Abstract
Several models have been proposed to suggest how the evolution of sex determining mechanisms might contribute to speciation. Here we describe the inheritance of sex in 19 fish species from the rapidly evolving flock of cichlids in Lake Malawi, Africa. We found that many of these species have a male heterogametic (XY) system on linkage group 7. Some species also segregate for a female heterogametic (ZW) system on linkage group 5 which is coincident with a dominant orange-blotch (OB) color pattern in females. The ZW system is epistatically dominant to the XY system when both are segregating within a family. Several lines of evidence suggest that additional sex-determining loci are segregating in some species. These results are consistent with the idea that genetic conflicts play an important role in the evolution of these species flocks and suggest that evolution of sex-determining mechanisms has contributed to the radiation of cichlid fishes in East Africa.
Keywords: sex determination, sexually antagonistic selection, speciation
INTRODUCTION
Because of their spectacular diversity, the cichlid fishes of East Africa have figured prominently in debates about the mechanisms of speciation. Independent radiations have produced morphologically and behaviorally diverse flocks of several hundred species in each of the major rift lakes (Turner et al. 2001). The extraordinary radiation of haplochromine cichlids in lakes Malawi and Victoria has occurred in the last million years (Meyer et al. 1990; Kocher et al. 1995, Verheyen et al. 2003, Genner et al. 2007). These species flocks provide a rare opportunity to study speciation occurring in nearly historical time (Kocher 2004).
For much of the last century, studies of the mechanisms of speciation focused on the gradual accumulation of Dobzhansky-Muller incompatibilities in allopatry. Classical allopatric speciation has clearly been important in the radiation of many groups (Mayr 1963). A high degree of population structure, together with a long history of lake-level fluctuations, has likely created opportunities for microallopatric speciation in African cichlids. Surveys of microsatellite variation have shown that Malawi cichlid populations are structured at extremely fine (<1km) geographic scales and typically exchange only one migrant per generation (van Oppen et al. 1997, Arnegard et al. 1999, Danley et al. 2000, Rico and Turner 2002). However, accumulating evidence suggests that natural selection also plays an important role in speciation (Schluter 2000). There is substantial evidence that strong ecological selection has created specialized feeding morphologies in African cichlids (Albertson et al. 2005), and may have contributed to the emergence of new species (Danley and Kocher 2001).
Sexual selection
Sexual selection is another powerful force which has been postulated to contribute to speciation (Lande 1981, Ritchie 2007). Sexual selection is particularly strong in haplochromine cichlids because of an unequal investment in parental care. Females lay large eggs and after mating do not feed for three weeks while they mouthbrood their embryos and larvae without help from the male. This skewed parental investment leads to strong intersexual selection as females carefully select their mates. It also leads to strong intrasexual selection on males, who compete fiercely for mating territories. The intense competition for mates may be the reason why males have evolved diverse gaudy color patterns, while females are typically inconspicuous. A recent phylogenetic analysis suggested that there have been more than 250 evolutionary transitions among a handful of core color patterns in Lake Malawi cichlids (Allender et al. 2003). Fish with similar color patterns collected from different regions of Lake Malawi are not closely related, indicating multiple regional radiations of the same color forms (Smith & Kornfield 2002). Sensory drive has been implicated in the evolution of male nuptial colors during speciation of the closely related flock of cichlids in Lake Victoria (Seehausen et al. 2008).
Stimulated by empirical results on the segregation of sex and color patterns in Lake Victoria cichlids, Lande et al. (2001) proposed two models of speciation based on sexual selection and evolution of sex determining mechanisms. The two models postulate the invasion of an XY sex determination system by a novel dominant female allele (W). The models also included an unlinked recessive suppressor of the W, an unlinked locus controlling mating preference, and a novel color mutation linked to the W chromosome. Their analytical results suggested possible mechanisms for repeated sympatric speciation through the rapid evolution of color, mate preferences and sex determining genes.
Sexual antagonism
Sexually antagonistic coevolution can arise over numerous traits related to courtship, mating and parental investment, when the phenotypic optima are different for males and females (Chapman 2006). Experiments have shown that intersexual genetic conflicts are numerous (Rice 1992; 1998). Sexual antagonism is clearly a major force shaping the evolution of the genome (Jin et al. 2001). So far, relatively little work has explored the impact of sexual antagonism in the evolution of cichlid fishes.
Theoretical work has explored the conditions under which sexual conflict can promote the evolution of new sex chromosomes (Bull 1983). These models begin with an autosomal locus segregating alleles which have different relative fitness in males and females. Selection will favor an increase in frequency of any new sex determining loci that are tightly linked to these autosomal alleles. Under some conditions both old and new sex-determining loci remain polymorphic in the population (van Doorn and Kirkpatrick 2007).
Goals of this study
The first goal of this study was to determine whether Lake Malawi cichlids have a predominantly genetic mechanism for sex determination. There is evidence for both environmental (Romer and Beisenherz 1996, Baroiller et al. 2009) and genetic (Devlin and Nagahama 2002, Cnaani et al. 2008) sex determination in tilapiine cichlids. In Malawi cichlids, published data have not provided evidence for morphologically distinct sex chromosomes (Kornfield, 1984), but there is some evidence for genetic sex determination in a few species. Albertson (2002) identified a dominant male sex-determining locus on LG7 in an intergeneric hybrid cross. Streelman and colleagues (2003) mapped a dominant female sex-determining locus linked to the orange-blotch color polymorphism on LG5.
The second goal was to determine whether the sex-determining mechanism differs among closely related species. We surveyed a number of species, focusing on the diverse genus Metriaclima. We wanted to know if both male (XY) and female (ZW) heterogametic sex determination systems exist, and whether closely-related species differ in their mechanism of sex determination, consistent with a possible role in speciation.
MATERIALS AND METHODS
Species and stocks
Single pair crosses were made for 19 species of Lake Malawi cichlid in our tropical aquaculture facility (Table 1). Each aquarium contained a single male and between one and fifteen females. Because Lake Malawi cichlids are maternal mouthbrooders, it was possible to collect the progeny of individual females one to five days post fertilization. Caudal fin samples were clipped from the parents at the time of embryo collection for subsequent extraction of DNA.
Table 1.
Sex ratios for each Lake Malawi cichild species.
Sex ratios for each family of Lake Malawi cichlid. The sex ratios (total # of males/total # of males + total # of females) were calculated for each family along with the average sex ratio for each species
| Species | Family | Males | Females | Unknown | Total | Sex Ratio | Binomial | Average sex ratio in species |
|---|---|---|---|---|---|---|---|---|
| Aulonocara baenschi | Aube-A002/A003/A005/A007 | 5 | 7 | 2 | 14 | 0.417 | 0.774 | 0.507 |
| Nkhomo Reef | Aube-A009 | 2 | 4 | 2 | 8 | 0.333 | 0.688 | |
| Aube-A013 | 12 | 3 | 2 | 17 | 0.800 | 0.035 | ||
| Aube-A014 | 6 | 15 | 2 | 23 | 0.286 | 0.078 | ||
| Aube-A022 | 6 | 4 | 10 | 0.600 | 0.754 | |||
| Aube-A023 | 7 | 4 | 11 | 0.636 | 0.549 | |||
| Cynotilapia ‘dwarf afra’ | Dcafra-A002 | 11 | 15 | 2 | 28 | 0.423 | 0.557 | 0.688 |
| Hai Reef | Dcafra-A019 | 22 | 0 | 22 | 1.000 | *** | ||
| Labeotropheus trewavasae | RltrMR009 | 10 | 7 | 17 | 0.588 | 0.629 | 0.515 | |
| Mapanga | RltrMR011 | 7 | 5 | 12 | 0.583 | 0.774 | ||
| RLtrMR012 | 6 | 9 | 15 | 0.400 | 0.607 | |||
| RltrMR015 | 11 | 11 | 22 | 0.500 | 1 | |||
| Metriaclima barlowi | MebaMBJ-A003 | 6 | 2 | 1 | 9 | 0.750 | 0.289 | 0.539 |
| Mbenjii Island | MebaMBJ-A006 | 3 | 7 | 10 | 0.300 | 0.344 | ||
| MebaMBJ-A009 | 5 | 3 | 8 | 0.625 | 0.727 | |||
| MebaMBJ-A010 | 0 | 11 | 11 | 0.000 | *** | |||
| MebaMBJ-A016 | 18 | 5 | 23 | 0.783 | * | |||
| MebaMBJ-A019 | 12 | 11 | 23 | 0.522 | 1 | |||
| MebaMBJ-A026 | 11 | 8 | 19 | 0.579 | 0.648 | |||
| Metriaclima benetos | MebeMZ-A002 | 11 | 10 | 21 | 0.524 | 1 | 0.410 | |
| Mazinzi Reef | MebeMZ-A006 | 6 | 13 | 10 | 29 | 0.316 | 0.167 | |
| MebeMZ-A007 | 11 | 26 | 3 | 40 | 0.297 | * | ||
| MebeMZ-A010 | 20 | 20 | 1 | 41 | 0.500 | 1 | ||
| Metriaclima callainos | Meca-A001 | 2 | 13 | 15 | 0.133 | ** | 0.137 | |
| Choifu Bay | Meca-A004 | 2 | 17 | 19 | 0.105 | *** | ||
| Meca-A005 | 1 | 18 | 19 | 0.053 | *** | |||
| RMca012 | 5 | 13 | 18 | 0.278 | 0.096 | |||
| RMca016 | 3 | 21 | 24 | 0.125 | *** | |||
| Metriaclima ‘daktari’ | Psda-A005 | 6 | 5 | 3 | 14 | 0.545 | 1 | 0.545 |
| Hai Reef | ||||||||
| Metriaclima fainzilberi | MefaLI-A002 | 5 | 12 | 17 | 0.294 | 0.143 | 0.140 | |
| Lundo Island | RMFI010 | 1 | 25 | 2 | 28 | 0.038 | *** | |
| Metriaclima fainzilberi | MefaLP-A001 | 1 | 21 | 22 | 0.045 | *** | 0.070 | |
| Lundo Point | MefaLP-A003 | 2 | 19 | 1 | 22 | 0.095 | *** | |
| Metriaclima ‘kompakt’ | MeblN-A001 | 10 | 13 | 1 | 24 | 0.435 | 0.678 | 0.488 |
| Southern Mbamba Bay | MeblN-A003 | 34 | 18 | 52 | 0.654 | * | ||
| Rmbl007 | 7 | 24 | 31 | 0.226 | ** | |||
| Rmbl004 | 8 | 8 | 16 | 0.500 | 1 | |||
| Metriaclima lombardoi | Melo-A001 | 2 | 15 | 2 | 19 | 0.118 | ** | 0.412 |
| Mbenjii Island | Melo-A003 | 0 | 5 | 5 | 0.000 | 0.063 | ||
| Melo-A015A | 5 | 8 | 13 | 0.385 | 0.581 | |||
| Melo-A015B | 3 | 5 | 8 | 0.375 | 0.727 | |||
| Melo-A016 | 7 | 3 | 1 | 11 | 0.700 | 0.344 | ||
| Melo-A017 | 2 | 6 | 8 | 0.250 | 0.289 | |||
| Melo-A019 | 10 | 13 | 23 | 0.435 | 0.678 | |||
| Melo-A026 | 18 | 12 | 30 | 0.600 | 0.362 | |||
| Metriaclima mbenji | Memb-A003 | 2 | 3 | 5 | 0.400 | 1 | 0.277 | |
| Mbenjii Island | Memb-A004 | 5 | 27 | 3 | 35 | 0.156 | *** | |
| RMmb002 | 5 | 12 | 17 | 0.294 | 0.143 | |||
| RMmb004 | 14 | 26 | 40 | 0.350 | 0.081 | |||
| Metriaclima msobo | Mems-A001/A002 | 15 | 14 | 2 | 31 | 0.517 | 1 | 0.517 |
| Lundo Island | ||||||||
| Metriaclima ‘mustardi’ | RMmus002 | 7 | 10 | 17 | 0.412 | 0.629 | 0.412 | |
| Metriaclima phaeos | MephNA-A001 | 14 | 18 | 3 | 35 | 0.438 | 0.597 | 0.504 |
| Undu Point | MephNA-A006 | 16 | 15 | 3 | 34 | 0.516 | 1 | |
| MephNA-A007 | 10 | 11 | 2 | 23 | 0.476 | 1 | ||
| Meph-A008 | 4 | 7 | 3 | 14 | 0.364 | 0.549 | ||
| Meph-A009 | 6 | 8 | 14 | 0.429 | 0.791 | |||
| Meph-A014 | 14 | 4 | 18 | 0.778 | * | |||
| Metriaclima pyrsonotus | Mepy-A002 | 14 | 5 | 19 | 0.737 | 0.064 | 0.187 | |
| Nakantenga Island | Mepy-A006 | 2 | 12 | 14 | 0.143 | * | ||
| Mepy-A009 | 2 | 26 | 1 | 29 | 0.071 | *** | ||
| Mepy-A010 | 1 | 21 | 22 | 0.045 | *** | |||
| Mepy-A013 | 0 | 29 | 29 | 0.000 | *** | |||
| Mepy-A014 | 4 | 24 | 28 | 0.143 | *** | |||
| Mepy-A017 | 8 | 17 | 25 | 0.320 | 0.108 | |||
| Mepy-A022 | 3 | 28 | 1 | 32 | 0.097 | *** | ||
| Mepy-A025 | 5 | 19 | 24 | 0.208 | ** | |||
| Mepy-A027 | 3 | 20 | 23 | 0.130 | *** | |||
| Mepy-A030 | 9 | 11 | 20 | 0.450 | 0.824 | |||
| RMpy007 | 5 | 10 | 3 | 18 | 0.333 | 0.302 | ||
| RMpy011 | 0 | 11 | 11 | 0.000 | *** | |||
| RMpy012 | 0 | 19 | 19 | 0.000 | *** | |||
| RMpy013 | 0 | 15 | 15 | 0.000 | *** | |||
| RMpy018 | 9 | 20 | 29 | 0.310 | 0.061 | |||
| RMpy021 | 9 | 35 | 44 | 0.205 | *** | |||
| Metriaclima saulosi | Mesa-A001 | 6 | 1 | 7 | 0.857 | 0.125 | 0.706 | |
| Taiwan Reef | MesaNA-A005 | 0 | 12 | 12 | 0.000 | *** | ||
| MesaNA-A009 | 38 | 3 | 2 | 43 | 0.927 | *** | ||
| MesaNA-A12 | 4 | 4 | 8 | 0.500 | 1 | |||
| Metriaclima zebra Mazinzi Reef | MezeMZR-A003 | 12 | 15 | 1 | 28 | 0.444 | 0.701 | 0.305 |
| Mazinzi Reef | MezeMZR-A004 | 4 | 2 | 6 | 0.667 | 0.688 | ||
| MezeMZR-A005 | 2 | 3 | 2 | 7 | 0.400 | 1 | ||
| MezeMZR-A006 | 1 | 24 | 25 | 0.040 | *** | |||
| MezeMZR-A007 | 4 | 8 | 12 | 0.333 | 0.388 | |||
| MezeMZR-A009 | 7 | 18 | 25 | 0.280 | * | |||
| MezeMZR-A010 | 6 | 16 | 22 | 0.273 | 0.052 | |||
| MezeMZR-A011 | 14 | 28 | 42 | 0.333 | * | |||
| Metriaclima zebra ‘Nankoma blue’ | Mena-A006 | 21 | 12 | 6 | 39 | 0.636 | 0.163 | 0.440 |
| Maleri Islands Ribbinck Nakoma G | Mena-A7/21 | 21 | 34 | 1 | 56 | 0.382 | 0.105 | |
| Mena-A8/17 | 31 | 51 | 82 | 0.378 | * | |||
| Mena-A018 | 23 | 25 | 48 | 0.479 | 0.885 | |||
| Pseudotropheus polit | Pspo-A003 | 7 | 9 | 2 | 18 | 0.438 | 0.804 | 0.511 |
| Lions Cove | Pspo-A004 | 5 | 5 | 10 | 0.500 | 1 | ||
| PspoLC-A005-B003 | 12 | 9 | 21 | 0.571 | 0.664 | |||
| Totals | Males | Females | Unknown | Individuals | Average Sex Ratio | |||
| 746 | 1250 | 70 | 2066 | 0.375 | ||||
| Mean | 8 | 13 | 2 |
P <0.05
P <0.01
P <0.001
Rearing and sexing
The embryos from each female were reared together in a small container until they had absorbed their yolk. They were then transferred to increasingly larger tanks and grown to sexual maturity. To maximize the number of fish raised in our facility, families of two to six species with distinct color patterns were sometimes grown together in the same tank. We added dither fish (e.g. Oreochromis mossambicus) to aquaria containing the more pugnacious species to diffuse aggressive interactions.
At maturity, each family was anesthetized using tricaine methanesulfonate (MS-222) and sacrificed to obtain phenotypic data. The standard length, color, and sex of each individual was noted. Gonads were examined using an acetocarmine squash procedure to identify spermatocytes or oocytes (Guerrero et al., 1974). Finally, a caudal fin clip was taken from each individual and stored in 100% EtOH at −20°C until processing for DNA extraction.
Microsatellites
Previous work in our lab detected linkage of sex to markers on linkage groups (LG) 5 and 7 in an intergeneric cross among Lake Malawi cichlids (Albertson et al. 2003). We therefore focused our genotyping effort on microsatellite markers from these two linkage groups (Table 2). The markers on LG5 (UNH2139, c-Ski, GM264a) span a region of ~12.2cM, and those on LG7 (UNH973, UNH2095, UNH2086, UNH2031) span a region of ~18.5cM (Albertson et al. 2003; Lee et al. 2005).
Table 2.
Fisher’s exact test for association of markers on LG5 and LG7 with gender. The segregation of dam and sire alleles were tested separately for markers on LG5 and LG7. Significant associations are indicated in bold letters. When a significant association was found for the ZW locus on LG5, we also tested the individuals not carrying the W chromosome (Blue or BB) for association of the LG7 XY locus. Some parents were not informative (NI) either because they were homozygous for a particular marker, or because they were heterozygous for the same alleles as the other parent. ND = not genotyped
| Species | Family | LG5 | LG7 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UNH2139 | cski | Gm264a | UNH2139 | cski | Gm264a | UNH2095 | UNH2086 | UNH2031 | UNH2095 | UNH2086 | UNH2031 | Interpretation | ||||
| Dam | Sire | Dam | Sire | |||||||||||||
| Aulonocara baenschi | Aube-A2/A3/A5/A7/A14/A23 | X2 | 0.660 | NI | 0.229 | NI | NI | 0.180 | 3.260 | 3.740 | 5.530 | 35.200 | 35.000 | 33.400 | ||
| P | 0.416 | Het | 0.632 | Homo | Het | 0.671 | 0.071 | 0.053 | 0.019 | <.0001 | <.0001 | <.0001 | LG 7 XY | |||
|
|
|
|||||||||||||||
| Labeotropheus trewavasae | RLtrMR012 | 15.000 | 11.200 | 15.000 | 0.185 | 0.714 | NI | NI | NI | NI | NI | NI | NI | |||
| <.0001 | 0.001 | <.0001 | 0.667 | 0.398 | Homo | Homo | Homo | Homo | Homo | Homo | Homo | LG 5 ZW | ||||
| RLtrMR015 | NI | 16.4 | ND | 0.188 | 1.82 | ND | NI | NI | NI | NI | NI | NI | ||||
| Homo | <.0001 | 0.665 | 0.178 | Homo | Homo | Homo | Homo | Homo | Homo | LG 5 ZW | ||||||
|
|
|
|||||||||||||||
| Metriaclima barlowi Mbenji Is | MebaMBJ-A009 | 0.036 | 0.036 | NI | 1.600 | 0.036 | 0.900 | 0.533 | 0.533 | 0.533 | NI | 3.730 | 8.000 | |||
| 0.850 | 0.850 | Homo | 0.206 | 0.850 | 0.343 | 0.465 | 0.465 | 0.465 | Homo | 0.053 | 0.005 | LG 7 XY | ||||
| MebaMBJ-A016 | NI | 2.650 | NI | 0.123 | 1.170 | NI | 4.110 | 0.0407 | NI | 0.157 | 0.509 | 0.0020 | ||||
| Homo | 0.104 | Homo | 0.726 | 0.280 | Homo | 0.043 | 0.8400 | Homo | 0.692 | 0.476 | 0.964 | ND | ||||
| MebaMBJ-A019 | 0.006 | 0.034 | ND | 0.354 | 0.381 | ND | 0.135 | 1.150 | 0.379 | 3.630 | 12.700 | 9.760 | ||||
| 0.937 | 0.855 | 0.552 | 0.537 | 0.714 | 0.283 | 0.538 | 0.057 | <.0001 | 0.002 | LG 7 XY | ||||||
|
|
|
|||||||||||||||
| Metriaclima benetos | MebeMZ-A002 | 0.800 | 0.202 | 3.330 | 3.200 | NI | 0.220 | NI | 0.800 | NI | NI | 16.400 | 16.400 | |||
| 0.371 | 0.653 | 0.068 | 0.074 | Homo | 0.639 | Homo | 0.371 | Homo | Homo | <.0001 | <.0001 | LG 7 XY | ||||
| Mebe-A006 | 0.277 | 0.277 | 0.652 | 0.277 | 0.024 | 0.024 | 0.903 | 0.277 | 1.530 | 0.903 | 12.100 | 15.000 | ||||
| 0.599 | 0.599 | 0.419 | 0.599 | 0.876 | 0.876 | 0.342 | 0.599 | 0.216 | 0.342 | 0.001 | <.0001 | LG 7 XY | ||||
| Mebe-A007 | 0 | 0.184 | 0.419 | 1.300 | 0.116 | 0.749 | 0.344 | 0.717 | 0.134 | 0.344 | 7.280 | 20.700 | ||||
| 1 | 0.668 | 0.517 | 0.253 | 0.734 | 0.387 | 0.556 | 0.397 | 0.714 | 0.556 | 0.007 | <.0001 | LG 7 XY | ||||
|
|
|
|||||||||||||||
| Metriaclima callainos | Rmca012 | Wh&Bl | 3.46 | 4.41 | NI | 0.055 | 0.554 | 1.270 | Wh&Bl | 0.019 | 0.382 | 0.019 | 0.259 | 0.019 | 0.780 | |
| 0.063 | 0.036 | Homo | 0.814 | 0.457 | 0.261 | 0.889 | 0.536 | 0.889 | 0.611 | 0.889 | 0.377 | LG 5 ZW | ||||
| Blue | 0 | 0.052 | 0.052 | 0.476 | 0.052 | 0.782 | ||||||||||
| 1 | 0.819 | 0.819 | 0.490 | 0.819 | 0.376 | ND | ||||||||||
| Rmca016 | Wh&Bl | 4.130 | 8.330 | NI | 0.556 | 0.599 | 0.206 | Wh&Bl | 2.200 | 0.423 | 0.046 | 0.200 | 0.093 | 0.005 | ||
| 0.042 | 0.004 | Homo | 0.456 | 0.439 | 0.650 | 0.138 | 0.515 | 0.830 | 0.639 | 0.761 | 0.943 | LG 5 ZW | ||||
| Blue | 0.467 | 2.920 | 2.920 | 0.058 | 0.058 | 0.058 | ||||||||||
| 0.495 | 0.088 | 0.088 | 0.809 | 0.809 | 0.809 | ND | ||||||||||
|
|
|
|||||||||||||||
| Metriaclima ‘daktari’ | Psda005 | 3.600 | 7.540 | NI | 4.290 | 5.240 | NI | NI | 0.110 | 0.400 | NI | 7.640 | 10.00 | |||
| 0.058 | 0.006 | Het | 0.038 | 0.022 | Het | Homo | 0.740 | 0.527 | Homo | 0.006 | 0.002 | LG 7 XY | ||||
|
|
|
|||||||||||||||
| Metriaclima fainzilberi Lundo Is | MeFaLI-A002 | OB&BB | 8.730 | 8.730 | 13.000 | 0.004 | 0.476 | 0.476 | OB&BB | 2.440 | 1.070 | 0.027 | 0.069 | 0.004 | 0.726 | |
| 0.003 | 0.003 | <.0001 | 0.949 | 0.490 | 0.490 | 0.119 | 0.301 | 0.870 | 0.793 | 0.949 | 0.394 | LG 5 ZW | ||||
| BB | 0.600 | 0.600 | 0.240 | 0.600 | 0.600 | 0.240 | ||||||||||
| 0.439 | 0.439 | 0.642 | 0.439 | 0.439 | 0.624 | ND | ||||||||||
|
|
|
|||||||||||||||
| Metriaclima ‘kompakt’ | Mebl-A001 | OB&BB | 7.080 | 9.670 | 12.400 | 1.310 | 0.619 | 0.619 | OB&BB | 2.390 | 0.419 | 3.490 | NI | 5.490 | 5.490 | |
| 0.008 | 0.002 | <.0001 | 0.253 | 0.431 | 0.431 | 0.122 | 0.518 | 0.062 | Homo | 0.019 | 0.019 | LG 5 ZW | ||||
| BB | 0 | 1.320 | 1.930 | NI | 2.930 | 2.930 | ||||||||||
| 1 | 0.251 | 0.165 | Homo | 0.087 | 0.087 | |||||||||||
| Mebl-A003 | OB&BB | 9.110 | 19.000 | 20.800 | 0.041 | 1.100 | 1.030 | OB&BB | 2.680 | 5.070 | 4.110 | NI | 0.157 | 0.628 | ||
| 0.003 | <.0001 | <.0001 | 0.839 | 0.294 | 0.311 | 0.101 | 0.024 | 0.043 | Homo | 0.692 | 0.428 | LG 5 ZW | ||||
| BB | 2.570 | 2.570 | 2.340 | NI | 1.090 | 1.090 | ||||||||||
| 0.109 | 0.109 | 0.126 | Homo | 0.296 | 0.296 | |||||||||||
| Rmbl007 | OB&BB | 4.210 | 4.210 | ND | 0.392 | 0.189 | ND | OB&BB | NI | NI | 0.305 | 0.215 | 0.003 | 0.003 | ||
| 0.040 | 0.040 | 0.531 | 0.664 | Homo | Homo | 0.580 | 0.643 | 0.995 | 0.995 | LG 5 ZW | ||||||
| BB | NI | NI | 0.782 | 0.010 | 0.010 | 0.010 | ||||||||||
| Homo | Homo | 0.376 | 0.921 | 0.921 | 0.921 | |||||||||||
|
|
|
|||||||||||||||
| Metriaclima lombardoi | Melo-A015A | 0.627 | NI | 5.080 | 2.440 | 0.008 | 1.170 | 1.590 | 0.627 | NI | 2.24 | 3.610 | 3.610 | |||
| 0.429 | Homo | 0.024 | 0.118 | 0.928 | 0.279 | 0.207 | 0.429 | Homo | 0.135 | 0.057 | 0.057 | ND | ||||
| Melo-A019 | 0.646 | NI | 0.053 | 1.310 | 0.006 | 0.032 | NI | 0.087 | 0.016 | NI | 19.000 | 18.300 | ||||
| 0.421 | Homo | 0.819 | 0.253 | 0.937 | 0.858 | Het | 0.768 | 0.899 | Het | <.0001 | <.0001 | LG 7 XY | ||||
| Melo-A026 | 5.69 | NI | 1.300 | 0.556 | 0.012 | 0.556 | NI | 0.052 | 1.800 | NI | 28.00 | 24.20 | ||||
| 0.017 | Homo | 0.255 | 0.456 | 0.913 | 0.456 | Het | 0.820 | 0.180 | Het | <.0001 | <.0001 | LG 7 XY | ||||
|
|
|
|||||||||||||||
| Metriaclima mbenjii | Memb-A004 | 0.399 | 0.292 | 0.039 | 0.943 | 1.000 | 1.640 | 0.476 | 1.010 | 0.294 | 0.011 | NI | 0.063 | |||
| 0.527 | 0.589 | 0.844 | 0.331 | 0.316 | 0.200 | 0.490 | 0.315 | 0.588 | 0.918 | Homo | 0.802 | ND | ||||
|
|
|
|||||||||||||||
| Metriaclima phaeos | Meph-A001 | 0.098 | 0.794 | 0.794 | NI | NI | 0.653 | 0 | 2.330 | 2.330 | 8.130 | 9.740 | NI | |||
| 0.755 | 0.373 | 0.373 | Homo | Homo | 0.419 | 1 | 0.127 | 0.127 | 0.004 | 0.002 | NI | LG 7 XY | ||||
| Meph-A006 | 0.735 | 0.386 | 0.220 | 0.280 | 0.540 | 0.489 | 0.017 | 0.043 | 1.510 | 29.00 | 29.00 | 29.00 | ||||
| 0.391 | 0.534 | 0.639 | 0.597 | 0.462 | 0.484 | 0.897 | 0.837 | 0.219 | <.0001 | <.0001 | <.0001 | LG 7 XY | ||||
|
|
|
|||||||||||||||
| Metriaclima pyrsonotus | Mepy-A002 | 2.54 | 0.798 | 0.168 | 0.012 | 0.516 | 0.018 | 2.460 | 0.787 | 0.064 | NI | 0.018 | 0.000 | |||
| 0.111 | 0.372 | 0.682 | 0.912 | 0.473 | 0.893 | 0.116 | 0.375 | 0.800 | Homo | 0.893 | 1 | ND | ||||
| Mepy-A014 | 0.362 | 0.169 | 0.024 | 0.266 | 2.650 | 0.059 | 0.112 | 0.482 | 0.097 | 0.059 | 0.862 | 0.233 | ||||
| 0.547 | 0.681 | 0.877 | 0.606 | 0.103 | 0.809 | 0.738 | 0.488 | 0.755 | 0.809 | 0.353 | 0.629 | ND | ||||
| Mepy-A017 | 0.005 | NI | 0.019 | 4.920 | NI | 1.720 | 0.0996 | 0.202 | 0.202 | 4.540 | 3.440 | 5.940 | ||||
| 0.949 | Het | 0.891 | 0.026 | Het | 0.189 | 0.752 | 0.653 | 0.653 | 0.033 | 0.064 | 0.015 | LG 7 XY | ||||
| Rmpy007 | OB&BB | 5.000 | NI | 3.000 | 0.150 | 0.150 | 3.000 | OB&BB | NI | 0 | NI | NI | 5.000 | 5.000 | ||
| 0.025 | Homo | 0.083 | 0.699 | 0.699 | 0.083 | Het | 1 | Homo | Het | 0.025 | 0.025 | LG 5 ZW/LG7 XY | ||||
| BB | NI | 1.100 | NI | NI | 9.000 | 9.000 | ||||||||||
| Het | 0.294 | Homo | Het | 0.003 | 0.003 | |||||||||||
| Rmpy018 | OB&BB | NI | 14.000 | 10.800 | 0.938 | 0.277 | 0.016 | OB&BB | 0.087 | 0.297 | 0.520 | 3.670 | 6.750 | 6.750 | ||
| Homo | <.0001 | 0.001 | 0.333 | 0.599 | 0.899 | 0.769 | 0.586 | 0.471 | 0.053 | 0.009 | 0.009 | LG 5 ZW/LG7 XY | ||||
| BB | 0 | 0.034 | 0.171 | 0.643 | 8.780 | 8.780 | ||||||||||
| 1 | 0.853 | 0.679 | 0.011 | 0.003 | 0.003 | |||||||||||
| Rmpy021 | OB&BB | 9.960 | 11.300 | 10.900 | 0.277 | 0.966 | 1.980 | OB&BB | 0.289 | 0.091 | 1.370 | 0.424 | 0.618 | 1.260 | ||
| 0.002 | 0.001 | 0.001 | 0.599 | 0.326 | 0.160 | 0.591 | 0.764 | 0.242 | 0.515 | 0.432 | 0.262 | LG 5 ZW | ||||
| BB | 1.350 | 0.556 | 1.350 | 0.016 | 0.357 | 0.016 | ||||||||||
| 0.245 | 0.456 | 0.245 | 0.899 | 0.550 | 0.899 | |||||||||||
|
|
|
|||||||||||||||
| Metriaclima saulosi | Mesa-A009 | 0.001 | NI | 0.044 | 3.090 | NI | 0.309 | NI | 1.220 | NI | NI | 2.550 | NI | |||
| 0.972 | Homo | 0.834 | 0.079 | Homo | 0.578 | Het | 0.270 | Homo | Het | 0.110 | Homo | ND | ||||
|
|
|
|||||||||||||||
| Metriaclima zebra Mazinzi Reef | Meze-A03 | NI | 0.016 | 0.181 | 1.330 | NI | NI | ND | 0.465 | 0.046 | ND | 7.800 | NI | |||
| Homo | 0.899 | 0.671 | 0.249 | Homo | Homo | 0.495 | 0.829 | 0.005 | Homo | LG 7 XY | ||||||
| Meze-A009 | 0.005 | NI | 1.190 | NI | NI | NI | NI | 0.006 | 0.121 | NI | 1.190 | 2.610 | ||||
| 0.943 | Homo | 0.276 | Homo | Homo | Homo | Homo | 0.939 | 0.728 | Homo | 0.276 | 0.106 | ND | ||||
|
|
|
|||||||||||||||
| Metriaclima zebra ‘Nankoma blue’ | Mena-A006 | 0.050 | NI | NI | 3.250 | 6.420 | 4.540 | NI | 0.426 | NI | NI | 15.200 | NI | |||
| 0.823 | Homo | Homo | 0.071 | 0.011 | 0.033 | Homo | 0.514 | Het | Homo | <.0001 | Het | LG 7 XY | ||||
|
|
|
|||||||||||||||
| Pseudotropheus polit | Pspo-A003 | NI | NI | 0.004 | NI | 1.400 | NI | NI | NI | NI | NI | 8.91 | NI | |||
| Homo | Homo | 0.949 | Homo | 0.237 | Homo | Het | Homo | Homo | Het | 0.003 | Homo | LG 7 XY | ||||
Not Informative (NI)
Not Determined (ND)
PCR amplification of each marker was performed on DNA extracted from caudal fin clips. The size of the amplified product was determined on an Applied Biosystems 377 DNA sequencer using GeneScan 3.1.2 software. We routinely multiplexed the separation of several markers on a single gel. Statistical analyses were performed using Fisher’s exact test of independence.
RESULTS
Sex ratios of broods
A total of 2066 individuals in 93 broods representing 19 species in five genera were produced for analysis (Table 1). The average brood size at the time of sacrifice was 22.2 animals. Offspring were sacrificed at 4–26 months of age to determine gonadal sex. The sex of 70 offspring could not be unambiguously determined. Among 613 offspring phenotyped at 4 to 6 months of age, 44 (7.18%) could not be definitively sexed. Among 1431 offspring phenotyped at 7 or more months of age, only 26 (1.82%) could not be definitively sexed. Therefore, the optimal age for determining phenotypic sex appears to be older than six months.
Sex ratios (as fraction of males) were calculated for each family (Table 1). The average sex ratio across all broods was 0.38 male. Binomial tests identify many significant deviations from the expectation of a 1:1 male:female sex ratio. One family contained only male offspring, and seven families contained only female offspring. In many cases, highly-skewed sex ratios prevented genetic analysis for sex-linkage. Families tested for genetic associations are listed in the Supplemental Tables.
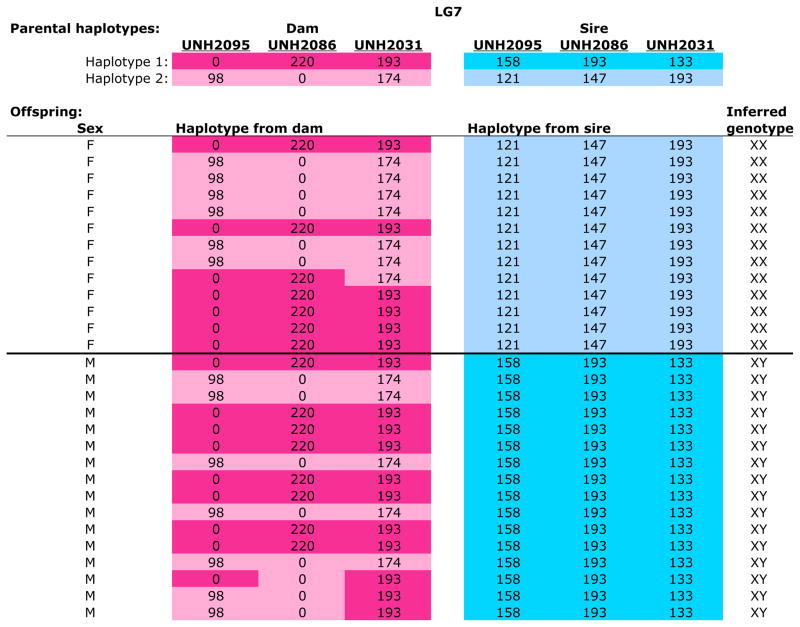
An XY system on LG7
We found strong evidence for a male heterogametic (XY) sex determination system in many of the families. Of the 44 families genotyped, 14 showed evidence for a pure XY system on LG7 (Table 2 and Figure 4). To take an example, we examine the data for a Metriaclima phaeos family (Figure 1). The 13 females all inherited the same LG7 haplotype from their father, while the 16 males all inherited the other paternal LG7 haplotype. The maternal haplotypes on LG7 were distributed equally to males and females. Both maternal and paternal haplotypes on LG5 were distributed equally to males and females. A second M. phaeos family also showed evidence of a male heterogametic sex-determining locus on LG7 (Table S12.1).
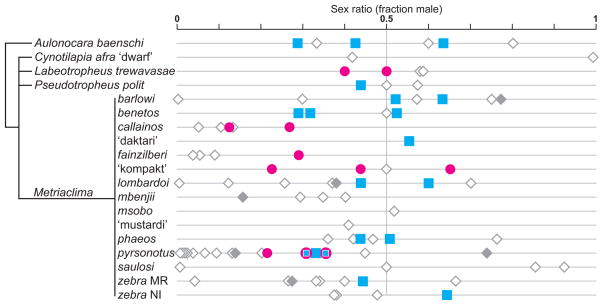
Figure 4.
Summary of the diverse sex determining systems found in Lake Malawi cichlids. Symbols indicate the sex ratio of each family studied. Filled symbols indicate families genotyped for ma rkers on LG5 and LG7. Blue squares indicate families with a significant association for an XY sex-determining locus on LG7. Pink circles indicate families with a significant association with a ZW sex-determining locus on LG5. The two families of M. pyrsonotus segregating both sex determining loci are indicated by the pink circle containing a small blue square. Grey diamonds indicate the family was genotyped, but no associations were found with either LG5 or LG7. Open diamonds indicate genotyping was not performed, or that statistical association was not possible due to highly skewed sex ratios. The taxonomic relationships of the species are shown in the tree on the left.
Figure 1.
Metriaclima phaeos family (Meph-A006) showing segregation of an XY sex-determining locus on LG7. The two haplotypes of the dam are shown in shades of pink, those of the sire in blue. Numbers indicate the length of the microsatellite allele for each locus and zero indicates a null allele. The inferred genotype at the sex-determining locus is shown in the last column.
We observed XY systems on LG7 in seven other species of Metriaclima (Table 2). In these species, segregation of alleles at this locus explains the sex of > 90% of the individuals. For example, in Metriclima benetos, 94% of the male progeny inherited a Y chromosome from their father (Table S4.1-2). In two of four M. lombardoi families a significant majority of males inherited the Y chromosome, while most females inherited the entire paternal X chromosome across the sex determination interval (Tables S10.3-4). The male heterogametic (XY) sex determination system was also found in the outgroups Aulonocara baenschi and Pseudotropheus polit (Tables S1.1, 17.1). All females in both of these species inherited the entire paternal X haplotype.
A ZW system on LG5
Four species of Metriaclima showed evidence of a female heterogametic (ZW) sex determination system on LG5 (Table 2 and Figure 4). This sex determination system is associated with a female color morph, orange blotch (OB), which is inherited as a dominant allele on the W chromosome. As an example, consider the family of Metriaclima fainzilberi ‘Lundo Island’ in Figure 2. This family has a female-biased sex ratio, but still shows strong associations between sex and maternally inherited markers on LG5. Markers UNH2139, c-Ski, and GM264a all showed significant associations with sex. All but one female expressed the OB phenotype and inherited the entire maternal W chromosome across the sex determination interval.
Figure 2.
Metriaclima fainzilberi family (MeFaLI-A002) showing segregation of a ZW sex-determining locus on LG5. The two haplotypes of the dam are shown in shades of pink, those of the sire in blue. Numbers indicate the length of the microsatellite allele for each locus. The dam had a color phenotype of orange-blotch (OB), while the male had the standard blue-black (BB) color, and OB segregates as a dominant Mendelian allele tightly linked to gender. The inferred genotype at the sex-determining locus is shown in the last column.
A female heterogametic sex determination system was identified also in the outgroup Labeotropheus trewavasae (Table S2.1-2) and families of several other Metriaclima species that segregate the OB color polymorphism. For example, in Metriaclima ‘kompakt‘, 13 of 18 females inherited a complete W haplotype, while most males inherited the Z allele on LG5 (Table S9.2). Metriaclima callainos segregates for white-blotch, an OB-like color polymorphism, which is also associated with the ZW system on LG5 (Tables S5.1, S5.2).
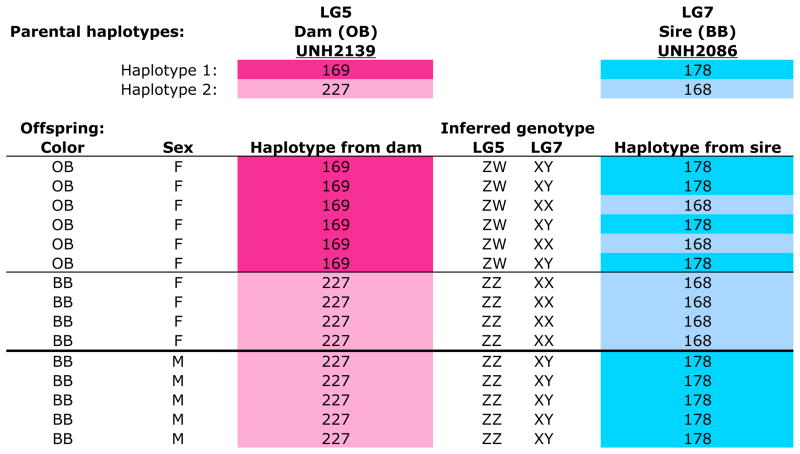
Epistasis
In M. pyrsonotus we found both of these sex determination systems segregating in the same family. Figure 3 shows the haplotype reconstruction for the offspring of a cross of an OB female to a normally colored blue-barred (BB) male M. pyrsonotus. Both parents are heterozygous at marker UNH2139 on LG5. The maternal alleles are 169/227, and the paternal alleles are 196/202. Six of 10 female offspring inherited the maternal 169bp allele and are OB in color. Four females, and all of the males, are BB in color and inherited the maternal 227bp allele. We infer that the 169bp allele marks an OB-associated W chromosome and that the 227bp allele labels a Z chromosome.
Figure 3.
Metriaclima pyrsonotus family (RMpy007) showing segregation of both a ZW sex determining locus on LG5 and an XY sex determining locus on LG7. The haplotypes of the dam on LG5 are shown in shades of pink. The haplotypes of the sire on LG7 are shown in blue. Numbers indicate the length of the microsatellite allele for each locus. The inferred genotypes at the two, epistatic, sex-determining loci segregating in this family are shown in the middle columns.
Further analysis controlling for the effects of the W locus on LG5 reveals that this family is also segregating the XY system on LG7. Here we ignore the progeny identified above as having a ZW genotype on LG5 and focus on the ZZ progeny. Both parents are heterozygous at marker UNH2086, with maternal alleles 136/206, and paternal alleles 178/168. All the ZZ females inherited a paternal X chromosome with a haplotype 143-144-168-193 at UNH973, UNH2095, UNH2086, and UNH2031. All of the males inherited a Y chromosome with a haplotype of 182-154-178-137 from their father.
Cross-tabulation of these results identifies strong epistatic interactions between these loci. Individuals that inherited the W chromosome on LG5 (the 169bp allele at marker UNH2139) are always female, regardless of the genotype for the XY locus on LG7. These females have a genotype of ZWXX or ZWXY and express the OB pigmentation phenotype. Individuals lacking the W chromosome on LG5, but carrying a Y chromosome on LG7 (marked by the 178bp allele at UNH2086), are invariably male. These males have a ZZXY genotype and the BB pigmentation phenotype. Finally, individuals with the ZZXX genotype are mostly female and are BB in color. Interestingly, while most individuals with the ZZXX genotype are female, ZZXX males have also been identified. Similar epistatic interactions were observed in a second M. pyrsonotus family (Table S13.11). Note that when both sex determining systems are segregating within a single family, the expected Mendelian sex-ratio becomes 1:3 (male:female).
Multifactorial sex determination
In several families, the association between phenotypic sex and the known genetic sex determining loci on LG 5 and LG 7 breaks down. Comparisons among eight families of M. pyrsonotus all sired by the same male are particularly illustrative of this point (Tables S13.5-S13.12). This male (#2005-5049) has a Y sex-determining locus on LG7 marked by the alleles 154-178-137 for markers UNH2095-UNH2086-UNH2031 (Table S13.6). This haplotype also behaves as a Y in two other broods from this male (Tables S13.7, S13.11) in which the epistatically dominant LG5 W is segregating.
In the other families sired by the same male, the effect of the LG7 Y is not as clear. In these families, male progeny generally carry the sire’s Y haplotype. But the offspring in three of these families were all female, despite the segregation of the sire’s Y haplotype (Tables S13.8–S13.10). Two other families (Tables S13.5-6) are highly female biased and show a pattern of inheritance similar to those segregating the LG5 ZW system. However, in these two families the epistasis is not attributable to segregation of the LG5 ZW system, suggesting the presence of still another dominant ZW sex determination locus elsewhere in the genome. The last female-biased family (Table S13.12) shows a clear segregation of the LG5 ZW system epistatic to the LG7 XY locus. But it also includes some females that carry the Y haplotype but not the W.
In three families of M. ‘kompakt’ (Tables S9.1-3) a portion of the phenotypic males have a ZW genotype at LG5 (OB males). The idea that some M. ‘kompakt’ males are genetically female at the LG5 locus, is confirmed by a family in which both parents are heterozygous for the LG5 ZW (OB) genotype (Table S9.3). Both parents of this family have a LG5 ZW (OB) genotype. Some of the female offspring are homozygous for the W sex determining locus, demonstrating viability of this genotype. All offspring that inherit a W allele are OB, whether they inherited the W from the male (marked by a 206 allele at UNH2139) or the female (213 allele at UNH2139). Most of these W-carrying offspring are female, regardless of whether they inherited the W from the sire or the dam. However, three OB individuals are male, either due to environmental effects or an additional XY locus not linked to the XY system on LG7.
Sex-reversed individuals create opportunities to produce individuals homozygous for dominant sex determination alleles, such as the LG5 WW individuals we identified in M. ‘kompakt’ (Table S9.3). A parent homozygous for a dominant sex determination allele would produce offspring of only one sex, a possible explanation for the single-sex families in our study (Tables S3.2, S13.3, S13.4, S13.8-13.10). Additionally, the presence of multiple independent sex determining loci could skew sex ratios within individual families. For example, in M. pyrsonotus we identify a ZW system, and infer another ZW system, both dominant to the LG7 XY system. If a female is heterozygous for both female sex determining loci, we would predict a sex ratio approaching 1:9 (male:female). The sex ratios of many of our individual families in that species fall near that prediction (Table 1, Figure 4). Highly skewed sex ratios in some families make it difficult to detect genetic linkage. A family of M. saulosi family was very male-biased (38M:3F) and did not show evidence for linkage of sex with markers on either LG5 or LG7 (Table S14.1). Three of four families of M. fainzilberi were strongly female-biased and show no linkage with either LG5 or LG7 (Tables S7.2, S8.1-2). We suspect that some of these families are homozygous for LG5 ZZ and LG7 XX genotypes, and/or that one or more loci on another chromosome are contributing to determining sex.
In a few cases, the strongly skewed sex ratios are associated with unequal transmission of alternative alleles from the female parent. Segregation distortion of maternal alleles on LG7 was observed in M. barlowi (Table S3.4), M. fainzilberi (Table S7.1), M. lombardoi (Table S10.1), and M. pyrsonotus (Tables S13.2 and 13.10). The only male-biased family of M. pyrsonotus shows strong segregation distortion for one of the female haplotypes on LG5 (Table S13.1). No similar distortions of paternal segregation were observed.
Finally, we have evidence that suggests species differ in whether males or females are the heterogametic sex on LG5. Two families showed evidence of an XY, rather than a ZW, system on LG5. A family of M. zebra ‘Nankoma’ is segregating for XY systems on both LG7 and LG5 (Table S16.1). A Y allele from either locus appears sufficient to determine a male fate. Metriaclima ‘daktari’ also appears to be segregating XY systems on both LG5 and LG7 (Table S6.1), although the result is not statistically significant in this small family. In these cases it is difficult to definitively attribute XY or ZW systems to each locus, perhaps because the phenotypic sex of a parent does not match their genetic sex for a particular locus. Analyses of additional families from these species are needed to establish a clear pattern.
DISCUSSION
Genetic sex determination
This survey identified at least two distinct genetic sex determination systems in the Lake Malawi cichlid flock: a male heterogametic (XY) sex determination system on linkage group 7, and a female heterogametic (ZW) sex determination system on linkage group 5. The XY system is widespread among the species we surveyed. Many Metriaclima species, as well as the outgroup species Aulonocara baenschi and Pseudotropheus polit, show XY sex determination linked to markers on LG7. The ZW sex determination system on LG5 has thus far only been found in Metriaclima and Labeotropheus species that exhibit orange- or white-blotch female phenotypes.
In M. pyrsonotus the two sex determining systems segregate within single families. When both dominant sex determination loci (W and Y) are present in a single individual, the ZW system is epistatically dominant to the XY system. Some individuals in these families inherit neither W nor Y. These ZZXX individuals are usually female but can also differentiate as males, possibly due to the segregation of additional sex determining loci.
Although it is not yet possible to definitively determine which sex determining system is ancestral, evidence points towards the LG7 XY system. In our sample, more species have a LG7 XY system than a LG5 ZW system (10 vs. 5 species) and two of three outgroup species (Aulonocara baenschi, Pseudotropheus polit) are LG7 XY. In the third outgroup species, Labeotropheus trewavasae, we were only able to test families from an OB dam segregating the LG5 ZW system. As non-OB females are common in this species in the wild, it is likely that the LG5 ZW locus is not the sole sex determination system in the species. Another species in the genus, L. fuelleborni, appears to segregate the LG7 XY system (Albertson, 2002). On this basis, we postulate that the XY system on LG7 is the ancestral system of sex determination.
Evidence from several species strongly suggests the presence of additional sex determination loci and cryptic epistatic interactions. Several families showed no linkage of sex to markers on either LG5 or LG7, suggesting that sex determination is controlled by genes on other chromosomes. Known sex determination loci are differentially penetrant across families. Multiple families from single sires produce different genotype-phenotype associations with different dams. Inheritance patterns across these families are strongly indicative of unknown, epistatically dominant sex determination loci segregating in the population in a Mendelian manner. Numerous families with highly skewed sex ratios also support the existence of epistatic interactions between additional sex determining loci segregating in these species. Our sampling of species and genera was focused on the genus Metriaclima, and we have limited information on sex determination loci segregating in other genera. We expect to find evidence for additional sex determining loci as we broaden our sampling to related genera.
These interactions among known and unknown sex determining loci complicate the genetic analysis of sex determination. The presence of a sex determination locus on LG5 or LG7 in a family may be masked if another locus is overriding its effects in a subset of the progeny. Similarly, the phenotypic sex of an individual may be determined by one locus, but it may also be segregating another sex determination locus. For example, in M. pyrsonotus we regularly identify females with a LG5 ZW, LG7 XY genotype. These females will contribute the LG7 Y to a portion of their progeny, determining male phenotypic sex. Finally, if both the dam and the sire are segregating the same sex determination allele, it confounds the identification of the association and direction of sex determination.
Although we believe that sex determination in Malawi cichlids is predominantly genetic, there remains room to hypothesize a role for environmental factors. These might be suspected to account for the highly skewed sex ratios in some families. However, the temperatures at which we raised the fish were not close to the temperatures known to affect sex determination in tilapiine cichlids (Bezault et al. 2007; Baroiller et al. 2009a). The variable sex of ZZXX individuals in some families might also be interpreted as evidence of environmental influence. While we do not deny the extensive evidence for environmental effects on sex determination in fishes (Baroiller et al. 2009b), we expect that further mapping efforts will identify additional genetic loci contributing to sex determination in these species.
Evolution of sex determination
We have now identified at least four distinct sex chromosomes in cichlid fishes. Some species of tilapiine cichlids have an XY system on linkage group 1, while others have a ZW system on linkage group 3 (Cnaani et al. 2008). Some families of the blue tilapia have been found segregating both loci, and in these cases the W chromosome trumps the Y (ZWXY individuals are female) (Lee et al. 2004). Extensive comparative mapping, using common marker sets, has demonstrated that the genetic maps for tilapia cichlids and Malawi cichlids are almost perfectly colinear. Therefore the sex determining loci we have mapped in Lake Malawi cichlids (the ZW system on linkage group 5 and the XY system on linkage group 7) are clearly distinct from the loci identified in tilapia. Although this pattern might have been created by the movement of sex determining genes among chromosomes (e.g. associated with transposons), we think it more likely that different genes have been recruited on each chromosome for a new role in sex determination. The haplochromine cichlids of East Africa diverged from the tilapiine cichlid lineage only 10 – 20 million years ago (Genner et al. 2007). The presence of at least four sex determining loci, and the relatively undifferentiated state of the sex chromosomes, make this group of fishes an excellent model for studying the early steps in the evolution of sex chromosomes (Charlesworth 1991), the mechanisms by which sex determination pathways evolve (Wilkins 1995), and the role of sex chromosomes in speciation (Presgraves 2008). Our data adds to the growing body of evidence that the mechanisms of sex determination are highly labile in many fish lineages. For example, in the medaka (Oryzias latipes) a recent duplication of dmrt1 has created a Y chromosome from an ancestral autosome (LG1) (Matsuda et al. 2002, Kondo et al. 2004). XY sex reversed females are observed as the result of structural and regulatory mutations of the dmrt1bY gene (Otake et al. 2006; 2008). There is also evidence for autosomal modifiers that create XX males (Nanda et al. 2003). Closely related species of Oryzias have XY sex determining loci on LG2, LG8, LG10 and LG12 (Nagai et al. 2008). Oryzias hubbsi has a ZW sex determining system on LG5 which has evolved since its divergence from O. dancena (Takehana 2007). Rapid evolution of sex determination systems has been observed also in poeciliids (Volff and Schartl 2001), salmonids (Woram et al. 2003), sticklebacks (Ross et al. 2009) and tilapias (Cnaani et al. 2008). Sex determination in the frog, Rana rugosa, is ancestrally an XY system, but ZW systems have evolved twice on the islands of Japan (Ogata et al. 2008). Vertebrate sex determination is more labile than it would appear from studies of avian and mammalian lineages.
The evolution of sex determining mechanisms is fundamentally constrained by natural selection on sex ratios (Fisher 1958). Multiple sex determination loci can coexist in a population if their fitness is equal. The different monogenic systems are then connected by paths of equilibria along lines of equal sex ratio (Bull 1983). In natural populations however, the fitness of these genotypes probably are not equal, and the polymorphisms are unlikely to persist (Rice 1986). The consensus of theoretical work is that multifactorial systems for sex determination are usually transient (Bull and Charnov 1977; Karlin and Lessard 1986) and will rapidly evolve toward a monogenic system with an equal sex ratio. Still, multifactorial sex determination has been described in several systems, including Xiphophorus (Kallman 1965) and Musca (Kozielska et al. 2006). Our results demonstrate that via dominant epistasis, one sex-determining locus can mask other underlying sex determination systems. Sex determination hierarchies like these are probably not unique to cichlid fishes. Reexamination of species in which sex determining loci already have been identified may reveal the segregation of additional genetic sex determination systems.
The existence of these complex systems of sex determination also requires that we reconsider the population genetics of sex determination. Most of the existing theoretical treatments consider the evolution of multiple sex determining alleles at a single locus (Orzack et al. 1980; Lande et al. 2001; Vuilleumier et al. 2007). Clearly, models in which multiple unlinked loci interact to determine sex are more relevant to the situation in Lake Malawi cichlids (Bull and Charnov 1977; van Doorn and Kirkpatrick 2007).
Sexually antagonistic selection
What selective forces can account for the rapid evolution of new sex determining loci in these fishes? A general theory is that new sex determination loci are fixed in response to sexually antagonistic selection (Rice 1986). Alleles that increase the fitness of one sex, but decrease the fitness of the opposite sex, create a genetic conflict which can be resolved by linkage to a nearby sex determining locus (van Doorn and Kirkpatrick 2007). If linkage disequilibrium between the sex determining locus and the sexually antagonistic allele can be maintained, the deleterious effects of sexually antagonistic selection will be reduced.
The ZW system on LG5 may have evolved more recently as a result of sexually antagonistic selection for the OB color pattern (Roberts et al. 2009). Orange-blotch provides advantages of crypsis in females, but reduces the mating success of males because it disrupts species-specific color patterns important in mate recognition. The OB allele is in strong linkage disequilibrium with the W sex determination locus on LG5, providing an effective mechanism to reduce the deleterious effects of sexually antagonistic selection on this color pattern. This is analogous to the situation in guppies, where genes for male ornamental coloration are primarily found on the Y chromosome (Lindholm and Breden 2002; Tripathi et al. 2009).
Post-zygotic isolation
The diversity of sex determining loci, and the inferred rapid evolution of gene regulatory networks underlying sexual differentiation, may promote speciation by contributing to post-zygotic reproductive isolation. Hybrids between species with different sex determining systems may produce intersex individuals with reduced viability or fertility, directly contributing to post-zygotic isolation. Anecdotal results in our lab suggest that some hybrid crosses do have reduced viability, although this has not yet been traced to the incompatibilities among the sex determining loci. Now that we have information on the mode of sex determination in each species it will be important to systematically examine the viability and fertility of hybrid crosses. Reduced fitness of some hybrid genotypes might in turn create selection for reinforcement of pre-mating isolating mechanisms (Lemmon and Kirkpatrick 2006).
An evolving model of speciation
The unexpected diversity of sex determining mechanisms, coupled with several other recent observations, lead us to propose a new model for speciation of the colorful cichlids of Lake Malawi. We begin with the observation that male-male competition for breeding territories is intense. Males defend permanent territories of a few square meters, and direct their strongest aggression toward other males. While conspecific territories do not overlap, the territories of congeners with different color patterns frequently do show significant overlap. Mutations which significantly alter the color pattern reduce the aggression a male receives from other males, and give him an enormous advantage in acquiring and/or maintaining a breeding territory (Seehausen and Schluter 2004; Pauers et al. 2008).
Given the strong role that color patterns play in mate recognition and female preference, one might expect this mutant male to suffer reduced opportunities for mating. However female mate preferences may not be hard-wired. Recent work suggests that fry imprint on olfactory cues while being brooded in the mother’s mouth for the first weeks of life (Verzijden and ten Cate 2007). The implication is that as females mature, they may use this imprint to learn the color patterns of those males emitting the appropriate olfactory cues. The females could then use the color cues to recognize males at a distance, as shown in many laboratory experiments (Couldridge and Alexander 2002; Jordan et al. 2003; Kidd et al. 2006). Males with new color mutations are thus able to breed with an existing population of females.
We next postulate that the male color mutation has negative fitness consequences when expressed in females. Bright color elements that improve male mating success might disrupt the cryptic patterns of females, setting up a pattern of sexually antagonistic selection on the pigmentation locus. There is as yet little data on the penetrance of male color mutations in females, but anecdotal evidence from our lab suggests that hybrid females do show limited expression of male color elements. One mechanism by which sexually antagonistic selection on the color pattern might be resolved is through the recruitment of new sex-determining loci. Selection might favor the recruitment of a new Y allele tightly linked to the male color mutation, that would restrict expression of the color to males. Alternatively, a mutation that suppressed the color element in females might become linked to a novel W allele in females. The result is a population segregating multiple sex determiners in linkage disequilibrium with genes controlling the color patterns. This would sex the stage for reinforcement of pre-mating isolating mechanisms.
While this model is speculative, it is consistent with published data, with the exceptionally strong forces of sexual selection acting on the rock-dwelling ‘mbuna’ cichlids, and with the breeding biology of these fishes in the wild. It suggests a variety of experiments to confirm the olfactory imprinting hypothesis, study the penetrance of novel male color mutations in females, and evaluate the linkage of color pattern genes with sex-determining loci.
In conclusion, we have uncovered a surprising diversity of sex determining systems among Lake Malawi cichlids. This diversity has evolved recently, since the radiation of this species flock occurred over the last million years. Our results suggest that attention should focus understanding the role of genetic conflicts in the evolution of new sex determination loci in this lineage. It will be interesting to learn how the evolution of sex determination may have contributed to the remarkable radiation of Lake Malawi cichlids.
Supplementary Material
Acknowledgments
We dedicate this paper to the memory of Stuart Grant (1937–2007), who facilitated so much scientific work on Lake Malawi cichlids. We work in Lake Malawi under permits from the Malawi Department of Parks and Wildlife, with the collaboration of the Department of Biology at Chancellor College, University of Malawi and with the cooperation of the Malawi Department of Fisheries. This material is based upon work supported by the National Science Foundation (DEB-0445212) and the National Institutes of Health (R01HD058635).
LITERATURE CITED
- Albertson RC. PhD thesis. University of New Hampshire; Durham. New Hampshire, USA: 2002. Genetic basis of adaptive radiation in East African cichlid fishes. [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD. Directional selection has shaped the oral jaws of Lake Malawi cichlid fishes. Proc Natl Acad Sci USA. 2003;100:5252–7. doi: 10.1073/pnas.0930235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc Natl Acad Sci USA. 2005;102:16287–92. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allender CJ, Seehausen O, Knight ME, Turner GF, Maclean N. Divergent selection during speciation of Lake Malawi cichlid fishes inferred from parallel radiations in nuptial coloration. Proc Natl Acad Sci USA. 2003;100:14074–9. doi: 10.1073/pnas.2332665100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnegard ME, Markert JA, Danley PD, Stauffer JR, Ambali AJ, Kocher TD. Population structure and colour variation of the cichlid fishes Labeotropheus fuelleborni Ahl along a recently formed archipelago of rocky habitat patches in southern Lake Malawi. Proc Biol Sci. 1999;266:119–130. [Google Scholar]
- Baroiller JF, D’Cotta H, Bezault E, Wessels S, Hoerstgen-Schwark G. Tilapia sex determination: Where temperature and genetics meet. Comp Biochem Physiol A Mol Integr Physiol. 2009a;153:30–8. doi: 10.1016/j.cbpa.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Baroiller JF, D’Cotta H, Saillant E. Environmental effects on fish sex determination and differentiation. Sex Dev. 2009b;3:118–135. doi: 10.1159/000223077. [DOI] [PubMed] [Google Scholar]
- Bezault E, Clota F, Derivaz M, Chevassus B, Baroiller JF. Sex determination and temperature-induced sex differentiation in three natural populations of Nile tilapia (Oreochromis niloticus) adapted to extreme temperature conditions. Aquaculture. 2007;272(S1):S3–S16. [Google Scholar]
- Bull JJ. Evolution of Sex Determining Mechanisms. Benjamin Cummings Publishing; Menlo Park, California: 1983. [Google Scholar]
- Bull JJ, Charnov EL. Changes in the heterogametic mechanism of sex determination. Heredity. 1977;39:1–14. doi: 10.1038/hdy.1977.38. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–3. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Chapman T. Evolutionary conflicts of interest between males and females. Current Biology. 2006;16:R744–R754. doi: 10.1016/j.cub.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Cnaani A, Lee BY, Zilberman N, Ozouf-Costaz C, Hulata G, Ron M, D’Hont A, Baroiller JF, D’Cotta H, Penman DJ, Tomasino E, Coutanceau JP, Pepey E, Shirak A, Kocher TD. Genetics of sex determination in tilapiine species. Sex Dev. 2008;2:43–54. doi: 10.1159/000117718. [DOI] [PubMed] [Google Scholar]
- Couldridge VCK, Alexander GJ. Colour patterns and species recognition in four closely related species of Lake Malawi cichlid. Behav Ecol. 2002;13:59–64. [Google Scholar]
- Danley PD, Markert JA, Arnegard ME, Kocher TD. Divergence with gene flow in the rock-dwelling cichlids of Lake Malawi. Evolution. 2000;54:1725–37. doi: 10.1111/j.0014-3820.2000.tb00716.x. [DOI] [PubMed] [Google Scholar]
- Danley PD, Kocher TD. Speciation in rapidly diverging systems: lessons from Lake Malawi. Mol Ecol. 2001;10:1075–86. doi: 10.1046/j.1365-294x.2001.01283.x. [DOI] [PubMed] [Google Scholar]
- Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influence. Aquaculture. 2002;208:191–364. [Google Scholar]
- Fisher RA. The Genetical Theory of Natural Selection. Dover Publications; NY: 1958. [Google Scholar]
- Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, Turner GF. Age of cichlids: new dates for ancient lake fish radiations. Mol Biol Evol. 2007;24:1269–82. doi: 10.1093/molbev/msm050. [DOI] [PubMed] [Google Scholar]
- Guerrero RD, Shelton WL. An aceto-carmine squash method for sexing juvenile fishes. Prog Fish Cult. 1974;36:55–56. [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet. 2001;29:389–95. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- Jordan R, Kellogg K, Juanes F, Stauffer JR., Jr Evaluation of female mate choice cues in a group of Lake Malawi mbuna (Cichlidae) Copeia. 2003;2003:181–186. [Google Scholar]
- Kallman KD. Genetics and geography of sex determination in the poeciliid fish, Xiphophorus maculatus. Zoologica. 1965;50:151–190. [Google Scholar]
- Karlin S, Lessard S. Sex Ratio Evolution. Princeton University Press; Princeton NJ: 1986. [Google Scholar]
- Kidd MR, Danley PD, Kocher TD. A direct assay of female choice in cichlids: all the eggs in one basket. J Fish Biol. 2006;68:373–384. [Google Scholar]
- Kocher TD, Conroy JA, McKaye KR, Stauffer JR, Lockwood SF. Evolution of NADH dehydrogenase subunit 2 in East African cichlid fish. Mol Phylogenet Evol. 1995;4:420–32. doi: 10.1006/mpev.1995.1039. [DOI] [PubMed] [Google Scholar]
- Kocher TD. Adaptive evolution and explosive speciation: the cichlid fish model. Nat Rev Genet. 2004;5:288–98. doi: 10.1038/nrg1316. [DOI] [PubMed] [Google Scholar]
- Kondo M, Nanda I, Hornung U, Schmid M, Schartl M. Evolutionary origin of the medaka Y chromosome. Curr Biol. 2004;14:1664–9. doi: 10.1016/j.cub.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Kornfield IL. Descriptive genetics of cichlid fishes. In: Turner BJ, editor. Evolutionary Genetics of Fishes. Plenum Publishing Corporation; New York: 1984. pp. 591–616. [Google Scholar]
- Kozielska M, Pen I, Beukeboom LW, Weissing FJ. Sex ratio selection and multifactorial sex determination in the housefly: a dynamic model. J Evol Biol. 2006;19:879–888. doi: 10.1111/j.1420-9101.2005.01040.x. [DOI] [PubMed] [Google Scholar]
- Lande R. Models of speciation by sexual selection on polygenic traits. Proc Natl Acad Sci USA. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Seehausen O, van Alphen JJ. Mechanisms of rapid sympatric speciation by sex reversal and sexual selection in cichlid fish. Genetica. 2001;112–113:435–43. [PubMed] [Google Scholar]
- Lemmon AR, Kirkpatrick M. Reinforcement and the genetics of hybrid incompatibilities. Genetis. 2006;173:1145–1155. doi: 10.1534/genetics.105.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm A, Breden F. Sex chromosomes and sexual selection in poeciliid fishes. Am Nat. 2002;160:S214–S224. doi: 10.1086/342898. [DOI] [PubMed] [Google Scholar]
- Lee B-Y, Hulata G, Kocher TD. Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus) Heredity. 2004;92:543–549. doi: 10.1038/sj.hdy.6800453. [DOI] [PubMed] [Google Scholar]
- Lee BY, Lee WJ, Streelman JT, Carleton KL, Howe AE, Hulata G, Slettan A, Stern JE, Terai Y, Kocher TD. A second-generation genetic linkage map of tilapia (Oreochromis spp.) Genetics. 2005;170:237–44. doi: 10.1534/genetics.104.035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, Hori H, Hamaguchi S, Sakaizumi M. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–63. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal Species and Evolution. Harvard University Press; Cambridge USA: 1963. [Google Scholar]
- Meyer A, Kocher TD, Basasibwaki P, Wilson AC. Monophyletic origin of Lake Victoria cichlid fishes suggested by mitochondrial DNA sequences. Nature. 1990;347:550–3. doi: 10.1038/347550a0. [DOI] [PubMed] [Google Scholar]
- Nagai T, Takehana Y, Hamaguchi S, Sakaizumi M. Identification of the sex-determining locus in the Thai medaka, Oryzias minutillus. Cytogenet Genome Res. 2008;121:137–42. doi: 10.1159/000125839. [DOI] [PubMed] [Google Scholar]
- Nanda I, Hornung U, Kondo M, Schmid M, Schartl M. Common spontaneous sex-reversed XX males of the medaka Oryzias latipes. Genetics. 2003;163:245–51. doi: 10.1093/genetics/163.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hasegawa Y, Ohtani H, Mineyama M, Miura I. The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog, Rana rugosa. Heredity. 2008;100:92–99. doi: 10.1038/sj.hdy.6801068. [DOI] [PubMed] [Google Scholar]
- Orzack SH, Sohn JS, Kallman KD, Levin SA, Johnston R. Maintenance of the three sex chromosome polymorphism in the platyfish, Xiphophorus maculatus. Evolution. 1980;34:663–672. doi: 10.1111/j.1558-5646.1980.tb04005.x. [DOI] [PubMed] [Google Scholar]
- Otake H, Shinomiya A, Matsuda M, Hamaguchi S, Sakaizumi M. Wild-derived XY sex-reversal mutants in the medaka, Oryzias latipes. Genetics. 2006;173:2083–90. doi: 10.1534/genetics.106.058941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otake H, Hayashi Y, Hamaguchi S, Sakaizumi M. The Y chromosome that lost the male-determining function behaves as an X chromosome in the medaka fish, Oryzias latipes. Genetics. 2008;179:2157–62. doi: 10.1534/genetics.108.090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauers MJ, Kapfer JM, Fendos CE, Berg CS. Aggressive biases towards similarly coloured males in Lake Malawi cichlid fishes. Biol Lett. 2008;4:156–9. doi: 10.1098/rsbl.2007.0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–43. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. On the instability of polygenic sex determination: the effect of sex-specific selection. Evolution. 1986;40:633–639. doi: 10.1111/j.1558-5646.1986.tb00514.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sexually antagonistic genes – experimental evidence. Science. 1992;256:1436–1439. doi: 10.1126/science.1604317. [DOI] [PubMed] [Google Scholar]
- Rice WR. Male fitness increases when females are eliminated from gene pool: implications for the Y chromosome. Proc Natl Acad Sci USA. 1998;95:6217–21. doi: 10.1073/pnas.95.11.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico C, Turner GF. Extreme microallopatric divergence in a cichlid species from Lake Malawi. Mol Ecol. 2002;11:1585–90. doi: 10.1046/j.1365-294x.2002.01537.x. [DOI] [PubMed] [Google Scholar]
- Ritchie MG. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 2007;38:79–102. [Google Scholar]
- Roberts RB, Kocher TD. Genetic basis of a sexually dimorphic color pattern in cichlid fishes. 2009. (submitted) [Google Scholar]
- Römer U, Beisenherz W. Environmental determination of sex in Apistogramma (Cichlidae) and two other fresh-water fishes (Teleostei) Journal of Fish Biology. 1996;48:714–725. [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genetics. 2009:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation. Oxford University Press; Oxford, UK: 2000. [Google Scholar]
- Seehausen O, Schluter D. Male-male competition and nuptial-colour displacement as a diversifying force in Lake Victoria cichlid fishes. Proc Biol Sci. 2004;271:1345–53. doi: 10.1098/rspb.2004.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O, Terai Y, Magalhaes IS, Carleton KL, Mrosso HDJ, Miyagi R, van der Sluijs I, Schneider MV, Maan ME, Tachida H, Imai H, Okada N. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- Smith PF, Kornfield I. Phylogeography of Lake Malawi cichlids of the genus Pseudotropheus: significance of allopatric colour variation. Proc Biol Sci. 2002;269:2495–502. doi: 10.1098/rspb.2002.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streelman JT, Albertson RC, Kocher TD. Genome mapping of the orange blotch colour pattern in cichlid fishes. Mol Ecol. 2003;12:2465–71. doi: 10.1046/j.1365-294x.2003.01920.x. [DOI] [PubMed] [Google Scholar]
- Takehana Y, Naruse K, Hamaguchi S, Sakaizumi M. Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma. 2007;116:463–470. doi: 10.1007/s00412-007-0110-z. [DOI] [PubMed] [Google Scholar]
- Tripathi N, Hoffmann M, Weigel D, Dreyer C. Linkage analysis reveals the independent origin of poeciliid sex chromosomes and a case of atypical sex inheritance in the guppy (Poecilia reticulata) Genetics. 2009;182:365–374. doi: 10.1534/genetics.108.098541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner GF, Seehausen O, Knight ME, Allender CJ, Robinson RL. How many species of cichlid fishes are there in African lakes? Mol Ecol. 2001;10:793–806. doi: 10.1046/j.1365-294x.2001.01200.x. [DOI] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909–912. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- van Oppen MJH, Turner GF, Rico C, Deutsch JC, Ibrahim KM, Robinson RL, Hewitt GM. Unusually fine-scale genetic structuring found in rapidly speciating Malawi cichlid fishes. Proc Biol Soc. 1997;264:1803–1812. [Google Scholar]
- Verheyen E, Salzburger W, Snoeks J, Meyer A. Origin of the superflock of cichlid fishes from Lake Victoria, East Africa. Science. 2003;300:325–9. doi: 10.1126/science.1080699. [DOI] [PubMed] [Google Scholar]
- Verzijden MN, ten Cate C. Early learning influences species assortative mating preferences in Lake Victoria cichlid fish. Biol Lett. 2007;3:134–6. doi: 10.1098/rsbl.2006.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Schartl M. Variability of genetic sex determination in poeciliid fishes. Genetica. 2001;111:101–10. doi: 10.1023/a:1013795415808. [DOI] [PubMed] [Google Scholar]
- Vuilleumier S, Lande R, van Alphen JJM, Seehausen O. Invasion and fixation of sex-reversal genes. J Evol Biol. 2007;20:913–920. doi: 10.1111/j.1420-9101.2007.01311.x. [DOI] [PubMed] [Google Scholar]
- Wilkins AS. Moving up the hierarchy: a hypothesis on the evolution of a genetic sex determination pathway. Bioessays. 1995;17:71–7. doi: 10.1002/bies.950170113. [DOI] [PubMed] [Google Scholar]
- Woram RA, Gharbi K, Sakamoto T, Hoyheim B, Holm LE, Naish K, McGowan C, Ferguson MM, Phillips RB, Stein J, Guyomard R, Cairney M, Taggart JB, Powell R, Davidson W, Danzmann RG. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 2003;13:272–80. doi: 10.1101/gr.578503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.