y367C mutation causes hearing loss and inner ear defect in a mouse model of chondrodysplasia. Biochim Biophys Acta. 2009;1792:140–7. doi: 10.1016/j.bbadis.2008.11.010. [DOI] [PubMed] [Google Scholar]
- Pasqualetti M, Neun R, Davenne M, Rijli FM. Retinoic acid rescues inner ear defects in Hoxa1 deficient mice. Nat Genet. 2001;29:34–9. doi: 10.1038/ng702. [DOI] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–40. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–72. doi: 10.1016/j.ydbio.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Wu H, Gudas LJ. Molecular cloning and analysis of a group of genes differentially expressed in cells which overexpress the Hoxa-1 homeobox gene. Exp Cell Res. 2000;259:274–83. doi: 10.1006/excr.2000.4963. [DOI] [PubMed] [Google Scholar]
- Tam PP. The control of somitogenesis in mouse embryos. J Embryol Exp Morphol. 1981;65(Suppl):103–28. [PubMed] [Google Scholar]
- Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–24. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield MA, Bosley TM, Salih MA, Alorainy IA, Sener EC, Nester MJ, Oystreck DT, Chan WM, Andrews C, Erickson RP, Engle EC. Homozygous HOXA1 mutations disrupt human brainstem, inner ear, cardiovascular and cognitive development. Nat Genet. 2005;37:1035–7. doi: 10.1038/ng1636. [DOI] [PubMed] [Google Scholar]
- Tvrdik P, Capecchi MR. Reversal of Hox1 gene subfunctionalization in the mouse. Dev Cell. 2006;11:239–50. doi: 10.1016/j.devcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelarayan LC, Vendrell V, Alvarez Y, Dominguez-Frutos E, Theil T, Alonso MT, Maconochie M, Schimmang T. Differential requirements for FGF3, FGF8 and FGF10 during inner ear development. Dev Biol. 2007;308:379–91. doi: 10.1016/j.ydbio.2007.05.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rank Products analysis revealed 137 down-regulated (green) and 162 up-regulated genes (red) in Hoxa1Δ/Δ samples. Column names are explained in the table of contents tab.
All primer pairs, except for β-actin, were obtained from the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/).