Summary
Hyper-activation of the PI 3-Kinase/AKT pathway is a driving force of many cancers. Here we identify the AKT-inactivating phosphatase PHLPP1 as a prostate tumor suppressor. We show that Phlpp1-loss causes neoplasia and, upon partial Pten-loss, carcinoma in mouse prostate. This genetic setting initially triggers a growth suppressive response via p53 and the Phlpp2 ortholog, and reveals spontaneous Trp53 inactivation as a condition for full-blown disease. Surprisingly, the co-deletion of PTEN and PHLPP1 in patient samples is highly restricted to metastatic disease and tightly correlated to deletion of TP53 and PHLPP2. These data establish a conceptual framework for progression of PTEN-mutant prostate cancer to life-threatening disease.
INTRODUCTION
With an annual average of 200,000 cases in the United States (US) alone, cancer of the prostate (CaP) is the most commonly diagnosed malignancy in western men and the second most common cause of US male cancer deaths (ACS, 2007–2009). Due to effective CaP screening programs, an increasing number of men is diagnosed and treated for clinically localized CaP even if the majority will not progress to life-threatening disease. Therefore, identification of men who will suffer disease recurrence constitutes the great challenge for CaP therapy (Shariat et al., 2008).
Recent advances in whole genome analysis are affording us with a look at the bulk of alterations that occur in cancer tissue and in commonly used cancer cell lines. In prostate, several such whole genome efforts have identified and validated commonly observed events, such as PTEN deletions, ERG fusion genes and chromosome 8 aberrations (Lapointe et al., 2007; Saramaki and Visakorpi, 2007). Furthermore, comparative copy number analysis of metastatic CaP has demonstrated how this technology can be used to trace the process of disease dissemination (Liu et al., 2009). Most recently, the integration of gene copy number, gene mutation and transcriptome analysis has provided a comprehensive look at the nature of the changes that separate indolent from aggressive disease, since the extent of copy number alteration could be linked to a patient’s risk of disease recurrence after prostatectomy (Taylor et al., 2010). Collectively, these studies demonstrate that advances in comprehensive cancer analysis could soon afford us with the catalog of changes associated with lethal disease progression in a patient – a prerequisite for the goal of effective patient-based target therapy (Schreiber et al., 2010). However, extraction of the relevant alterations that constitute a driving force for disease still poses a major challenge (Chin et al., 2011). Aberrant PI 3-Kinase pathway signaling is common in CaP and its specific targeting holds great therapeutic potential (Majumder and Sellers, 2005; Taylor et al., 2010; Wong et al., 2010). Therefore, it is paramount to understand which pathway players, together or alone, can be regarded as sentinels of pathway deregulation when they are found to be mutated.
Modeling the relevance of disease associated genes in Genetically Engineered Mice (GEMs) has proven to be the gold standard for establishing causality in cancer (Frese and Tuveson, 2007). Research using Pten-mutant GEM models of CaP has revealed that genetic context dictates disease outcome through both extent and cellular distribution of Akt activity (Carver et al., 2009b; Di Cristofano et al., 2001; Majumder et al., 2003; Trotman et al., 2006; Trotman et al., 2003). Thus, these studies highlighted the role of gene alteration events that cooperate to enhance AKT/PI 3-Kinase signaling in prostate cancer. PTEN prevents activation of AKT by dephosphorylating the membrane phospholipid PIP3 (Maehama and Dixon, 1998). Thus, loss of PTEN results in increased AKT recruitment to the plasma membrane (PM), where it is activated by PDK-1 kinase through phosphorylation on the threonine-308 (Thr308) residue. The second Akt activating kinase consists of the mTOR complex 2 (mTORC2), which phosphorylates serine-473 (Ser473) on AKT (Sarbassov et al., 2005). In vitro results have led to the conclusion that dual activation of AKT is essential for activity, a notion that has been challenged by recent results obtained in vivo using genetic interference with the mTORC2 complex (as recently reviewed (Alessi et al., 2009)).
Recently, PHLPP1 and PHLPP2 have been identified through a search for genes that combine a phosphatase with a PH domain, reasoning that such a design would counteract PH domain containing kinases like AKT. Indeed the PHLPP1/2 serine/threonine phosphatases have been shown to directly inactivate AKT and PKC (Brognard et al., 2007; Gao et al., 2008; Gao et al., 2005). In addition to their roles in growth control, they have been implicated in memory formation and maintenance of circadian rhythms in mice (Masubuchi et al., 2010; Shimizu et al., 2010).
Recent genetic studies in mice have shown that neoplasia and cancer in Pten-deficient prostate depend on Akt activation on Ser473 by mTorc2 (Guertin et al., 2009). Since PHLPP1/2 are the two known phosphatases to specifically revert this activation, we sought to determine the relevance of PHLPP1 as a tumor suppressor by combining GEM modeling with patient whole genome analysis.
RESULTS
Phlpp1 is a tumor suppressor in mouse prostate
By crossing Pten+/− with Phlpp1−/− mice we generated a cohort containing more than 400 mice of six genotypes (note that Pten−/− mice are embryonic lethal). As shown in Figure 1A, complete loss of Phlpp1 in Pten+/− mice caused strong reduction of overall lifespan. Pathology analysis of these cohorts revealed acceleration of the polyclonal autoimmune disorder, which constitutes the major cause of death in our Pten+/− animals (Di Cristofano et al., 1999) (not shown). Loss of Phlpp1 on its own also significantly reduced overall lifespan, presumably due to the observed lymphadenopathy (not shown). These observations prompted us to determine the tumor suppressor role of Phlpp1 in greater detail. We focused on the role of Phlpp1 in prostate tumorigenesis, a tissue that is very sensitive to the degree of Akt pathway activation (Trotman et al., 2003).
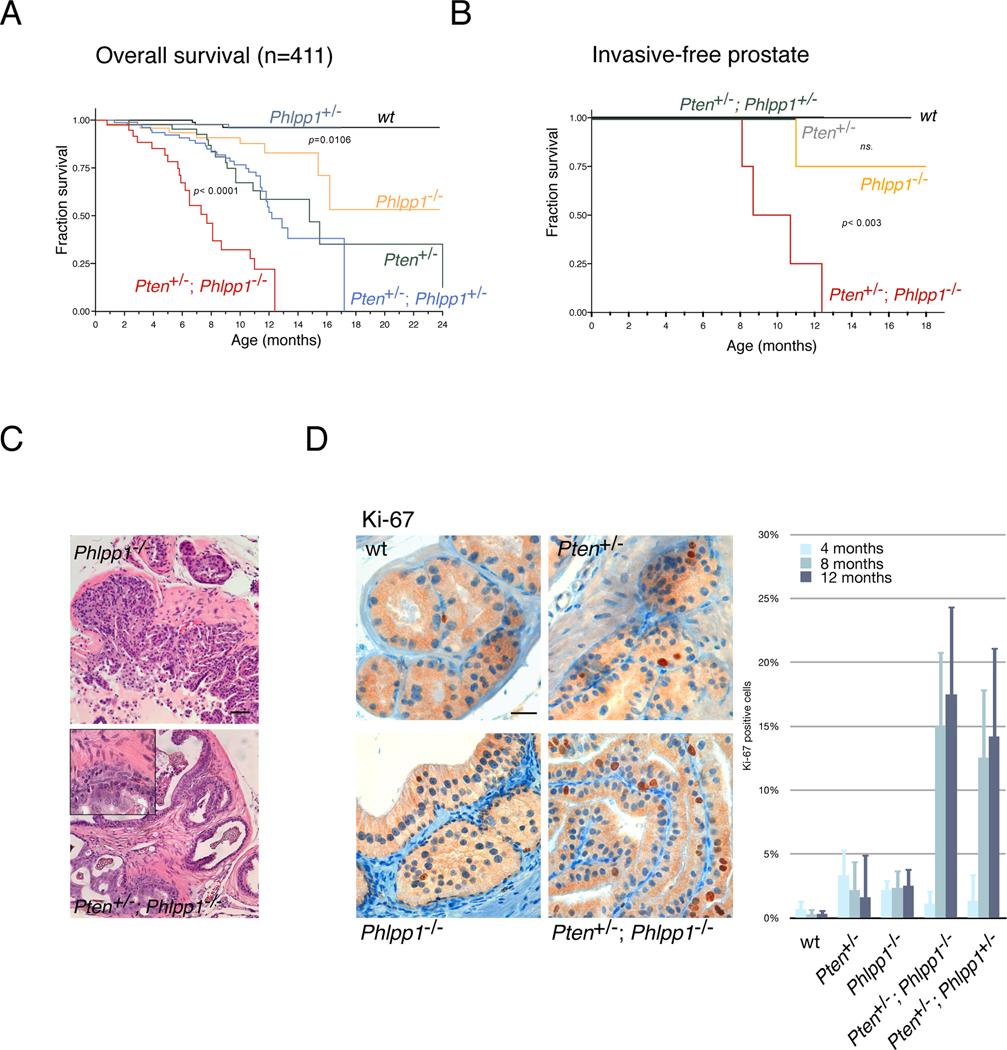
Figure 1. Phlpp1 is a tumor suppressor and dictates carcinogenesis in Pten heterozygous mice.
(A) Kaplan-Meier plot for overall survival. Number of mice (n) and statistical significance (p) is given for comparison between Pten+/− and Pten+/−;Phlpp1−/− mice and between WT and Phlpp1−/− mice. Numbers in cohort arms are: WT (98), Phlpp1+/− (97), Phlpp1−/− (49), Pten+/− (45), Pten+/−; Phlpp1+/− (81), and Pten+/−; Phlpp1−/− (41) for a total of 411 animals.
(B) Kaplan-Meier plot for invasive-free survival showing complete penetrance of prostate cancer in Pten+/−;Phlpp1−/− animals. p-value for Pten+/−;Phlpp1−/− comparison with WT animals is shown. Numbers in cohort arms are: WT (6), Pten+/− (5), Phlpp1−/− (9), Pten+/−;Phlpp1+/− (6), Pten+/−;Phlpp1−/− (11) for a total of 37 animals (ns., not significant).
(C) Microscopic analysis of 8 month prostate lesions reveals high grade PIN in Phlpp1−/− and adenocarcinoma in Pten+/−; Phlpp1−/− prostates, as indicated. Insert shows high magnification. Scale bar, 100µm. (See also Figures S1A, B).
(D) Analysis of cell proliferation in prostate at 8 months using Ki-67 immunohistochemistry (left panels, scale bar, 50µm) reveals increased proliferation in compound mutant. Progression of disease over time as shown by average Ki-67 positive cells per 100 in neoplastic foci (graph, see also Experimental Procedures). p-values are < 0.0001 and < 0.004 for comparison of compound mutant prostates (Pten+/−;Phlpp1−/−) with Phlpp1−/− or Pten+/−, respectively at 8 months and p < 0.0006 and < 0.003, respectively at 12 months. Error bars are s.d. See also Figures S1C, D.
As shown in Figures 1B and 1C (bottom panel), prostates of Pten+/−; Phlpp1−/− animals revealed regions of high grade prostatic intraepithelial neoplasia (HGPIN), which is characterized by an intraglandular proliferation of crowding cells with atypia, enlarged nuclei and prominent nucleoli, and invasive adenocarcinoma (characterized by the proliferation of atypical cells which break the basal membrane and invade through the prostatic stroma) at full penetrance with onset at 8 months (see also summary in Figure S1A). In contrast, prostates of Pten+/− mice only suffered hyperplasia (characterized by the proliferation of luminal with no cytological atypia) and HGPIN with onset after 8 months (Figure S1A), as previously published (Di Cristofano et al., 2001; Podsypanina et al., 1999; Trotman et al., 2003). While the Pten+/−; Phlpp1+/− mice did not develop foci of invasive adenocarcinoma (Figure 1B), they showed hyperplasia and low grade PIN, starting as early as 4 months and HGPIN at 8 and 12 months (see Figure S1A and also Figure 2C). Collectively, these data were consistent with a gene dose-to-effect relation of progression previously identified for Pten alone (Trotman et al., 2003). Importantly, we found that Phlpp-loss on its own triggered HGPIN at full penetrance within 9 months of age (Figure 1C and Figure S1A) and one case of invasive adenocarcinoma at 12 months (Figure 1B, Figure S1A, 12 months). This was confirmed by immunohistochemistry (IHC) for the basal cell marker cytokeratin 5, which is lost in the invasive malignant glands (Figure S1B, left panels, red arrows). Furthermore by staining early neoplasia for p63, we found no expansion of basal cells (Figure S1B, right panels).
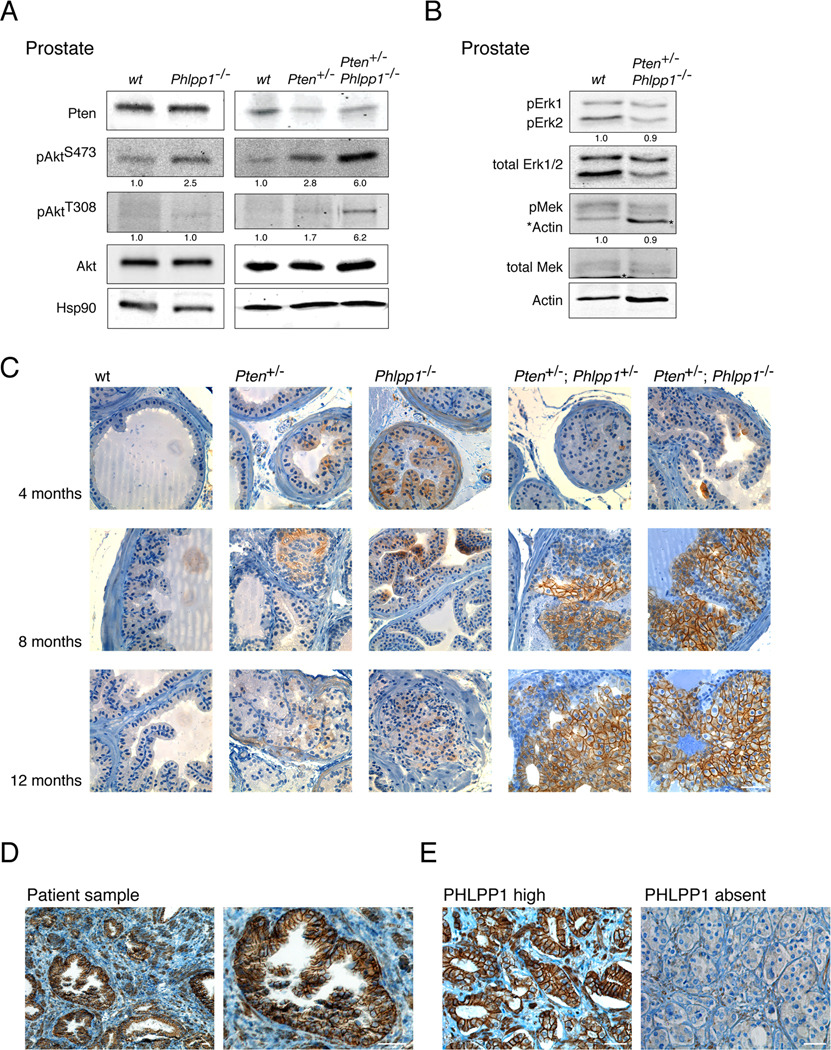
Figure 2. Phlpp1-loss in prostate triggers Akt Ser473 activation and localization to plasma membrane.
(A) Western blotting of Akt activation in 8 month old prostates and quantification of pAkt Thr308: Akt and pAkt Ser473: Akt ratios reveals strong phosphorylation on the hydrophobic Ser473 motif. See also Figures S2A–D.
(B) Western blotting and quantification of Erk and Mek kinase activation (Asterisks denote actin leftover staining on blot) reveals no activation of this signaling axis in Pten/Phlpp1-mutant prostate.
(C) time course of signal activation by IHC staining for pAkt Ser473 demonstrates activation and strong membrane localization in compound mutant genotypes after 4 months of age. Scale bar, 100 µm.
(D) PHLPP1-IHC staining in human prostate cancer sample reveals membrane localization in epithelium. Scale bar, 50 µm.
(E) Examples of differential PHLPP1 levels in human prostate cancers as detected by PHLPP1-IHC staining of the Tumor Tissue Microarray. Scale bar, 50 µm.
Pten-alteration in mouse prostate leads to increased proliferation, so we assessed Ki-67 staining over time. As shown and quantified in Figure 1D, the Pten+/− and Phlpp1−/− tissue showed similar rates of proliferation in the few earliest hyperplastic regions (4 months) and in the neoplasms at 8 months and 12 months. In contrast, we observed a sharp increase in proliferation upon combined loss but only after 4 months, thus quantitatively demonstrating that the two genes synergize in a time-dependent manner. Next, we quantified malignancy by measuring average lesion sizes of the 5 cohort arms at 3 time points. As shown (Figure S1C), combined gene loss strongly cooperated to produce disproportionately larger lesions over time. Moreover, we found that changes in apoptotic rates could not account for tumorigenesis as they slightly increased (not decreased) in Phlpp1- and compound mutants, nor could we detect inflammatory responses as a cause of prostate malignancy (Figure S1D). Collectively, our results demonstrate that Phlpp1 acts as a tumor suppressor in mouse prostate, and that its loss is synergistic with partial Pten-loss, but in a time-dependent manner.
Phlpp1 blocks Akt activation at the epithelial plasma membrane
Since Phlpp1 has been shown to directly dephosphorylate Ser473 of Akt (Gao et al., 2005), we sought to determine the phosphorylation of Akt (pAkt) in prostates of our mice. As shown in Figure 2A, loss of Phlpp1 affected pAkt activation on Ser473. We have been unable to detect significant above background phosphorylation on Thr308 in Phlpp1−/− prostates. While these data are consistent with the published Ser473 specificity of Phlpp1 we cannot exclude additional (especially indirect) activation of Akt on Thr308 in Phlpp1−/− prostate (see below). In the context of Pten-heterozygosity, Phlpp1-loss also increased Ser473 phosphorylation and consistently showed a marked effect on Thr308 (see Figure 2A and Figures S2A, B for double heterozygous prostate and further quantification, respectively).
PHLPP1 has also been implicated in MAP-kinase and PKC signaling (Gao et al., 2008; Shimizu et al., 2003), yet in the prostates of compound mutant mice we did not find an increase in MAP-kinase pathway signaling (Figure 2B) and no consistent effects on PKC-beta levels (not shown) but confirmed Gsk3-beta, 4Ebp1 and S6 phosphorylation, while Foxo3a and Pras40 where not detectably affected (Figure S2C). Moreover, loss of Phlpp1 had no general effect on Pten-levels (See Figure 2A, top panels). However, microscopic analysis revealed the presence of some glands with spontaneously reduced Pten protein and correlating pAkt Ser473 activation in the Pten+/−; Phlpp1−/− lesions (Figure S2D). In the absence of an equally effective IHC antibody for pAkt Thr308 activation, we assume that this reduction in Pten levels is contributing to the observed activation of pAkt Thr308 above background levels in the Pten+/−; Phlpp1−/− tumors (Figure 2A, last lane).
We have previously shown that the cellular localization of activated Akt greatly varies in different CaP models: strong PM association is seen in the conditional Pten-null model (Trotman et al., 2003), while tumors in Pten+/−; Pml−/− animals show strong nuclear pAkt localization and activity (Trotman et al., 2006) causing female sterility via Foxo3a inhibition (Castrillon et al., 2003; Trotman et al., 2006). The membrane associated pAkt activation after complete Pten-loss correlates with senescence arrest in vitro and in vivo (Alimonti et al., 2010; Chen et al., 2005). Using immunohistochemistry time course analysis at 4, 8 and 12 months (Figure 2C), we found patchy above-background cytoplasmic pAkt Ser473 localization in Pten+/− prostates. In Phlpp1−/− prostate, we observed a similar stain, also at all time points, confirming comparable overall Akt activation levels seen in Figure 2A. At 4 months, the compound mutant prostates revealed no major difference compared to the single mutant mutant tissues. However, a striking shift of pAkt signal to the PM was observed in 8 month and 12 month compound mutant prostates. At these two time points, we found strong foci of Akt-activation, which over time increased both in size (Figure 2C, two right most columns, 8 months and 12 months) and number (not shown). Moreover, we found that these pAkt-foci typically correlated with patches of slightly reduced Pten protein (Figure S2D).
Together, our findings are consistent with a Phlpp1-dose dependent phenotype as the activation foci were more dominant in Pten+/−; Phlpp1−/− than in double heterozygous mice. Therefore, the degree of Phlpp1 activity in the prostate epithelium controls pAkt concentrations at the PM after partial Pten-loss. Finally, we determined if this pAkt localization was due to the cellular distribution of Phlpp1 itself. As shown (Figure S2E), the Phlpp1 protein has several domains that could direct it to the PM. Using a human PHLPP1 antibody, we confirmed strong membrane localization of PHLPP1 in human prostate epithelium (Figure 2D) and readily identified differential PHLPP1 expression levels on a human Tumor Tissue Microarray (Figure 2E). Thus, we conclude that Phlpp1 acts at the prostate epithelial membrane to antagonize Ser473-activation of Akt and that its loss, especially after Pten deregulation, can direct pAkt signaling to the plasma membrane.
Phlpp1-loss triggers p53 activation in Pten+/− prostate
The above data revealed focal full-blown pathway activation that occurred only after a certain latency and in compound mutants, consistent with the delayed onset maximal proliferation observed in the double-mutant genotypes quantified in Figure 1D. Since Pten-loss triggers PM Akt activation and p53-mediated senescence (Alimonti et al., 2010; Chen et al., 2005; Kim et al., 2006) (as recently reviewed (Collado and Serrano, 2010)), we next determined if inactivation of Phlpp1 in Pten+/− mice could affect p53. As shown in Figure 3A, Phlpp1−/− and also Pten+/−; Phlpp1−/− prostates showed elevated p53 levels at 8 months. This increase was consistently observed in p53 protein while p53 transcript levels remained the same (Figure S3A). However, we failed to detect a corresponding increase in p21 transcription. On the contrary, we found lower p21 transcription in these same prostate samples (Figure 3B) suggesting spontaneous p53 loss-of-function.
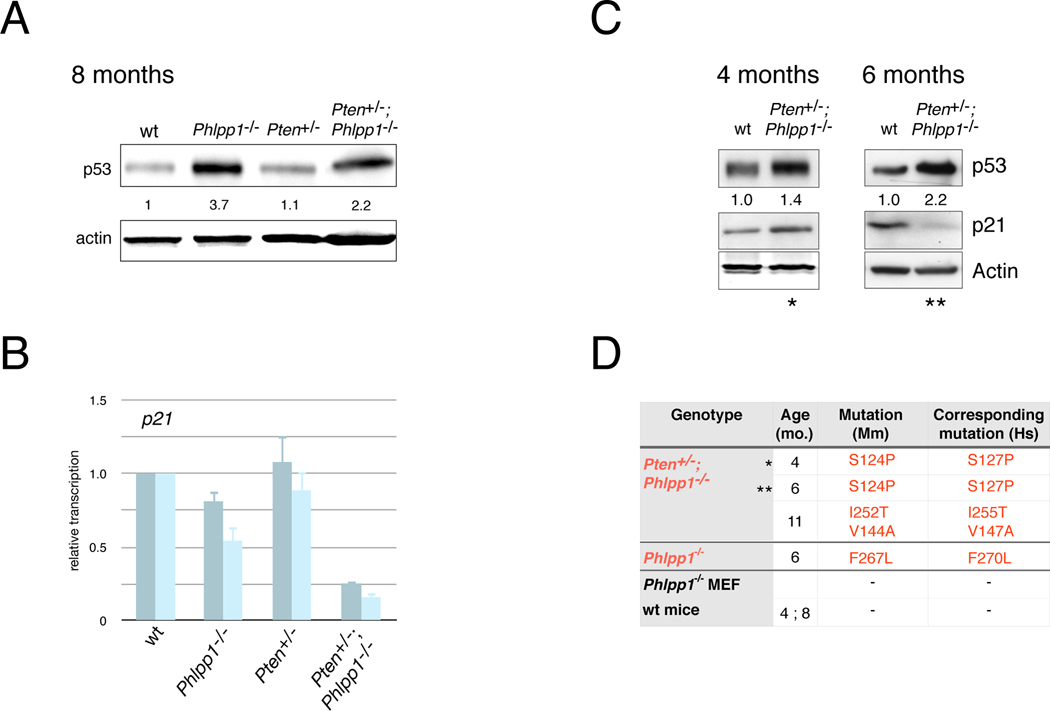
Figure 3. p53 activation and mutation occurs in Pten/Phlpp1-mutants.
(A) Western blotting and quantification of p53 shows increase in p53 levels in the Phlpp1 mutant prostates at 8 monhts of age. See also Figure S3A.
(B) RT-qPCR of p21waf1 transcripts extracted from the same lysates as shown in (A) reveals lack of p21 activation in spite of p53 protein increase in the Phlpp1 mutant tissues. Bars represent different primer pairs. Error bars are s.d. of triplicates.
(C) Western blotting of p53 and p21 induction in prostates from indicated ages and genotypes confirms loss of p53 activity at 6 months. See also Figures S3B and S6F.
(D) Summary of p53 inactivating mutations identified in above mouse prostates of indicated genotypes and ages. Asterisks denote that analyses were derived from identical samples (see also Experimental Procedures).
To explore the state and activity of p53 during the neoplastic process in more detail, we studied prostates of WT and Pten+/−; Phlpp1−/− mice at 4, and 6 months of age and found high p53 levels in the double mutants already at 4 months (see Figure 3C). Yet, the Pten+/−; Phlpp1−/− prostate did show a p21 response at 4 months relative to WT prostate, which was absent from the 6 month time point, suggesting that p53 was transiently active. Next, we validated this p53 activation and inactivation scenario between 4 and 6 month samples by transcriptome analysis for p53 activation signatures in the same two compound mutant samples shown in Figure 3C (see Experimental Procedures). As shown in Figure S3B, we found p53 target gene upregulation (including p21) at 4 months. This however was absent from the 6 month old prostate sample, confirming lack of p53 activity at this time point, in spite of high protein levels in the same sample. We also noted elevated transcripts of the prostate senescence associated SerpinE1/Pai-1 gene (Chen et al., 2005) at 6 months (see Figure S3B) and validated elevated Pai-1 transcripts by qPCR (Figure S3C, 6 months). However, we found Pai-1 strongly reduced in the 8 month old double mutant prostate (Figure S3C, 8 months), similar to the loss of Cdkn1a mRNA shown in Figure 3B at that time point. In sum, (in spite of a 4–6 week variability between animals of the same genotype), we found that p53 was transiently activated, peaking at around 4 months of age just before overt focal lesions are forming.
Since the above findings strongly suggested that the resulting neoplastic glands harbored mutant, inactive p53, we turned to sequencing the above prostate samples for Trp53-mutations at the level of the cDNA transcripts, which were abundant based on our Western results. As summarized in Figure 3D, we successfully isolated mutant forms of p53 (that were conserved in human and confirmed to be functionally inactivating, see Experimental Procedures) from 3 out of 5 Pten+/−; Phlpp1−/− prostates analyzed (Figures 3C–D, see asterisks for Western and sequencing correlations). This was neither seen in the WT, or Pten+/− control prostates nor in the primary Phlpp1−/− MEFs (see Experimental Procedures).
Collectively, these data suggested that increased p53 activity could be observed around 4 months and was then lost at the 6 month and 8 month time points. They were also consistent with the notion that compound mutant prostates were under pressure to mutate p53. Paradoxically, these findings suggested that, at 4 months, p53 activation (as revealed by transcription profiling) and inactivating mutation could occur in the same tissue and might give rise to tissue heterogeneity. Since this notion was consistent with the the focal nature of tumorigenesis that we had already observed, we next explored the histological features of this process in more detail.
p53 inactivation drives compound mutant escape from senescence
Based on the above findings we sought to visualize the molecular events in early Pten+/−; Phlpp1−/− prostate at the microscopic level. We therefore tested the 4 month old compound mutant prostate that scored positive for p53 activation (Figure 3C and S3B, 4 months) and also harbored a p53 mutation (Figure 3D, 4months) for features of cellular senescence. Senescence associated β-galactosidase (SABG) staining revealed the presence of distinct positive blue areas in neoplastic glands, absent from the normal surrounding and from WT glands, and intriguingly also absent from the dysplastic core of the gland (Figure 4A). To confirm that this differential staining was due to molecular differences we tested pAkt activation by immunofluorescence and found an inverse staining pattern for pAkt (Ser473) activation in an adjacent section of the same gland (Figure 4B, area 1 vs. 2). Moreover the SABG positive peripheral area 1 showed few Ki-67 positive cells compared to the pAkt positive luminal area 2, consistent with cellular senescence (Figure 4B).
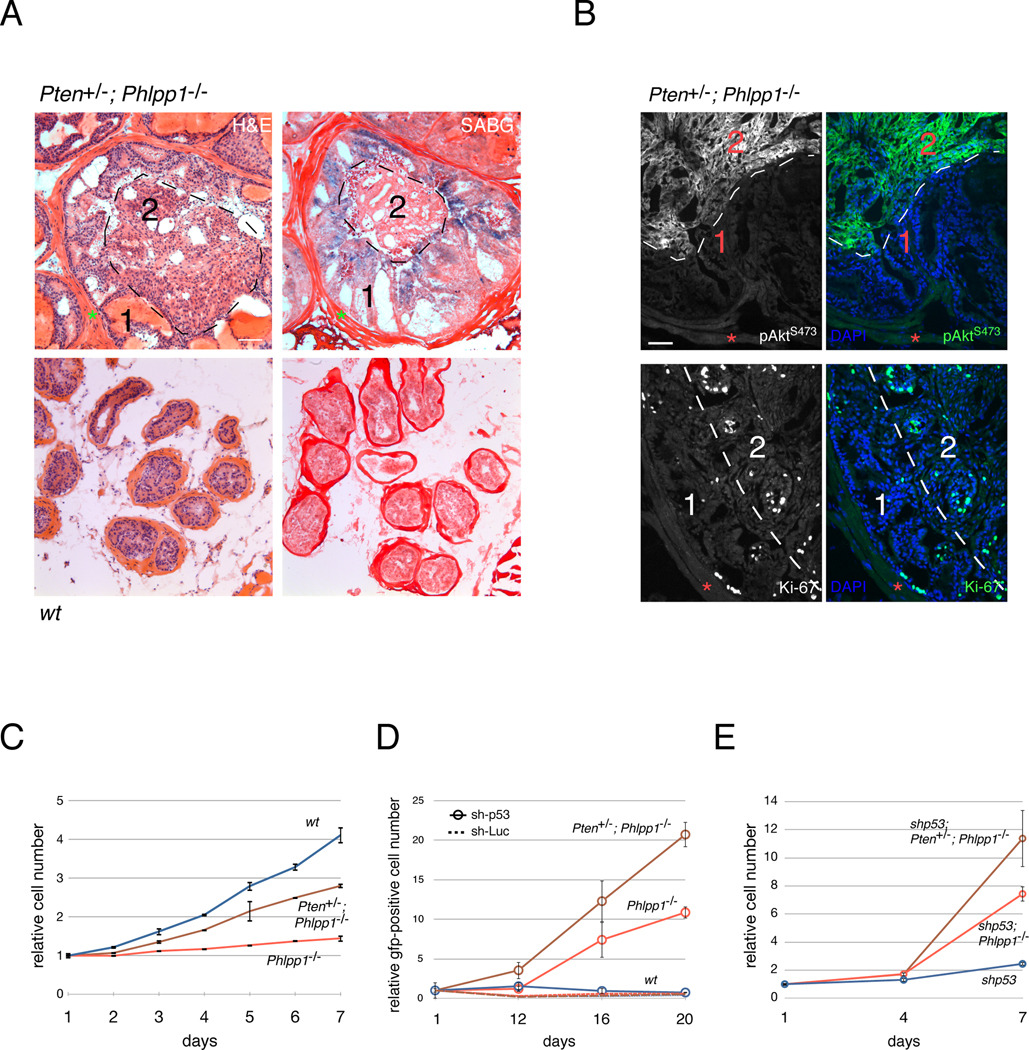
Figure 4. p53 inactivation and escape from senescence.
(A) H&E (left) and corresponding adjacent section staining for senescence-associated beta-galactosidase (SABG, right) in 4 month old prostates of indicated genotypes. Note that the peripheral area 1 is SABG positive, and the central luminal area 2 is SABG negative, indicative of tissue heterogeneity. Red asterisks denote basal lamina. Scale bar, 200 µm.
(B) Immunofluorescence on adjacent sections of Pten+/−; Phlpp1−/− prostate gland from (A) reveals strong pAkt Ser473 activation (top) and proliferation as measured by Ki-67 (bottom) in the SABG-negative luminal area 2. In contrast, the SABG-positive area 1 shows few Ki-67 positive cells, and weak pAkt. Red asterisks denote basal lamina. Scale bar, 100 µm.
(C) Growth curve of WT, Phlpp1−/−, and Pten+/−;Phlpp1−/− MEFs reveals suppressed proliferation in compound mutant genotypes. Error bars are s.d. of triplicates.
(D) p53-knockdown bypasses growth suppression in the Pten+/−; Phlpp1−/− and Phlpp1−/− mutant primary MEFs leading to clonal outgrowth of the sh-p53 positive cells (See also Figure S3E). Genotypes of primary MEFs are WT (blue), Pten+/−; Phlpp1−/− (brown), Phlpp1−/− (red). Hatched lines denote control shRNA-Luciferase infections (same color code).
(E) Growth curves of Phlpp1−/−; shp53 and Pten+/−; Phlpp1−/−; shp53, and shp53 MEFs confirm results from (D) after selection of stably transduced cells. Error bars are s.d. of triplicates.
Thus, our data suggested that tumorigenesis in the Pten+/−; Phlpp1−/− model requires the inactivation of p53 to escape from senescence (through spontaneous or pre-existing alterations). We found that Phlpp1−/− and Pten+/−; Phlpp1−/− primary MEFs arrested in a growth assay when compared to WT MEFs (see Figure 4C). Indeed, we found increased p53 in these cells, correlating to pathway activation (see Figure 7A). Upon infection of these single and double mutant MEFs with a virus-based anti-p53 short hairpin (sh-p53 with Gfp marker) and Gfp-based tracking of the infected population we found that these cells underwent clonal expansion (see Figure 4D and Figure S3E) after 12 days post infection (without antibiotic selection, see Experimental Procedures) and we confirmed these results after deriving the stable anti-p53 hairpin expressing MEFs (Figure 4E). Of these, only (Pten/) Phlpp1-mutant cells could form colonies in agar (Figure S3G). We confirmed Akt-dependence of these cells using an AKT-inhibitory drug resulting in growth suppression, decreased cell viability and increased apoptosis (Figures S3H–I).
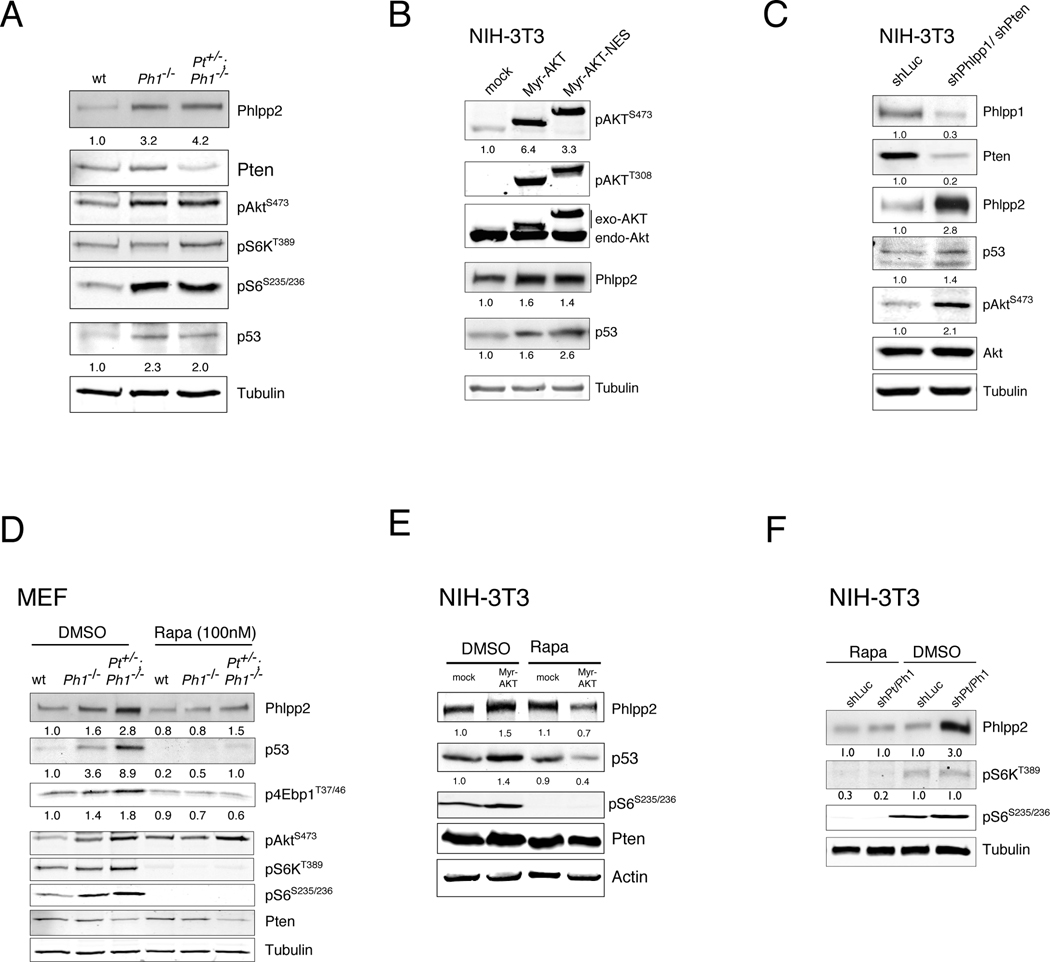
Figure 7. Pten/Phlpp1 co-deletion triggers rapamycin-sensitive p53 and Phlpp2 activation.
(A) A surge of steady state Phlpp2 and p53 levels is seen in the primary Phlpp1−/− and the Pten+/−;Phlpp1−/− mutant MEFs. See also Figure S6B, C, F.
(B) Activated cytoplasmic AKT overexpression recapitulates the Phlpp2 and p53 activation from (A) in NIH-3T3 cells. See also Figure S6A.
(C) The Phlpp2/p53 response is also found after shRNA-mediated Phlpp1/Pten knockdown in NIH-3T3 cells. See also Figure S6D, G.
(D) The surge of Phlpp2/p53 in double mutant primary MEFs from (A) is blocked by rapamycin suggesting mTor dependent translation (see also Figure S6D, E).
(E) Phlpp2/p53 activation driven by myrAKT overexpression in NIH-3T3 cells is blunted by rapamycin.
(F) Phlpp2/p53 activation after Pten/Phlpp1 knockdown in NIH-3T3 cells is reverted by rapamycin. See also Figures S6D, E, H, I)
These data demonstrated that loss of p53 in a (Pten/) Phlpp1-mutant setting could switch cells from arrest into proliferation. Note that consistent with the previous reports on senescence after Pten-loss (Alimonti et al., 2010; Chen et al., 2005), we found no phospho-Ser15 phosphorylation in the increased p53 of Pten+/−; Phlpp1−/− prostate (Figure S3F) nor changes in phosphorylation on a panel of other p53 modification sites (see Experimental Procedures) in Phlpp1 knockdown cells (not shown).
To summarize, our findings revealed that loss of Phlpp1 in the context of partial Pten-loss causes p53 activation and cellular senescence in the prostate and in MEFs. While this fail-safe response can delay disease progression, we did observe that it is invariably overcome in the prostates of our mice. Our data thus suggested a general conceptual paradigm of triggering and breaking senescence as a condition for Pten/Phlpp1-mutant prostate tumorigenesis. However, in contrast to the natural human disease, every prostate cell in our engineered model suffers Pten and Phlpp1-loss, thus increasing the likelihood of spontaneous escape from arrest through either pre-existing or spontaneous p53-mutations. To test if and at what stage breaking of the p53 senescence response is important in the human disease, we validated this genetic progression scheme in a comprehensive human prostate cancer database.
PHLPP1 is a tumor suppressor in human prostate
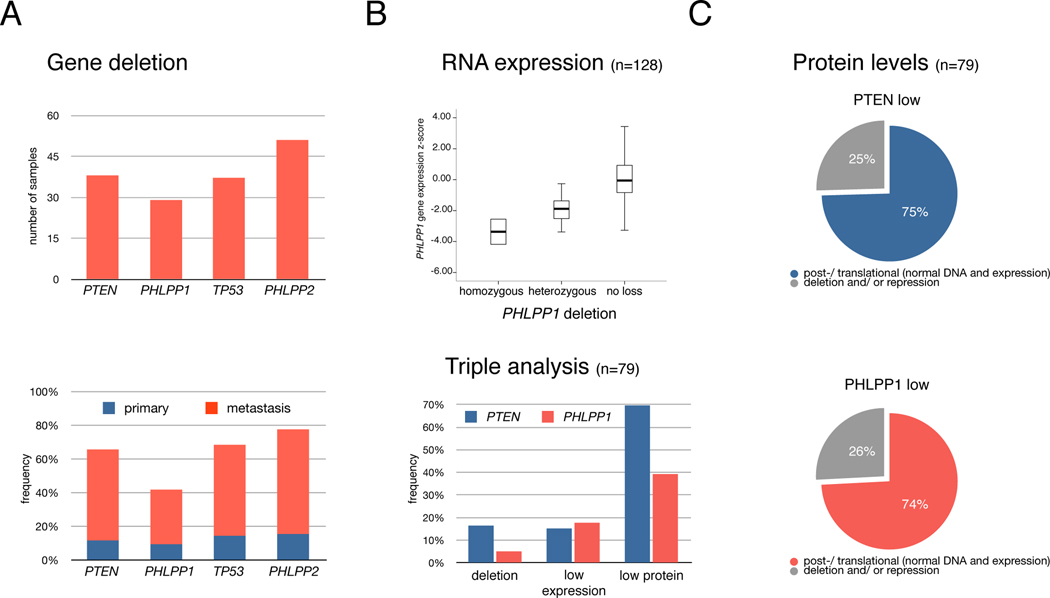
To validate our findings in human prostate cancer, we studied a data set of 218 tumor samples (181 primary and 37 metastatic CaP specimens) from patients with clinical and pathologic annotation (Taylor et al., 2010). Of these, 128 samples were also profiled with gene expression arrays (see Experimental Procedures Section). We found that PHLPP1 (18q21) was indeed lost at the genomic level in 29 cases (Figure 5A, top). As shown (Figure S4), 22 of 29 samples exhibited broad heterozygous loss of PHLPP1 that often included the recently validated prostate cancer gene SMAD4 (Ding et al., 2011). For comparison, 38 and 37 samples had either broad heterozygous or focal homozygous deletions involving PTEN or TP53, respectively. In addition, PHLPP2 (on chromosome 16), the paralog of PHLPP1, was found in a region of frequent broad heterozygous loss (Figure S4). Moreover, these genes were often lost in the samples from metastatic sites (see Figure 5A, bottom).
Figure 5. PHLPP1 is a tumor suppressor of human prostate cancer.
(A) Upper panel: copy number loss of PTEN, PHLPP1, TP53 and PHLPP2 in the data set of 218 human prostate cancer samples (see Experimental Procedures). Lower panel: deletion frequency of above genes in the 181 primary tumors and 37 metastases. See also Figure S4.
(B) PHLPP1 copy number alterations correlate with gene expression profiling (top). Boxes contain expression values of 50% of cases, bars show the remaining 50% except for rare outliers. A bold line indicates the median expression value. Z-scores are relative to expression in normal prostate. Bottom graph: overview of alteration frequencies in 79 samples that were triple-analyzed at copy number, RNA expression and protein level for PTEN and PHLPP1.
(C) Breakdown of alterations in samples with low PTEN (top) and PHLPP1 (bottom) protein levels. Protein reduction (scored low or absent) in cancers is observed most frequently in spite of the presence of both gene copies and normal RNA levels.
We then tested for more specific PHLPP1 alteration, first at the transcript level. As shown in Figure 5B (top), we found that genomic loss was significantly associated with reduced PHLPP1 expression levels using statistical variance testing (p-value <0.001, see Experimental Procedures). We also identified eight human tumors in which PHLPP1 was apparently diploid, but harbored reduced transcript expression levels (more than 2 standard deviations below the mean expression of 29 normal adjacent prostate samples). Similarly, we found statistically significant association between PTEN expression levels and its copy number (Figure S5A). To further explore the extent of PHLPP1-specific alteration in CaP, we analyzed tumor tissue microarrays (TMAs) for PTEN and PHLPP1 proteins in the patients. As shown in Figure 5B (bottom), our analysis revealed that low or absent levels of PTEN or PHLPP1 were 3–5 times more frequent than alteration at either DNA or RNA level. We also found some overlap between PTEN and PHLPP1 protein loss (p=0.017). By integrating the TMA data with genomic and expression analysis on 79 samples we could demonstrate that the vast majority of PTEN and PHLPP1 protein loss happens in cases with normal DNA and/or expression of the genes (Figure 5C).
Thus, both PTEN and PHLPP1 are frequently and specifically targeted at the translational or post-translational level. Note that candidate exon re-sequencing of 80 tumors from this set identified few gene mutations (e.g. only two p53 mutant samples (Taylor et al., 2010)). We therefore chose not to re-sequence the full data set (no PHLPP1 mutations were found in 9 samples). Our analysis confirmed that PHLPP1 is frequently deleted in primary and advanced disease and also altered at the RNA- and, in particular, at the protein-level.
Co-deletion of PHLPP1 and PTEN is strongly associated with metastatic disease
Through mouse modeling we have discovered a relationship between triggering of a p53-fail-safe response by combined loss of Pten and Phlpp1 on the one hand, and breaking it through loss of p53-function on the other. If, as previously suspected (Chen et al., 2005), senescence arrest after complete PTEN/PHLPP1-loss would suppress the early, still benign stages of disease, then the escaping prostate cancers would frequently present with triple alteration of PTEN, PHLPP1 and TP53. Thus, we analyzed the human data set for co-alteration of these loci.
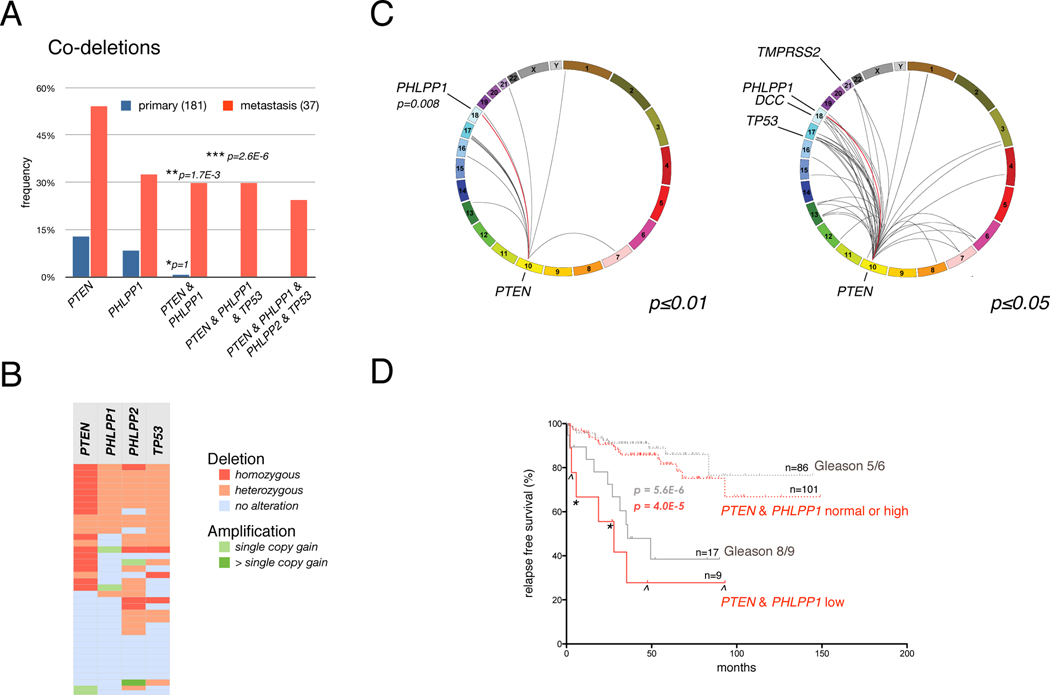
As shown (Figure 6A), primary samples showed no significant association between loss of PHLPP1 and PTEN– this occurred in only one of the 181 samples, which had no deletion of TP53. In contrast, we found statistically significant co-deletion of PTEN and PHLPP1 in metastatic samples. When testing for concomitant PTEN, PHLPP1, and TP53 copy number alteration, we found statistically significant association (p-value < 2.6e–6, see also Figure 6A, legend) and frequent broad 16q co-deletion events which harbor the PHLPP2 locus. Because the metastatic lesions harbor a greater number of alterations than the primary samples, we performed two independent statistical analyses. First, we determined the genome-wide deletions, which are associated with PTEN-loss in metastatic samples as a function of the association’s p-value. At p≤0.01 (Figure 6C, left plot) we find that PHLPP1 is in one of only 17 deletions containing 69 genes that are co-deleted with PTEN (p=0.008). At higher p-value (≤0.05) neighboring genes in the PHLPP1 locus also became statistically significant, consistent with the recent discovery of Pten/Smad4 cooperation in mouse (Ding et al., 2011). We indeed observed activation of Tgf-beta signaling in the Phlpp1-deficient mouse prostate (Figure S3D) and the human data showed significant association with further indicated events that were recently validated to strongly cooperate with loss of Pten in mouse (Carver et al., 2009a; Chen et al., 2005; King et al., 2009). Also, combined PTEN-PHLPP1 loss correlates significantly with loss of TP53 (Figure S5D).
Figure 6. Co-deletion of PHLPP1 and PTEN is restricted to the metastatic samples.
(A) Co-deletion analysis of PTEN, PHLPP1 and TP53 in primary and metastatic samples shows frequent co-deletion in metastatic samples. The p-values for the significance of association between both PTEN and PHLPP1 loss of any kind with primary (asterisk) or metastatic (double asterisk) samples are indicated. A triple asterisk denotes the p-value for the association of PTEN, PHLPP1 and TP53 triple-loss among all samples (no triple deletion occurred in primary samples). See also Figure S5A, B.
(B) Heat map of copy number alteration in the 37 metastatic samples shows association of PTEN, PHLPP1, PHLPP2 and TP53 alterations. Color code for deletion and amplification is indicated. See also Figure S5C.
(C) Circos plot of genome-wide co-deletion events in PTEN-mutant metastatic samples at p≤0.01. PHLPP1 (with two next neighbors, p=0.008) is in one of 17 PTEN-associated deletion regions that harbor a total of 69 genes. Right panel: at p≤0.05 a greater number of deletions are associated with PTEN-loss. See also Figure S5D.
(D) Kaplan-Meier outcome analysis for relapse after radical prostatectomy based on expression levels of PTEN and PHLPP1 mRNA. The same analysis based on Gleason-scoring is shown for reference. Patients with Gleason score 6 (asterisks) and 7 (arrowheads) are shown. See also Figure S5E.
Next, we performed an unbiased determination of significant co-deletions in metastases. To this end, we carried out a False Discovery Rate analysis by first performing a Monte Carlo permutation test of the significance of each possible co-deleted pair of genes in the metastatic genomes (total of >3000 genes giving 5 Mio pairs). The result is the histogram of p-values depicted in Figure S5C. As shown, the vast majority of these pairs are statistically consistent with absence of correlation: their p-values form a constant plateau in the graph. In contrast, there is a number of true positives, i.e. gene pairs that are significantly correlated, statistically. These are exhibited by the overrepresentation of low p-values to the left side of the graph (see also Experimental Procedures). This small subset of significantly correlated genes contains all the pertinent gene pairs formed by loss of PTEN, PHLPP1, PHLPP2 and TP53. Considering that these are physically independent genes, they are statistically likely to be biologically linked.
To summarize, our two additional analyses confirm that the metastatic samples show a statistically significant enrichment of co-deletion of these genes, while in contrast, the primary samples do not. When we analyzed the heat-map of inactivation of these genes among only metastatic samples (shown in Figure 6B), we indeed observed that co-deletion of PTEN, PHLPP1, PHLPP2 and TP53 clustered together. These data are consistent with a prostate cancer progression model, where loss of p53 is a prerequisite for the combined loss of PTEN and the PHLPP genes, which is found in metastasis.
Taken together, our analysis suggests that (i) PHLPP1 is a tumor suppressor in human prostate cancer, that (ii) strong activation of AKT signaling through loss of PTEN and PHLPP1 is associated with the loss of Tp53, and that (iii) the co-incidence of these three deletion events is frequent in metastatic samples but absent from primary tumors. These data are thus consistent with the notion that the p53-response acts as a barrier to prostate cancer progression, and not initiation. However, we note that our analysis does not allow us to conclude the order in which these three human deletion events occur.
To test if the latter finding could be of clinical value, we analyzed if combined alteration of PTEN and PHLPP1 at RNA or protein level could predict disease recurrence after prostatectomy. PTEN/PHLPP1 protein levels did not correlate with disease recurrence (not shown). Plotting the combined transcript levels of PTEN and PHLPP1 in prostatectomies (Figure 6D), we did observe a significant difference between the time to biochemical relapse in PTEN/PHLPP1 low mRNA samples and those with at least one normal transcript level (p=4E-5). This outcome analysis was similar to using the histopathology based Gleason score as predictor in this data set(Figure 6D). These results suggest that combining pathology based information with the PTEN/PHLPP1 transcript signature could identify patients that would benefit from PI 3-Kinase pathway therapy. Note that only 3 (of 37) samples with low PTEN/PHLPP1 transcripts had Gleason scores of 7. Thus, we could not test if the transcript levels could significantly stratify this important patient group. When performing this analysis with other pairs that are associated with low PTEN expression including cancer genes near the PHLPP1 locus (Figure S5B, bottom) only the PTEN/SMAD4 combination stood out with significant predictive power (Figure S5E) consistent with recent findings (Ding et al., 2011).
Phlpp2 and p53 show a concerted response to Pten/Phlpp1 status
Finally, we asked why late stage samples consistently showed loss of the PHLPP2 gene (Figure 6B, bottom) when PTEN/PHLPP1 deletion could already drive the pathway to prostate cancer, especially after loss of p53. To this end, we studied Phlpp2 regulation in the genetically defined setting of primary WT, Phlpp1−/− and Pten+/−; Phlpp1−/− double mutant MEFs. As shown in Figure 7A, both Phlpp1 mutant settings resulted in increased Phlpp2 as well as increased p53 levels. To test if this response was elicited by AKT activation, we used NIH-3T3 cells stably transfected with constitutively active AKT (via myristoylation, myr-AKT, Figure 7B). This experiment confirmed AKT dependent increase in Phlpp2 and p53 levels. Importantly, a myr-AKT plasmid that was targeted exclusively to the cytoplasm (myr-AKT-NES, see Figure S6A for localization) was also able to induce the two proteins consistent with a Phlpp2 and p53 response triggered by cytoplasmic AKT. To confirm that Phlpp2 and p53 levels are coupled to Pten and Phlpp1 levels we used RNAi in this cell line. As shown (Figures 7C and S6B), combined Pten/Phlpp1 knockdown resulted in increased Phlpp2 and p53 levels, confirming a putative negative feedback mechanism. Next, we tested this response in a controlled setting with a human PTEN knockout cell line, the HCT116 PTEN+/+ and PTEN−/− system (Kim et al., 2006). Figure S6C shows that the PTEN-deficient cells display activation of pAKT and p53, as published (Kim et al., 2006). At the same time these PTEN null cells also exhibited strong PHLPP2 activation, consistent with our findings in primary and immortalized mouse cells.
To better understand these findings mechanistically, we tested if the Phlpp2 protein response was caused by increased transcript levels or protein stability in the NIH-3T3 Pten/Phlpp1 knockdown cells. However (see Figure S6D–E), we found neither increased RNA levels and slightly decreased, not increased Phlpp2 stability after Pten/Phlpp1 knockdown. This transcript-independent principle was confirmed in the compound mutant prostate, and we confirmed that reversal of this surge by Phlpp2 knockdown results in Akt activation in compound mutant MEFs (Figure S6F–G). Since we found inactivation of the Tsc2-axis after Phlpp1-loss (FigureFigure S6H), we tested if the response was mediated by translation downstream of mTOR, as previously shown for p53 upon loss of Pten (Alimonti et al., 2010). Intriguingly, using the Pten+/−; Phlpp1−/− primary MEFs (Figure 7D), addition of rapamycin abrogated the Phlpp2 activation that persisted in the DMSO control treatment. Similarly (see Figure 7E), rapamycin also blunted the Phlpp2/p53 feedback in the NIH-3T3 cells, which were stably transfected with myrAKT- or the anti-Pten/Phlpp1 RNAi-plasmids (Figure 7E–F). Finally, we confirmed that genetic activation of mTorc1 by Tsc1−/− ablation also leads to a strong Phlpp2 surge without increasing its transcript (Figure S6I).
Taken together, our data show that in the genetically defined context of Pten+/−; Phlpp1−/− primary MEFs, Phlpp2 positively responds to pathway activation in an mTorc1-dependent manner, similar to p53. These findings suggest that the high co-deletion rate of the PHLPP1/2 and TP53 genes with PTEN in advanced cancer is required to break this feedback in late stage disease. Furthermore, our results suggest that clinically relevant pathway inhibitory drugs that target mTORC1 could negatively affect the levels of PHLPP2 and p53 in patients.
DISCUSSION
Cancer researchers are facing a deluge of genome data. Turning this powerful information into our advantage ultimately depends on highly reliable systems for experimental validation. Our study shows how data from large scale genomics can be put into a meaningful biological context and transformed into actionable information by comparison with results from hypothesis driven research in genetically engineered mice.
First, we have identified in PHLPP1, a ‘druggable’ suppressor of prostate cancer progression since its antagonist, mTORC2 can be pharmacologically inhibited. Most importantly, genetic mTORC2-inactivation has no adverse effects on adult prostate tissue (Guertin and Sabatini, 2009). The deletion involving PHLPP1 (18q21) contains still other suspected and confirmed tumor suppressors. The TGF-beta effectors SMAD2, SMAD4, SMAD7, as well as the DCC gene, are co-deleted in the majority of cases. The cooperation of Smad4 with complete Pten-loss in prostate cancer has recently been shown in knockout mice (Ding et al., 2011) and similarly, we find Smad-activation in response to Phlpp1-loss in prostate. The co-deletion of SMAD4 and PHLPP1 could thus conceivably exacerbate the consequences of PTEN-loss in human prostate.
Second, our study reveals a progression principle of PTEN-pathway driven aggressive prostate cancer. After its initial discovery (Chen et al., 2005), the senescence response in prostate was primarily thought to protect early hyperplastic precursor lesions from becoming clinically relevant cancer (reviewed in (Narita and Lowe, 2005)). In contrast, our genomic analysis reveals that strong activation of the pathway coincides with p53-deletion in metastasis, not cancer. Therefore we propose that primary prostate lesions must and do develop in PTEN haploinsufficiency (75% show reduction in protein) in order to fly below the radar of the p53 activation system unless a p53 alteration has already occurred.
Third, we find that low PTEN/PHLPP1 transcription correlates with biochemical relapse in patients after prostate surgery. If confirmed in expanded studies, this finding could yield important molecular information that might be used to stratify patients for PI 3-Kinase inhibitor trials. Importantly, recent results (Carver et al., 2011; Mulholland et al., 2011) have revealed that blockade of AR- or PI 3-Kinase signaling is mutually reinforcing, demonstrating the need for combined therapeutic pathway inactivation. Intriguingly, this research has shown that AR-mediated AKT-inhibition is carried out by PHLPP1 activation since PHLPP1 is degraded by AR blockade, a mainstay of advanced prostate cancer therapy. These results reinforce the crucial role of PHLPP1 status in prostate cancer progression.
Finally, we identify the PHLPP2 protein as part of a cell autonomous fail-safe mechanism, which in concert with p53 responds to excessive pathway signaling in prostate. The activation of these responses cannot prevent tumorigenesis in our animal system, yet they critically shape the disease time course in a model where, unlike in human, every prostate cell is engineered to suffer Pten/Phlpp1-loss. The PHLPP2-mediated negative pathway feedback represents another potential mechanism by which mTORC1 activation inhibits AKT activity. Since the pharmacological inhibition of mTorc1 is able to derail this response, our data argue for checking thisPHLPP2 activation in patients before they receive mTORC1-targeting pathway therapy.
Taken together, our results identify the critical role of the PHLPP proteins in prostate cancer and suggest that defining their status in relation to PTEN and p53 is important for understanding and combatting the disease.
Experimental Procedures
Mice
Generation of Phlpp1 knockout mice was recently described (Masubuchi S et al. Proc Natl Acad Sci USA. 2010 Jan 26;107(4):1642-7). Phlpp1 null mice (129 Sv/C57BL6) were crossed with wild-type mice (129 Sv/C57BL6, from our Pten+/− cohort) for two generations. Offspring were intercrossed for 6 generations to obtain the study cohort of over 400 animals. For genotyping, PCR primers 5’- TGGGAAGAACCTAGCTTGGAGG- 3’ and 5’-TTCCATTTGTCACGTCCTGCAC-3’ and 5’-ACTCTACCAGCCCAAGGCCCGG-3’ were used for Pten. Primers 5’- TAGGAGAGACTAGTGACATC -3’ and 5’-TGAGCT TATACGCTGTGATGC-3’ and 5’-AGCCGATTGTCTGTTGTGC-3’ were used for Phlpp1. Overall and disease-free survival curves were calculated by the Kaplan-Meier method and Log Rank (Mantel-Cox) testing with Prism 5.0 software for Apple Macintosh OS X. Cohort sizes were 411 mice for overall survival (WT (98), Phlpp1+/− (97), Phlpp1−/− (49), Pten+/− (45), Pten+/−;Phlpp1+/− (81), Pten+/−;Phlpp1−/− (41)), and 37 animals for prostate cancer free survival (WT (6), Pten+/− (5), Phlpp1−/− (9), Pten+/−;Phlpp1+/− (6), Pten+/−;Phlpp1−/− (11)). All work with animals was performed along the CSHL IACUC approved protocol # 11-08-3 "Tumor suppressor mutation in mouse models for cancer (11-08-3), Trotman, PI).
Western Blotting
Prostates from dissected animals of all genotypes were homogenized and protein was extracted simultaneously with DNA and RNA using the AllPrep DNA/RNA/Protein kit (Qiagen) per manufacturer’s instructions. Cells were lysed in 50nM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM sodium ortho-vanadate (Na3VO4), 10 mM NaF, protease inhibitor cocktail (Roche) and cleared by centrifugation; concentrations were determined by Bio-Rad Protein Assay (Bio-Rad Laboratories). Resulting samples were taken into an SDS sample loading buffer followed by brief sonication and centrifugation, then the supernatant was collected for western blotting. Prostate and MEFs were from at least two sets of mice of all genotypes and samples were analyzed three times or more for confirmation.
Cluster Analysis
Gene set or pathway analysis was done by using GAGE (Luo W et al. BMC Bioinformatics 2009, 10:161.), generally applicable gene set enrichment. The most differentially regulated KEGG pathways and GO groups were selected with FDR q-value < 0.1. In significant KEGG pathways or GO groups, genes with above noise-level expression changes are taken to be substantially perturbed. These data were then row/gene-wise normalized and visualized using heat-maps.
Plasmids and stable lines
pCDNA3-Myr-HA-AKT plasmid was obtained from Addgene (originally generated by William Sellers, Addgene plasmid 9008). pCDNA3-Myr-HA-AKT-NES was constructed by cloning synthetic oligos corresponding to the nuclear export signal (NES) of the PKI protein to the 3-prime end of Myr-HA-AKT. Plasmids were transfected into NIH3T3 cells using Lipofectamine 2000 (Invitrogen), and selected with 800 µg/ml neomycin 48 hours post-transfection to generate stable lines.
Human prostate cancer data
For the analysis of human prostate tumors, 218 frozen prostate cancer specimens and 149 matched normal tissue samples were procured from patients treated by radical prostatectomy at Memorial Sloan-Kettering Cancer Center. DNA from 194 tumor specimens were labeled and hybridized along with either their matched normal tissue or a pool of reference normal to Agilent 244K array comparative genomic hybridization arrays (aCGH) using the manufacturer’s protocol. Raw copy number profiles were normalized, segmented with Circular Binary Segmentation (Venkatraman and Olshen, Bioinformatics. 2007 Mar 15;23(6):657-63.) and analyzed with RAE as previously described in Taylor BS et al, PLoS One. 2008 Sep 11;3(9). Expression levels were determined for 128 of these tumors and 29 normal prostate tissues using Affymetrix Human Exon 1.0 ST arrays and tested for their association with copy number status by Anova testing as published (Taylor et al., 2010). For clinical evaluation, data from this work was previously analyzed by the Memorial Sloan Kettering Cancer Center (MSKCC) Prostate Cancer Oncogenome Group. Clinical and pathologic data from this patient cohort is maintained in a prospective fashion on the MSKCC prostate cancer clinical database and the published data on 181 primary and 37 metastatic tumors (Taylor et al., 2010) have been visualized using the Nexus Copy Number software v 5.1 (Biodiscovery, Inc.).All analyses for this publication were performed on de-identified patient data and material and thus qualified for exemption from human subjects statements under 45 CFR 46.101 (b) (4).
Highlights.
-
-
PHLPP1 is a prostate tumor suppressor that cooperates with PTEN
-
-
Phlpp1/Pten co-deletion requires p53 inactivation for senescence bypass
-
-
Co-deletion is associated with metastatic disease
-
-
Co-deletion triggers rapamycin-sensitive p53 and Phlpp2 activation
Significance.
Excessive AKT activity triggers p53-dependent growth arrest in mouse prostate. However, it has remained ill-defined if and at what stage this response acts in human prostate cancer. We now show that this surveillance mechanism forms a common barrier against prostate cancer progression. It antagonizes the co-deletion of the AKT suppressors PTEN and PHLPP1, whose deletions are mutually exclusive in primary cancers. Since rapamycin strongly inhibits the feedback activation of p53 and Phlpp2, our data call for checking the status of this fail-safe response before patients receive mTorc1-targeting therapy. Collectively, our findings identify the PHLPP proteins as key players in prostate cancer and reveal the tightly orchestrated nature of tumor suppressor activity in this disease.
Supplementary Material
Acknowledgements
We thank S. Lowe, S. Powers, M. Zhang, K. Maimer, J. Hicks, M. Spector, J. Zuber, and W. Xue for discussion, help with analyses, and reagents, L. Bianco, A. Nourjanova, K. Manova, and A. Barlas for help with histology procedures and analysis, R. McCombie and S. Muller for help with sequencing, M. Hammell, W. Luo, and C. Johns for discussion and help with RNA expression array production and evaluation, and J. Simon and M. Taylor for discussion of animal procedures. This work was supported by grants to L.C.T. from the Department of the Army (W81XWH-09-1-0557), the Starr Foundation (I3-A154), the V Foundation, the V Kann Rasmussen Foundation (VKRF), and the NIH (1R01CA137050-01A2), as well as by the MSKCC Prostate SPORE and NIH grant GM-43154 to A.C.N. L.C.T. is a Rita Allen Foundation Scholar and would like to dedicate this work to the memory of William L. Gerald.
Abbreviations
Gene/Protein species nomenclature convention was used. Example:
- PTEN
human gene
- PTEN
human protein
- Pten
murine gene
- Pten
murine protein
- H&E
Haematoxylin-Eosin
- pAkt
phospho-Ser473-Akt, unless otherwise noted
- PHLPP
PH domain Leucine-rich repeat Protein Phosphatase. Note that the PHLPP1 gene has been renamed from PHLPP, as has the closely related paralog PHLPP2 (formerly PHLPPL) gene
- PI
Propidium Iodide
- PIN
Prostatic Intraepithelial Neoplasia
- PIP3
Phosphatidylinositol (3,4,5)- trisphosphate
- PKC
Protein Kinase C
- PM
Plasma Membrane
- Trp53
Transformation Related Protein 53 gene, p53-gene
- s.d.
standard deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interests Statement The authors declare that they have no competing financial interests.
Accession numbers
Microarray data generated in this study have been deposited in the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE30987.
References
- ACS. The American Cancer Society; 2007–2009. Combined Cancer Statistics 2007 to 2009. [Google Scholar]
- Alessi DR, Pearce LR, Garcia-Martinez JM. New insights into mTOR signaling: mTORC2 and beyond. Sci Signal. 2009;2 doi: 10.1126/scisignal.267pe27. pe27. [DOI] [PubMed] [Google Scholar]
- Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, Cheng K, Varmeh S, Kozma SC, Thomas G, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–693. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brognard J, Sierecki E, Gao T, Newton AC. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell. 2007;25:917–931. doi: 10.1016/j.molcel.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Chen Z, Carracedo-Perez A, Alimonti A, Nardella C, Gopalan A, Scardino PT, Cordon-Cardo C, Gerald W, Pandolfi PP. ETS rearrangements and prostate cancer initiation. Nature. 2009a;457:E1. doi: 10.1038/nature07738. discussion E2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009b;41:619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534–555. doi: 10.1101/gad.2017311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2010;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Kotsi P, Peng YF, Cordon-Cardo C, Elkon KB, Pandolfi PP. Impaired Fas response and autoimmunity in Pten+/− mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- Gao T, Brognard J, Newton AC. The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem. 2008;283:6300–6311. doi: 10.1074/jbc.M707319200. [DOI] [PubMed] [Google Scholar]
- Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2 doi: 10.1126/scisignal.267pe24. pe24. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Saitoh M, Kinkel S, Crosby K, Sheen JH, Mullholland DJ, Magnuson MA, Wu H, Sabatini DM. mTOR complex 2 is required for the development of prostate cancer induced by Pten loss in mice. Cancer Cell. 2009;15:148–159. doi: 10.1016/j.ccr.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-Dependent Growth Suppression in Human Cells by Mutations in PTEN or PIK3CA. Mol Cell Biol. 2006 doi: 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–526. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–565. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Majumder PK, Sellers WR. Akt-regulated pathways in prostate cancer. Oncogene. 2005;24:7465–7474. doi: 10.1038/sj.onc.1209096. [DOI] [PubMed] [Google Scholar]
- Majumder PK, Yeh JJ, George DJ, Febbo PG, Kum J, Xue Q, Bikoff R, Ma H, Kantoff PW, Golub TR, et al. Prostate intraepithelial neoplasia induced by prostate restricted Akt activation: the MPAKT model. Proc Natl Acad Sci U S A. 2003;100:7841–7846. doi: 10.1073/pnas.1232229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masubuchi S, Gao T, O'Neill A, Eckel-Mahan K, Newton AC, Sassone-Corsi P. Protein phosphatase PHLPP1 controls the light-induced resetting of the circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1642–1647. doi: 10.1073/pnas.0910292107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP, Huang J, Graeber TG, Wu H. Cell Autonomous Role of PTEN in Regulating Castration-Resistant Prostate Cancer Growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Lowe SW. Senescence comes of age. Nat Med. 2005;11:920–922. doi: 10.1038/nm0905-920. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saramaki O, Visakorpi T. Chromosomal aberrations in prostate cancer. Front Biosci. 2007;12:3287–3301. doi: 10.2741/2312. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Shamji AF, Clemons PA, Hon C, Koehler AN, Munoz B, Palmer M, Stern AM, Wagner BK, Powers S, et al. Towards patient-based cancer therapeutics. Nat Biotechnol. 2010;28:904–906. doi: 10.1038/nbt0910-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariat SF, Scardino PT, Lilja H. Screening for prostate cancer: an update. Can J Urol. 2008;15:4363–4374. [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Mackenzie SM, Storm DR. SCOP/PHLPP and its functional role in the brain. Mol Biosyst. 2010;6:38–43. doi: 10.1039/b911410f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Okada M, Nagai K, Fukada Y. Suprachiasmatic nucleus circadian oscillatory protein, a novel binding partner of K-Ras in the membrane rafts, negatively regulates MAPK pathway. J Biol Chem. 2003;278:14920–14925. doi: 10.1074/jbc.M213214200. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441:523–527. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, Khoo AS, Roy-Burman P, Greenberg NM, Van Dyke T, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr Opin Genet Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.