Abstract
The KNOTTED-LIKE HOMEODOMAIN (KNOX) genes play a central role in maintenance of the shoot apical meristem. They also contribute to the morphology of simple and compound leaves. In this report we characterize the FaKNOX1 gene from strawberry (Fragaria spp.) and demonstrate its function in trasgenic plants. The FaKNOX1 cDNA was isolated from a cultivated strawberry (F.×ananassa) flower EST library. The sequence is most similar to Class I KNOX genes, and was mapped to linkage group VI of the diploid strawberry genome. Unlike most KNOX genes studied, steady-state transcript levels were highest in flowers and fruits. Transcripts were also detected in emerging leaf primordia and the apical dome. Transgenic strawberry plants suppressing or overexpressing FaKNOX1 exhibited conspicuous changes in plant form. The FaKNOX1 RNAi plants presented a dwarfed phenotype with deeply serrated leaflets and exaggerated petiolules. They also exhibited a high level of cellular disorganization of the shoot apical meristem and leaves. Overexpression of FaKNOX1 caused dwarfed stature with wrinkled leaves. These gain- and loss-of-function assays in strawberry functionally demonstrate the contributions of a KNOX domain protein in a rosaceous species.
Introduction
Over the past decade elegant studies in Arabidopsis thaliana have unveiled the intricate programs that govern leaf development and morphology. These findings have been translated to tomato and other species, where new interactions with the environment and expansion of the Arabidopsis model have been described [1], [2]. In general, leaves show shape and size variations that reflect their adaptation to various environmental conditions and ecological contexts. Leaves can be generally classified as either simple or compound based on their arrangement and complexity. A simple leaf has a single blade at a node and can be deeply lobed or dissected, whereas, a compound leaf consists of two or more separate blades (leaflets) borne from a common rachis [3], [4]. Various hypotheses have been presented to describe the development of compound leaves [3], [5], [6], [7]. One view suggests compound leaves as intermediate structures that share similarity with both shoots and leaves. Thus, the leaflets of compound leaves are equivalent to simple leaves [8]. The other view proposes that the entire compound leaf is a simple leaf with a highly-divided leaf blade [9].
Some of the first documented scientific inquiry exploring the variation between the simple and compound leaf came from diploid strawberry (F. vesca). A French teenager named Antoine Duchesne examined strawberries in the royal botanical gardens in Versailles. In 1763 he identified a variant possessing single-bladed leaves that produced plants presenting only single-bladed leaves when propagated by seeds or runners [10]. Later, in 1914, C. W. Richardson would cross this same variant line against trifoliate strawberry and demonstrate that the characteristic leaf phenotype was likely controlled by a recessive mutation [10]. Today strawberry is a potentially useful system to expand the understanding of compound leaf development. As the seedling grows, its first leaves emerge as single blades, followed by a transition to the familiar mature trifoliate pattern once several simple leaves have emerged [11]. Some Fragaria species (e.g. F. pentaphylla) possess three major leaflets and two reduced ones for a total of five. When flowering, a single-bladed leaf known as a flag leaf often emerges to accompany a floral pedicle. The changes in leaf complexity observed during early development suggest that strawberry maintains complex networks of regulators that may expand the understanding of leaf development in plants.
The KNOTTED-LIKE HOMEODOMAIN (KNOX) genes, known for their contribution to maintenance of the shoot apical meristem (SAM), have also been shown to function in leaf initiation and regulation of leaf form [1], [12], [13], [14], [15]. In simple-leafed plant species such as Arabidopsis and maize, Class I KNOX genes are exclusively expressed in the SAM and are down-regulated at the early stages of leaf development. Mutations of Arabidopsis Class I KNOX gene, SHOOTMERISTEMLESS (STM) affect embryonic shoot meristem formation [12], [16]. Overexpression of these genes generates pronounced leaf lobing and ectopic meristem formation [17], [18], [19], [20]. Contrary to the situation in simple leaves, the Class I KNOX genes are expressed in the compound leaves of species like tomato (Solanum lycopersicum), bittercress (Cardamine hirsuta) and pepperweed (Lepidium perfoliatum) [2], [21], [22], [23], [24]. Overexpression of Class I KNOX genes in tomato results in excessive proliferation of leaflets [21], [22]. Studies on simple and complex-leaved species of Lepidium indicate that the final leaf morphology does not always correlate with early Class I KNOX expression patterns [1], [2]. For example, Lepidium oleraceum generates simple leaves but the Class I KNOX expression is not down-regulated at the early leaf primordium stage. Detailed analyses revealed that leaves show a typical compound leaf development pattern during early development. Comparative investigation on Class I KNOX genes from tomato and Cardamine hirsuta suggests a highly balanced spatial and temporal action as well as differential activity generated by upstream gene regulatory sequences regulate the leaf development [23], [25]. Thus, changes in the expression of Class I KNOX accompany the progression of development in the leaf.
The role of Class I KNOX genes has been studied in species related to strawberry. The KNOPE1 and KNOPE3 genes have been identified as pivotal regulators of specific physiological processes in peach (Prunus persica). KNOPE1 transcripts are elevated in leaves infected with Taphrina deformans, leading to leaf curl [26]. Expression patterns of KNOPE3 suggest a role in carbohydrate translocation in addition to organ development [27]. Transgenic plum (Prunus domestica) plants overexpressing apple MdKN1 showed reduction in plant height and generated small malformed curly leaves [28].
In the present study, a KNOX gene (FaKNOX1) was isolated from a cultivated strawberry (F. × ananassa) flower EST library. The FaKNOX1 protein shows similarity to Class I KNOTTED-LIKE HOMEODOMAIN proteins, with 60% identity to maize LIGULELESS3. FaKNOX1 expression is mostly observed in the fruits and flowers, as well as in runner tips, single-bladed leaves and developing trifoliate leaves. Transgenic strawberry plants demonstrating the effects of loss and overexpression of this gene, and overexpression phenotypes in Arabidopsis are presented. This work demonstrates the in planta effect of a KNOX gene in strawberry leaf development and compares the findings to those observed in other species.
Results
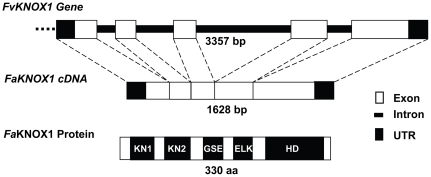
Structural organization of FaKNOX1
The full-length FaKNOX1 cDNA (accession number GQ465832) was isolated from a flower cDNA library prepared from F. × ananassa cv. Strawberry Festival. The associated genomic sequence (accession number GU339211) was PCR-amplified from the diploid F. vesca genome and showed 98.7% identity to the FaKNOX1 cDNA sequence. Comparison of the cDNA with genomic sequence revealed that the FaKNOX1 gene contains four introns and encodes a protein of 330 aa (Figure 1). FaKNOX1 shares 62% identity with TKN4 and NTH1, 52% with Arabidopsis KNAT2 and KNAT6 [29] and 60% with closely related monocot genes, ZmLG3 and OSH6 [30], [31]. FaKNOX1 maintained the same domain structure as other eudicot (AtSTM, KNAT1 and KNAT6) [12], [17], [29] and monocot species (OSH6, ZmLG3) [30], [31]. FaKNOX1 possesses all three highly conserved domains typical to the KNOX proteins: the MEINOX domain that is subdivided into KNOX1 and KNOX2; the ELK domain and the homeodomain (Figure 2A). FaKNOX1 also contained a GSE domain between the MEINOX and ELK domains, similar to previous reports [32]. Phylogenetic clustering of KNOX protein sequences from diverse species positioned FaKNOX1 with Class I KNOX proteins, within a subgroup containing LIGULELESS and OSH6 (Figure 2B).
Figure 1. Structural organization of FaKNOX1 gene.
The schematic diagram represents the exon/intron boundaries of FaKNOX1 cDNA and its corresponding genomic sequence. The FaKNOX1 cDNA encodes a protein containing four domains: MEINOX domain, the GSE domain, the ELK domain and the homeodomain.
Figure 2. Analyses of Class I KNOX genes.
(A) Alignment of FaKNOX1 amino acid sequence along with five representatives of Class I KNOX transcription factors. Black boxes indicate identical residues and the grey boxes indicate similar residues. The lines above the sequence mark the KNOX1, KNOX2, GSE and ELK domains and the homeodomain. As characteristic of TALE superclass transcription factors the homeodomain is represented by a three-amino-acid loop extension present between the first and second α-helices of the homeodomain. The letter ‘L’ and ‘T’ represent loop and turn, respectively. (B) The phylogenetic analysis of KNOX genes. Class II KNOX proteins were used as outgroup. Bootstrap values based on 1000 replicates are shown next to the branching points.
Linkage Mapping of FaKNOX1
Diploid strawberry possesses a well-covered linkage map [33]. Recent definition of a bin mapping set provides a means to rapidly assign a linkage position to a given gene [34]. The FaKNOX1 gene was mapped on the basis of DNA sequence differences between mapping parents Fn601 and Fv815. Comparison to the bin mapping progeny set indicated that FaKNOX1 be assigned to Linkage Group VI, Bin 14.
Expression pattern of FaKNOX1
FaKNOX1 transcripts are most abundant in the flower and fruit
RNA-gel-blot analysis and real-time PCR were employed to assess the tissue- specific accumulation pattern of FaKNOX1 mRNA. FaKNOX1 transcripts were more abundant in flower and fruit tissues than in leaves and roots (Figure 3A). Compared to the flower reference sample, relative real-time PCR results showed two- to three-fold increased FaKNOX1 transcript abundance in runner tips (containing vegetative shoot apex) and mature fruits, while FaKNOX1 transcript accumulation was considerably less in emerging and mature leaves (Figure 3B).
Figure 3. Steady-state FaKNOX1 transcript accumulation patterns in various organs.
(A) Results obtained by RNA-gel-blot analysis and hybridization. (B) A corresponding comparative quantitative real-time PCR analysis. The flower sample was used as reference tissue and EF1a was used as reference gene. Error bars represent standard error of the mean derived from three replicates.
FaKNOX1 transcripts are present in simple and compounds leaves
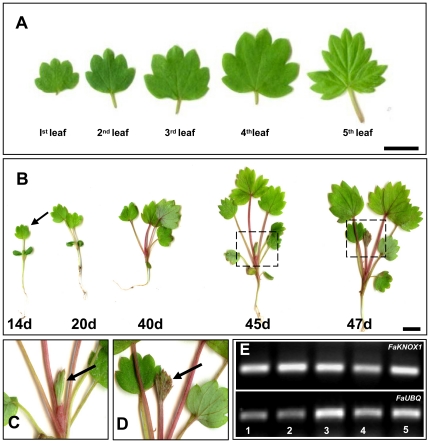
To understand the role, if any, of FaKNOX1 in the developmental or morphological changes in strawberry leaves, it was important to assess transcript abundance in different stages of leaf development. In strawberry the first four to five leaves are always simple, then the trifoliate pattern is followed [11]. The progression of strawberry leaf development from single-leaflet to trifoliate is depicted in Figure 4. In developing seedlings of F. vesca Hawaii-4, RT-PCR analyses revealed that FaKNOX1 transcripts were readily detected at approximately the same level from the early single-leaf form to an actively expanding compound leaf (Figure 4E). Transcript levels were low in these seedling leaf tissues as the FaKNOX1 amplicons were detected only after 38 cycles.
Figure 4. Leaf development pattern of strawberry seedlings.
(A) The first four to five leaves of strawberry seedlings are simple, and then follow the familiar trifoliate leaves. (B) Development of strawberry (F. vesca) seedling showing the progression of leaf morphogeneisis from simple leaf to compound trifoliate leaf form. (C and D) Represent an enlarged view of developing stages of emerging trifoliate leaves (dotted regions on panel B). The arrow indicates the specific portion of leaf used for semi-quantitative RT-PCR analysis. (E) A comparative semi-quantitative RT-PCR showing that FaKNOX1 transcript was readily detectable in leaf samples harvested from 14 d to 47–50-d-old strawberry plants. Lanes 1–3 represent expression of FaKNOX1 in simple leaves of 14-d to 40-d-old seedlings and lanes 4–5 represent expression of FaKNOX1 in emerging trifoliate leaves of 45- and 47-d-old seedlings. FaUBQ was used as an internal control gene. Bars = 5 mm.
To determine if FaKNOX1 is expressed in discrete regions of the SAM and leaf primordia during leaf development, in situ hybridization was performed. Two leaf developmental stages were tested: 20-day-old seedlings (with simple leaves) and 45-d-old seedlings (with trifoliate leaf emerging). At the two-leaf-stage, the FaKNOX1 transcript was detected at higher levels in the rib meristem region of SAM and lateral developing leaf (Figure 5A, B). As leaf development progressed to the trifoliate stage, the FaKNOX1 transcript became more abundant in the dome of the SAM and at the base and outer layers of the developing leaf. Low-level expression of FaKNOX1 was also observed within leaf primordia (Figure 5C, D, E).
Figure 5. In situ hybridization of FaKNOX1 in the vegetative meristem of strawberry seedlings.
The presence of FaKNOX1 is indicated by blue/black staining. Longitudinal section through the SAM of 2-leaf-stage seedling hybridized to a sense FaKNOX1 RNA probe (A) or antisense FaKNOX1 RNA probe (B). Longitudinal section through the SAM of 45-d leaf-stage seedling with emerging trifoliate leaf hybridized with a sense FaKNOX1 RNA probe (C) or an antisense FaKNOX1 probe (D and E). Bars = 1000 µm.
Analysis of FaKNOX1 RNAi transgenic plants
To study the function of FaKNOX1 in planta, loss- and gain-of-function transgenic plants were generated. For RNAi suppression constructs two copies of the full-length FaKNOX1 cDNA were placed under control of the cauliflower mosaic virus 35S (CaMV 35S) promoter in a head-to-head configuration. The RNAi construct was introduced into the diploid strawberry F. vesca Hawaii-4 using slight modifications of published protocols [35]. More than twenty independent T0 transgenic lines were analyzed. RT-PCR analysis from flower or runner tip tissues of RNAi plants showed no detectable transcript (several lines depicted in Figure S1). To verify the specificity of the RNAi construct, levels of other KNOX-encoding transcripts were tested. A computational analysis was performed to identify other KNOX-like sequences in the strawberry genome (www.strawberrygenome.org/www.rosaceae.org). Based on the BLASTn search, four related genes (noted as FvKNOX2 - FvKNOX5) were identified. However, these genes shared limited homology at a stretch of 80-30 nucleotides ranging from 86–96% identity (Figure 2B). Tissue-specific expression analysis indicates that transcripts for all of these genes were present in runner tips. FvKNOX2 and FvKNOX3 were abundantly expressed in mature fruits whereas barely detectable in leaf tissue. FvKNOX5, a member of Class II KNOX genes was present to a detectable label in all the tissue examined (Figure S4). To test if perturbation of FaKNOX1 expression affected other KNOX genes, accumulation of FvKNOX2 - FvKNOX5 transcripts was monitored using relative quantitative real-time PCR analysis with mRNA isolated from runner tips of four FaKNOX1 RNAi independent lines. The data show that expression of other KNOX family members is not increased significantly and that the slight repeatable differences observed are the partial suppression of the FvKNOX2 paralog transcripts (Figure S2). Thus the data indicate that the transgenic phenotypes observed may be mostly, if not completely, attributed to the suppression of the FaKNOX1 transcript with minimal changes in other transcript levels.
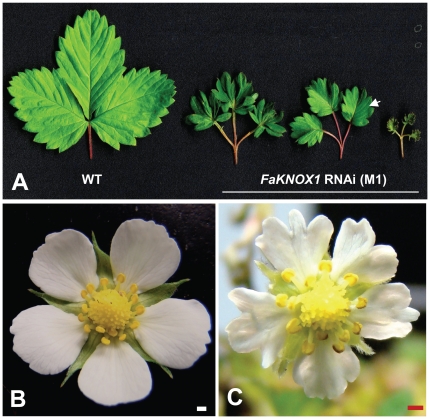
The effect of loss-of-function was evident early in plant regeneration. Whereas organogenesis from F. vesca callus first produces simple leaves, mostly the FaKNOX1 RNAi callus generated trifoliate leaves directly from the callus (not shown). The resulting transgenic plants were grouped into two classes based on severity of phenotype. These classes were designated as M1 (conspicuous effect) and M2 (severe effect) (Figure 6A). The RNAi plants from class M1 (Figure 6C) presented a dwarf stature with three types of leaf form as shown in Figure 7A. The M2 transgenic plants (Figure 6B) were extremely small in size with compact rosette-like stature and produced one form of thick, highly invaginated parsley-like leaves. This leaf form was common among both M1 and M2 plants. Compared to wild-type, the leaflets of RNAi plants exhibited elongated petiolules (Figure 7A) and plants occasionally produced small flowers with lobed petals (Figure 7B, 7C). While flowers were complete, they failed to produce fruits. These plants generated runners that gave rise to daughter plants exhibiting pale colored, narrow and elongated leaves. The developing clonal daughter plants emerging from the runner buds also presented leaves from two or three distinct phenotypic classes based on size, lobing and petiolule length.
Figure 6. FaKNOX1 RNAi mutants.
(A) Phenotypic comparison of wild-type F. vesca plants with two representatives of the severe (M2) and conspicuous (M1) categories of FaKNOX1 RNAi transgenic lines. Representative of (B) M2 FaKNOX1 RNAi plants and (C) M1 FaKNOX1 RNAi plants. Bars = 1 cm.
Figure 7. Leaf and flower morphology of a FaKNOX1 RNAi line.
(A) A wild-type leaf compared to different leaf forms of FaKNOX1 RNAi plants. The arrow indicates the leaf portion used for cross sections in Figure 7B. (B) Wild-type strawberry flower. (C) FaKNOX1 RNAi flower. Bars = 1 mm.
The macroscopic variation in leaf morphology suggested that fine cellular differences might present as well. To assess the FaKNOX1 RNAi effect on cellular organization, leaf cross sections from M1 and M2 plants were examined. Generally the wild-type leaf consists of adaxial and abaxial epidermis and middle mesophyll layer (Figure 8A). The mesophyll layer is subdivided into palisade and spongy layers. The palisade layer has tightly packed and elongated cells whereas the cells of the spongy layer are more rounded and loosely distributed. As mentioned earlier, the M1 RNAi plant shows three types of leaves. For anatomical studies only one type of leaf morphology was selected. In this class, the leaflets showed the least severe defect (Figure 7A) and except for small size appeared similar to wild-type. Leaves from M1 plants showed cellular structure similar to wild-type (Figure 8B). Examination of the M2 plants showed that differentiation of mesophyll layer was not apparent, and the space was occupied by big, round and tightly packed cells (Figure 8C).
Figure 8. Cellular organization of leaf of wild-type and FaKNOX1 RNAi plant.
Cross sections through the (A) wild-type expanded leaf, (B) M1 FaKNOX1 RNAi expanded leaf and (C) M2 FaKNOX1 RNAi expanded leaf to show the variation in the arrangement of different cell layers. AD, adaxial epidermis: PL, palisade layer; SL, spongy layer and AB, abaxial epidermis. Bars = 50 µm.
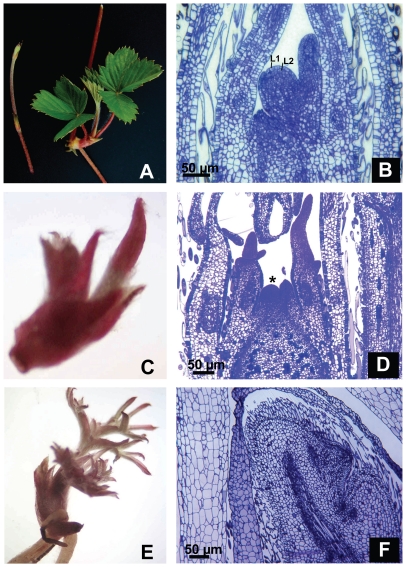
Expression analyses showed elevated transcript levels in the runner tip, a structure supporting rapid differentiation of new leaf materials. To investigate whether FaKNOX1 RNAi affected the development and/or topology of the shoot apical meristem, longitudinal sections of different developmental stages of runner tip were analyzed. It was observed that the SAM of wild-type F. vesca is dome-shaped and composed of several layers of cells with outermost layer tunica and innermost corpus. Developing leaf primordia were observed at the flanks of the SAM (Figure 9A, 9B). The SAM of FaKNOX1 RNAi young runner tips also showed a dome-like structure with developing leaves (Figure 9C, 9D). As the runner tip developed the SAM grew in size and small bulges corresponding to formation of leaves were observed (Fig. 9E, 9F). The cellular structure of the outer layer showed a high degree of disorganization. In place of round and compactly arranged cells, disorganized elongated cells were observed. Some cells also show hair-like structures. These data indicate that suppression of FaKNOX1 expression causes deviations from normal SAM development in strawberry.
Figure 9. Structural organization of the shoot apical meristem of wild-type and FaKNOX1 RNAi plants.
(A) Runner tip and emerging plantlet from the stolon of wild-type strawberry plants, (C) runner bud and (E) young developing plantlet from FaKNOX1 RNAi plants showing pale colored, narrow and elongated leaves. Transverse section through the shoot apical meristem of wild-type (B) and different stages from developing runner tip of FaKNOX1 RNAi plants (D and F). Asterisk indicates SAM. Bars = 50 µm.
Overexpression of FaKNOX1 in Arabidopsis and Strawberry
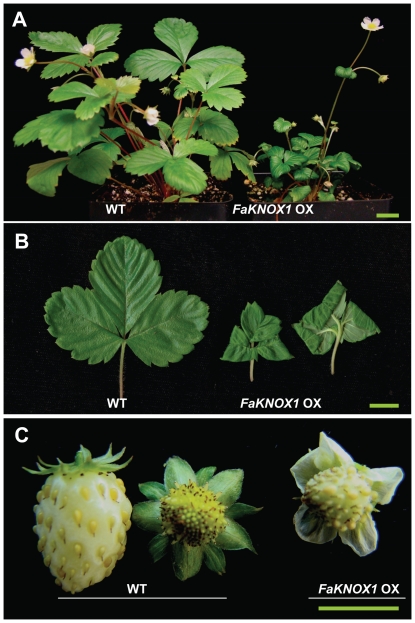
To determine the effect of ectopic expression of FaKNOX1 on the leaf growth and development, the full-length FaKNOX1 cDNA was expressed from a CaMV 35S promoter. The construct was first tested in Arabidopsis. Over twenty T0 independent Arabidopsis lines were selected as described in Materials and Methods. Ectopic overexpression of FaKNOX1 in Arabidopsis led to dwarfed rosettes and low fertility, producing approximately 1% the seed of normal plants. The leaves were highly lobed (Figure 10B, 10C) and the flowers presented short, thickened stamens (Figure 10D) that produced little pollen. The petals appeared normal but easily abscised following minimal contact. At least twenty T0 independent overexpression lines were also analyzed in strawberry. RNA-gel-blots were performed to verify overexpression in these lines (Figure S3). Overexpression of FaKNOX1 resulted in severely dwarfed plants with highly wrinkled and curled leaves, as the adaxial side was larger than the abaxial (Figure 11A, 11B). These plants produced prolific flowers on long pedicles but rarely generated fruits. Many of these plants generated seeds and T1 generation plants maintained the phenotype. Flowers appeared relatively normal with some flowers producing slight serrations on the petals. Some flowers exhibited a delay in, or complete lack of petal abscission (Figure 11C).
Figure 10. Arabidopsis plants overexpressing FaKNOX1.
Phenotype of wild-type Arabidopsis plants (A) and plants overexpressing FaKNOX1 (B and C). Overexpression lines present deeply lobed leaves, asymmetrical adaxial/abaxial growth, and a severe dwarf phenotype. (d) Flowers of FaKNOX1 overexpression lines show early abscission of petals and stamens that fail to develop (inset = wild-type flowers). Bars = 5 mm.
Figure 11. Strawberry plants overexpressing FaKNOX1.
(A) Comparison of phenotypes of wild-type and FaKNOX1 overexpression transgenic plants. (B) Phenotype of wild-type leaf with leaf forms obtained from FaKNOX1 overexpression plant. (C) Comparison of phenotype of wild-type fruits with FaKNOX1 overexpressing fruit. Note that the petals of wild-type fruit have abscised prior to fruit expansion whereas the FaKNOX1 overexpression plants produce fruits where petals fail to abscise. Bars = 1 cm.
Discussion
The understanding of the molecular control of leaf morphology has been greatly accelerated by the definition of genes that contribute to the formation of simple or compound leaves. In this report we present foundational analyses on a Class I KNOX gene that plays a role in regulation of plant stature and leaf morphology in strawberry. The strawberry system is a unique backdrop for this work. First, strawberry is a member of the Rosaceae family, a group of valuable fruit, nut and ornamental plants that rely heavily on organ shape and size to present marketable plant products. Second, strawberry plants have a distinctive leaf developmental pattern. Emerging strawberry leaves start out as simple leaves for the first weeks of plant development, but as the crown matures, the simple leaf form becomes increasingly complex, transitioning to the familiar three-lobed leaf. The present report provides groundwork to explore this process.
The FaKNOX1 sequence was obtained from a survey of F. × ananassa flower cDNA sequences that did not match any GenBank database sequences with high similarity at the time of identification. In an effort to understand the function of this novel sequence, loss- and gain-of-function transgenic plants were generated and yielded conspicuous phenotypes. Further investigation indicated that FaKNOX1 belongs to the Class I KNOTTED-LIKE HOMEODOMAIN group of proteins. FaKNOX1 contains the basic domain organization of proteins in this family, including a MEINOX domain, GSE domain, ELK domain and homeodomain with PYP three amino acid loops (Figure 2A). Genetic linkage analysis places FaKNOX1 on diploid (FV×FN) linkage group VI, bin 14.
The FaKNOX1 transcript was present to a detectable level in all the organs examined, including runner tip (containing vegetative shoot apex), leaf, flower, fruit and root (Figure 3A, 3B) of F. vesca plant. The transcript was more abundant in flowers and fruits, in contrast to earlier studies that show that expression of most of the Class I genes is generally restricted to the SAM and floral meristem [1]. The observation that these transcripts are present at substantially higher levels in the strawberry mature fruit and flower supports the hypothesis that FaKNOX1 is necessary for normal patterning in these tissues. This hypothesis is supported by analysis of transgenic plants, where defects in flower form, petal abscission, fruit set, and fertility were observed (Figures 4, 10 and 11b). Low levels of the transcript were detected in single- and multiple-leafleted leaves of F. vesca, indicating that FaKNOX1 is not solely responsible for the variations in leaf morphology (Figure 4).
However, RNAi suppression of FaKNOX1 resulted in dwarfed plants with conspicuous alterations in leaf morphology. Short stature is also observed in a loss-of-function mutations of OSH15 (rice) and KNAT1 (Arabidopsis) [36], [37]. Deeply serrated, parsley-like leaves with abnormally long petiolules (Figure 7A) were common among all RNAi lines. The leaf phenotypic effects generated by suppression of FaKNOX1 indicate that FaKNOX1 does not function identically to other Class I KNOX genes in other species. This difference may be attributed to the fact that FaKNOX1 is grouped with a separate subclass of KNOX proteins, in this case the OSH6 Class Ib KNOX proteins (Figure 2B). The functional comparisons between the roles of FaKNOX1 and other OSH6 Class Ib KNOX proteins indicate that while most of members in this class affect leaf development, they do so in different and in some cases contrasting ways. Ectopic expression of the monocot orthologs, ZmLG3 and OSH6 resulted in blade-to sheath transformation as well as loss or displacement of the ligule and auricles [30], [38]. Overexpression of NTH1 in tobacco resulted in curved leaves with a normal height [39] whereas overexpression of FaKNOX1 results in dwarf plants with wrinkled and curved leaves with asymmetrical growth between abaxial and adaxial layers (Figure 11B). Many reports indicate that down regulation of gibberellin biosynthesis leads to dwarf stature in KNOX overexpressing plants [19], [40]. Comparative studies in families like Papaveraceae, Gesneriacea, Brasicaceae and simple and complex leaf species of Lepidium also illustrate the contrasting patterns of Class I KNOX gene expression in the control of leaf morphology [2], [23], [41], [42], [43]. Thus the final leaf form obtained is dependent on Class I KNOX genes, but the roles and mechanisms differ between species. Here the loss-of-function effects on strawberry plantlets add another piece of information to this understanding.
Two distinct classes of phenotypic severity were observed among FaKNOX1 RNAi lines (Figure 6), even though suppression was complete in all the tested lines (Figure S1). While difficult to reconcile this finding, it is possible to speculate that the differences observed arose from variable RNAi suppression in specific cell types or more robust compensatory changes from paralogs that were confined to specific tissues (Figure S2). The mesophyll layer of severely affected RNAi leaves does not have distinguishable palisade and spongy layers (Figure 8). The cells are tightly arranged and are much larger in size than in comparable wild-type leaf tissues. A similar disorganization of mesophyll layer was observed with ectopic overexpression of POTH1 in potato and KNAT1 and KNAT2 in Arabidopsis. In contrast to FaKNOX1 RNAi effects, the leaves from these overexpression plants feature small cells or cells with a greater nucleus/cytoplasmic volume ratio [18], [20], [40], [44]. In another study, Hay and Tsiantis (2006) demonstrated that STM RNAi lines in C. hirsuta showed increased cell expansion and decreased expression of cell cycle markers. Thus during early stages of leaf initiation KNOX transcription factors drive leaf development in various ways in different species by controlling the temporal action of cellular growth and differentiation pathways.
In the present study the RNAi analyses were performed with the caveat that FaKNOX1 is a member of a gene family and RNAi suppression could affect other family members. Analysis of sequences present in EST collections, the recently sequenced diploid strawberry genome and contigs derived from high-throughput sequencing suggests that FaKNOX1 bears only 86–96% identity at stretch of 80-30 nucleotides with four paralogous sequences [45]. Analyses via relative real-time quantitative PCR of available KNOX sequences provided minor evidence of collateral suppression of only FvKNOX2 (Figure 2). The different classes of FaKNOX1 RNAi phenotypes may also arise due to the slight suppression of FvKNOX2 transcript.
Thus phenotypes observed across RNAi plants are likely due to direct suppression of FaKNOX1 transcripts. However, we cannot rule out a partial effect of compensatory changes from paralogous genes. Overexpression of other Class I or undescribed KNOX genes in response to suppression of FaKNOX1 could possibly underlie phenotypes observed, as these are reminiscent of overexpression phenotypes in other systems. While a formal possibility, we have no evidence to support this hypothesis. Such overexpression could be occurring in discrete cellular contexts or developmental stages that would have profound effects but would be difficult to detect.
FaKNOX1 RNAi strawberry plants rarely flowered. Flowering was generally suppressed with an occasional pedicle developing on some plants. Like the leaves, the petals were invaginated compared to those from a wild-type plant (Figure 7B, 7C) and these plants failed to produce fruits. Overexpression of FaKNOX1 in Arabidopsis led to flowers with normal floral organization that showed early abscission of petals (Figure 10). In strawberry a contrasting phenotype was observed, as overexpressing transgenic lines exhibited marked delay in petal abscission (Figure 11C). These findings indicate FaKNOX1 may also have a role in the abscission of petals, and which is regulated in different ways among diverse species. In tomato, KNOTTED TKN4 is expressed at a higher level in the abscission zone of pedicel compared to the non-abscission zone. Pretreatment with the ethylene action inhibitor 1-MCP completely inhibited pedicel abscission and led to reduction of expression of TKN4 [46]. Severe inflorescence and floral defects were also observed in a loss-of-function mutant of maize KNOTTED1 [47] and a gain-of-function mutant of soybean GmKNT1 [48]. Overexpression of OSH15 in tobacco generated malformed flowers with low fertility [49]. Thus along with leaf development KNOX plays a significant role in floral architecture and function, consistent with its relatively high level of expression in these tissues.
In Arabidopsis the STM gene is a central player in shoot meristem formation and maintenance [12], [16], [50], [51]. EMS mutants of STM fail to produce a SAM and gain-of-function mutants in tobacco produce ectopic shoots. Other subfamily members like KNAT1/BP and KNAT6 can function redundantly with STM for the SAM maintenance [18], [29], [52]. However, the loss of KNAT2 does not have an effect on the SAM maintenance and function [29]. Loss of FaKNOX1 perturbed the organization of the SAM (Figure 9D, 9F). The cellular structure of the outermost tunica layer showed elongated cells with hair-like structures. Small bulges corresponding to the formation of lateral organs were observed. These disturbances in the early SAM development likely contributed to the eventual altered leaf phenotypes. Meristems from strawberry RNAi runner tips exhibited alterations in early leaf morphology, supporting this interpretation. These results indicate a role of FaKNOX1 in maintenance and differentiation of the developing meristem that affects long-term leaf morphology and cellular organization.
Across plant species, expression of KNOX genes is a crucial factor in determination of simple or compound leaf morphology. Studies in simple-leaved species like Arabidopsis, tobacco, maize and rice indicate that at the early stage of leaf development KNOX expression is down-regulated [17], [32], [39], [53]. In contrast, the presence of KNOX expression in tomato leaves contributes to the compound leaf form [21], [22]. Recently, Muller et al. [54] has demonstrated that ectopic expression of KNOX results in a developmental shift from simple to compound leaf form in dandelion (Taraxacum officinale Web.). Similarly RNAi lines of the STM ortholog in Cardamine hirsuta show reduction in the number of leaf lobes whereas ectopic expression of KN1 leads to increased leaflets number [23], [24]. FaKNOX1 RNAi plants consistently produce few or no single-lobed leaves during early development. Three-lobed shoots emerged directly from callus-based organogenesis in RNAi plants, contrasting against wild-type shoot induction that only produced single-bladed leaves (not shown). Low levels of FaKNOX1 transcripts were always detected in all stages of leaf development, including in strawberry seedlings during trifoliate leaf morphogenesis from single-bladed leaf forms (Figure 4). Data obtained from in situ hybridization of the SAM and leaf primordium regions indicated the presence of FaKNOX1 at early stages in developing leaves and in the apical dome (Figure 5). These observations, in conjunction with loss- and gain-of-function studies in transgenic plants, supports the interpretation that FaKNOX1 participates in establishing normal cellular organization and leaf morphology, and that the roles are most pronounced during early leaf development. There are no data in this report that indicate a clear role for FaKNOX1 in the developmental progression from single-leaflet to trifoliate leaves, as the transcript is detected at low levels in leaves from both stages (Figure 4E).
In conclusion, the FaKNOX1 gene has clear roles in strawberry leaf development, cellular organization and eventual floral, fruit and leaf morphology. Additional genetic and genomic tools exist to further delineate the roles of FaKNOX1 genes in control of strawberry form. The complete set of KNOX genes may be analyzed in the agile F. vesca transformation system. During his time at the Versailles botanical gardens Duschene noted the unusual features of a plant now recognized as F. vesca monophylla, a line of F. vesca that always produces single-leafleted leaves. This strawberry variant also may become an important resource for further study. This first report presents the foundation for further studies leveraging the unique attributes of the strawberry system.
Materials and Methods
Isolation of FaKNOX1
The full-length FaKNOX1 transcript was isolated by random Sanger sequencing of cDNA clones from a Fragaria × ananassa flowering library. The genomic sequence of FvKNOX1 was obtained from F. vesca Hawaii-4 by PCR amplification using primer pair 5′-CCCACTGGGACTTCATTCTTCT-3′and 5′-GGCCCAAATAATGGAAATTTAGAG-3′. The sequence of the genomic and cDNA clones are available in the Genbank as GU339211 (Gene) and GQ465832 (cDNA).
Linkage Mapping of FaKNOX1
Linkage mapping was performed from DNA obtained from lyophilized F. vesca tissue. The Fv×Fn F2 generation of bin mapping plants [34] were used for these analyses. DNA was isolated using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA) and the TissueLyser (Qiagen, Valencia, CA) procedure for lyophilized tissue. Gene specific primers 5′-TCCACCAGAGATTAAAAGCCTACT-3′ and 5′-TCAATGTTGTTCAAGAATGTGGTT-3′ were used for amplification of a polymorphic region of FaKNOX1.
PCR reactions were prepared with 20 ng genomic DNA template with AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA), following the manufacturer's protocol. Reactions were amplified with an initial denaturation of 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, annealing at 53°C for 30 s and elongation at 68°C for 40 s, followed by a final elongation at 68°C for 10 min.
Genotypes of F2 plants were assigned on the basis of sequences obtained from direct sequencing of PCR products. Sequencing templates (10 ng each) and primers (2 µL at 2 µM each) were provided for sequencing using BigDye Terminator Cycle Sequencing Kits (ABI) on the ABI3130 DNA Analyzer at the Hubbard Center for Genome Studies (University of New Hampshire, Durham).
The FaKNOX1 gene was mapped on the basis of DNA sequence differences between mapping parents Fn601 and Fv815. When DNA chromatograms were visualized, clean unambiguous sequences were observed for plants 32, 42, 46, and 62, and these sequences exactly matched those of parent Fv815. Thus, these plants were homozygous for the Fv815 allele, and were coded “a”. Conversely, the chromatograms for plants 59 and 83 displayed superimposed peaks, yet through careful visual deconvolution it was possible to discern the presence of Fv815 and Fn601 alleles, both of which had been sequenced independently. Thus, plants 59 and 83 were heterozygous, and were coded “h”. The genotypes were compared with the FV × FN Bin Map code sheet provided by D. Sargent (East Malling Research, UK), allowing assignment of the gene to its linkage group and bin.
RNA isolation, RT-PCR and real-time PCR
Total RNA was isolated from various tissues using a modification of a CTAB-based method [55]. Tissue-specific expression analysis was performed using RNA-gel-blot analysis and quantative real-time PCR. Results obtained from both analytical methods, performed with two independent biological replicates produced nearly identical results. RNA-gel-blot analysis was performed as described [56].
Real-time PCR was performed using primer pair 5′FaKNOX1-AATGGTGAAGGACCAAGTT-3′, 5′FaKNOX1-TTCAACCTGCTCATTATTGCCTA-3′ and internal control (Elongation factor 1 alpha; EF1a) 5′EFL1A-GCCCATGGTTGTTGAAAGTTTC-3′ and 5′EFL1A-GGCGCATGTCCCTCACA-3′. The CT (cycle threshold) values for all genes in different RNA samples were normalized to the CT value of an internal control gene, EF1A. The relative mRNA levels for each candidate gene in different tissue samples were calculated using the ΔΔCT method (StepOnePlus Real-Time PCR System, Applied Biosystems, USA). Error bars in the figures indicate standard error of the mean.
For two-step RT-PCR a standard reverse transcription protocol was performed using ImProm-II reverse transcriptase (Promega, USA). The primer pair used was 5′FaKNOX1-GTAGGTCGGCTAAGCTTCCCA-3′ and 5′FaKNOX1- GCTGCCTGAAGGTCACAGTTG -3′. PCR conditions were: 2 min at 94°C; 38 cycles (30 s at 94°C; 30 s at 55°C; 20 s at 72°C); followed by a 10 min final extension step at 72°C. FaUBQ (the most similar strawberry homolog to Arabidopsis UBQ10) served as the internal control.
In situ hybridization
Shoot apices of 20-d-leaf and 45-d-leaf stage seedlings were dissected out and fixed in cold formalin acetic alcohol (FAA; 3.7% formaldehyde, 5% acetic acid and 50% ethanol) for 24 h, followed by dehydration through an ethanol series and embedded in Paraplast Plus paraffin. cDNA fragment specific to FaKNOX1 was amplified using primer pair 5′-AACCCACTGGGACTTCATTC-3′ and 5′-GGTAGCCACTGTACTTATGCA-3′. Sense and antisense RNA probes of FaKNOX1 were labeled using the DIG RNA labeling kit according to manufacturer's instructions (Roche Diagnostics GmbH, Mannheim, Germany). The probe was hydrolyzed using 0.2 M carbonate buffer at 60°C for 51 min. For detection of FaKNOX1 transcripts the sections were pre-treated, hybridized, washed and developed according to Jackson [57]. The sections were viewed and photo documented with a Nikon Digital Sight DS-Fi1 camera on a Nikon AZ100 light microscope.
RNAi and overexpression constructs
The full-length FaKNOX1 cDNA was shuttled into pK7GWIWG2D (for RNAi) and pH7WG2D (for overexpression) binary vectors [58] using standard clonase reaction conditions (Invitrogen, Carlsbad, CA). For the RNAi construct the full cDNA sequence was used because a substantial portion of the transcript lacked similarity to known transcripts at the time of cloning. Both vectors contain a GFP cassette on a separate promoter for identification of transgenic callus. The correct orientation and insertion sizes were confirmed by restriction digestion and by sequencing of recombination borders. Constructs were introduced into Agrobacterium strain GV3101 by electroporation.
Strawberry transformation for RNAi and overexpression transgenic plants
For transformation and regeneration, leaf sections and petioles from in vitro grown F. vesca plants (Hawaii-4 genotype) were co-cultivated with Agrobacterium harboring an RNAi or overexpression construct in media containing MS salts, Gamborg vitamins, 2% sucrose, 3 mg/L benzyladenine, 0.2 mg/L indole-3-butyric acid and 0.7% agar [35], [59]. After 2 d of co-cultivation, explants were washed and transferred to media containing 3 mg/L benzyladenine, 0.2 mg/L indole-3-butyric acid, 5 mg/L kanamycin or 5 mg/L hygromycin) and 250 mg/ml cefotaxime. Explants were subcultured every 14 days until shoots appeared, typically 60–90 d. Healthy shoots were transferred to rooting media (0.01 mg/L of IBA, 2% glucose, 0.5× MS salts, 0.7% phytoagar, pH 5.8). The GFP-positive rooted plants were transferred to soil, acclimated to ambient temperature and humidity, and then moved to greenhouse conditions.
More than 20 RNAi lines and overexpression lines were generated and were clearly discernable by the presence of GFP and alterations in leaf morphology. The presence of many independent lines and agreement between GFP and the leaf phenotype indicates that the phenotypes observed are related to successful expression of the target sequence.
Arabidopsis transformation for overexpression transgenic plants
The overexpression construct was transferred to Arabidopsis using the floral dip method [60]. Transgenic seedlings were selected on MS medium using hygromycin selection, and resistant/GFP-positive lines were selected. Further these lines were selfed for T1 and T2 generation.
Light Microscopy
Runner tips and leaf sections of strawberry were fixed in 3.0% gluteraldehyde in 0.1 M sodium cacodylate pH 7.4 overnight at 4°C. Following fixation, materials were rinsed with buffer and fixed again with 2% osmium tetroxide in 0.1 M sodium cacodylate. Samples were dehydrated through a graded ethanol series. The tissue was then transferred to acetone and embedded in spur resin and polymerized at 70°C. Semi thin sections of 0.5 micron were prepared using ultra microtome (Power Tome X, Boeckeler). The samples were stained with toluidine blue-O and examined with Olympus-BH2-RFCA microscope.
Phylogenetic analysis and accession numbers
The neighbor-joining method (Saitou and Nei, 1987) was used for phylogenic clustering which were conducted by MEGA4 [61]. Bootstrap values were based on 1000 replicates [62]. Accession numbers used in the relationship analysis are provided as a Table S1.
Supporting Information
RT-PCR analysis for detection of full-length FaKNOX1 transcript from four independent lines of FaKNOX1 RNAi transgenic plants. Flower and runner tip mRNA were prepared as tissue samples as FaKNOX1 is expressed in high abundance in these tissue. Based on availability of samples some lines were tested with flower and some with runner tip sample. ACTIN was used as an internal control.
(TIF)
Real-time PCR analysis for quantitation of transcript of different members of KNOX family members in WT and FaKNOX1 RNAi lines. Runner tip was used as tissue samples as KNOX is expressed in high abundance in this tissue. Data from four FaKNOX1 RNAi lines are presented here, three lines from M1 class and one from M2 class.
(TIF)
RNA-gel-blot analysis for quantitation of FaKNOX1 transcript from four independent lines of FaKNOX1 OX transgenic plants. RNA was prepared from mature leaves for overexpression lines.
(TIF)
A relative real-time PCR analysis to show tissue-specific expression of FvKNOX2 - FvKNOX5 transcripts. The flower sample was used as reference tissue. Error bars represent standard error of the mean derived from three replicates.
(TIF)
Accession numbers and their corresponding genes used for phylogenetic clustering.
(DOC)
Acknowledgments
We thank Melanie Shields for performing the DNA isolations and genotyping procedures associated with bin mapping. We are grateful to Dr. Prem Chourey and Dr. Mukesh Jain for providing useful suggestions and lab facility support for performing in situ hybridization. We appreciate Kathleen Ciola Evans for her assistance with tissue processing methods for in situ hybridization. The authors also recognize Sasha Ricuarte for her expert technical assistance in tissue culture and plant propagation.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The work was funded by National Science Foundation Grant IOS 0941335 (KMF) and in part by the University of Florida Open Access Publishing Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hake S, Smith HM, Holtan H, Magnani E, Mele G, et al. The role of knox genes in plant development. Annu Rev Cell Dev Biol. 2004;20:125–151. doi: 10.1146/annurev.cellbio.20.031803.093824. [DOI] [PubMed] [Google Scholar]
- 2.Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, et al. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 3.Champagne C, Sinha N. Compound leaves: equal to the sum of their parts? Development. 2004;131:4401–4412. doi: 10.1242/dev.01338. [DOI] [PubMed] [Google Scholar]
- 4.Pozzi C, Rossini L, Agosti F. Patterns and symmetries in leaf development. Semin Cell Dev Biol. 2001;12:363–372. doi: 10.1006/scdb.2001.0265. [DOI] [PubMed] [Google Scholar]
- 5.Bharathan G, Sinha NR. The regulation of compound leaf development. Plant Physiol. 2001;127:1533–1538. [PMC free article] [PubMed] [Google Scholar]
- 6.Piazza P, Jasinski S, Tsiantis M. Evolution of leaf developmental mechanisms. New Phytol. 2005;167:693–710. doi: 10.1111/j.1469-8137.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan DR. Fundamental concepts of leaf morphology and morphogenesis:a contribution to the interpretation of molecular genetic mutants. Int J Plant Sci. 2001;162:465–474. [Google Scholar]
- 8.Sattler R, Rutishauser R. Partial homology of pinnate leaves and shoots: orientation of leaflet inception. Bot Jahrb Syst Pflanzengesch Pflanzengeogr. 1992;114:61–79. [Google Scholar]
- 9.Kaplan DR. Comparative developmental evaluation of the morphology of Unifacial leaves in the monocotyledons. Bot Jahrb Syst. 1975;95:101–105. [Google Scholar]
- 10.Darrow GM. The Strawberry. New York: 1966. [Google Scholar]
- 11.Korona VV, Dubrovnaya SA. A Method for Evaluating the Morphological Diversity of Strawberry Leaves in Natural Populations Russian. Journal of Ecology. 2001;32:203–205. [Google Scholar]
- 12.Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 13.Vollbrecht E, Reiser L, Hake S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development. 2000;127:3161–3172. doi: 10.1242/dev.127.14.3161. [DOI] [PubMed] [Google Scholar]
- 14.Sinha N. Simple and compound leaves: reduction or multiplication? . Trends in Plant Science. 1997;2:396–402. [Google Scholar]
- 15.Sinha N. LEAF DEVELOPMENT IN ANGIOSPERMS. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:419–446. doi: 10.1146/annurev.arplant.50.1.419. [DOI] [PubMed] [Google Scholar]
- 16.Barton MK, Poethig R. Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development. 1993;119:823–831. [Google Scholar]
- 17.Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chuck G, Lincoln C, Hake S. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell. 1996;8:1277–1289. doi: 10.1105/tpc.8.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamaoki M, Kusaba S, Kano-Murakami Y, Matsuoka M. Ectopic expression of a tobacco homeobox gene, NTH15, dramatically alters leaf morphology and hormone levels in transgenic tobacco. Plant Cell Physiol. 1997;38:917–927. doi: 10.1093/oxfordjournals.pcp.a029252. [DOI] [PubMed] [Google Scholar]
- 20.Frugis G, Giannino D, Mele G, Nicolodi C, Chiappetta A, et al. Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 2001;126:1370–1380. doi: 10.1104/pp.126.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell. 1996;84:735–744. doi: 10.1016/s0092-8674(00)81051-x. [DOI] [PubMed] [Google Scholar]
- 22.Janssen BJ, Lund L, Sinha N. Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol. 1998;117:771–786. doi: 10.1104/pp.117.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet. 2006;38:942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 24.Canales C, Barkoulas M, Galinha C, Tsiantis M. Weeds of change: Cardamine hirsuta as a new model system for studying dissected leaf development. J Plant Res. 2009;123:25–33. doi: 10.1007/s10265-009-0263-3. [DOI] [PubMed] [Google Scholar]
- 25.Shani E, Burko Y, Ben-Yaakov L, Berger Y, Amsellem Z, et al. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell. 2009;21:3078–3092. doi: 10.1105/tpc.109.068148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Testone G, Bruno L, Condello E, Chiappetta A, Bruno A, et al. Peach [Prunus persica (L.) Batsch] KNOPE1, a class 1 KNOX orthologue to Arabidopsis BREVIPEDICELLUS/KNAT1, is misexpressed during hyperplasia of leaf curl disease. Journal of Experimental Botany. 2008;59:389–402. doi: 10.1093/jxb/erm317. [DOI] [PubMed] [Google Scholar]
- 27.Testone G, Condello E, Verde I, Caboni E, Iannelli MA, et al. The peach (Prunus persica [L.] Batsch) homeobox gene KNOPE3, which encodes a class 2 knotted-like transcription factor, is regulated during leaf development and triggered by sugars. Molecular Genetics and Genomics. 2009;282:47–64. doi: 10.1007/s00438-009-0445-7. [DOI] [PubMed] [Google Scholar]
- 28.Srinivasan C, Liu Z, Scorza R. Ectopic expression of class 1 KNOX genes induce adventitious shoot regeneration and alter growth and development of tobacco (Nicotiana tabacum L) and European plum (Prunus domestica L). Plant Cell Rep. 2011;30:655–664. doi: 10.1007/s00299-010-0993-7. [DOI] [PubMed] [Google Scholar]
- 29.Belles-Boix E, Hamant O, Witiak SM, Morin H, Traas J, et al. KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell. 2006;18:1900–1907. doi: 10.1105/tpc.106.041988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muehlbauer GJ, Fowler JE, Girard L, Tyers R, Harper L, et al. Ectopic expression of the maize homeobox gene liguleless3 alters cell fates in the leaf. Plant Physiol. 1999;119:651–662. doi: 10.1104/pp.119.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sentoku N, Sato Y, Kurata N, Ito Y, Kitano H, et al. Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell. 1999;11:1651–1664. doi: 10.1105/tpc.11.9.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagasaki H, Sakamoto T, Sato Y, Matsuoka M. Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell. 2001;13:2085–2098. doi: 10.1105/TPC.010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sargent DJ, Clarke J, Simpson DW, Tobutt KR, Arus P, et al. An enhanced microsatellite map of diploid Fragaria. Theor Appl Genet. 2006 doi: 10.1007/s00122-006-0237-y. [DOI] [PubMed] [Google Scholar]
- 34.Sargent DJ, Cipriani G, Vilanova S, Gil-Ariza D, Arus P, et al. The development of a bin mapping population and the selective mapping of 103 markers in the diploid Fragaria reference map. Genome. 2008;51:120–127. doi: 10.1139/g07-107. [DOI] [PubMed] [Google Scholar]
- 35.Oosumi T, Gruszewski HA, Blischak LA, Baxter AJ, Wadl PA, et al. High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta. 2006;223:1219–1230. doi: 10.1007/s00425-005-0170-3. [DOI] [PubMed] [Google Scholar]
- 36.Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, et al. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 1999;18:992–1002. doi: 10.1093/emboj/18.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell. 2002;14:547–558. doi: 10.1105/tpc.010391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sung HP, Chul MK, Byoung IJ, Su HP, Soon JP, et al. A Ds-insertion mutant of OSH6 (Oryza sativa Homeobox 6) exhibits outgrowth of vestigial leaf-like structures, bracts, in rice. Planta. 2007;2007:1–12. doi: 10.1007/s00425-007-0576-1. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura A, Tamaoki M, Sakamoto T, Matsuoka M. Over-expression of tobacco knotted1-type class1 homeobox genes alters various leaf morphology. Plant Cell Physiol. 2000;41:583–590. doi: 10.1093/pcp/41.5.583. [DOI] [PubMed] [Google Scholar]
- 40.Rosin FM, Hart JK, Horner HT, Davies PJ, Hannapel DJ. Overexpression of a knotted-like homeobox gene of potato alters vegetative development by decreasing gibberellin accumulation. Plant Physiol. 2003;132:106–117. doi: 10.1104/pp.102.015560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groot EP, Sinha N, Gleissberg S. Expression patterns of STM-like KNOX and Histone H4 genes in shoot development of the dissected-leaved basal eudicot plants Chelidonium majus and Eschscholzia californica (Papaveraceae). Plant Mol Biol. 2005;58:317–331. doi: 10.1007/s11103-005-4548-1. [DOI] [PubMed] [Google Scholar]
- 42.Harrison CJ, Corley SB, Moylan EC, Alexander DL, Scotland RW, et al. Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature. 2005;434:509–514. doi: 10.1038/nature03410. [DOI] [PubMed] [Google Scholar]
- 43.Harrison J, Moller M, Langdale J, Cronk Q, Hudson A. The role of KNOX genes in the evolution of morphological novelty in Streptocarpus. Plant Cell. 2005;17:430–443. doi: 10.1105/tpc.104.028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pautot V, Dockx J, Hamant O, Kronenberger J, Grandjean O, et al. KNAT2: evidence for a link between knotted-like genes and carpel development. Plant Cell. 2001;13:1719–1734. doi: 10.1105/TPC.010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, et al. The genome of woodland strawberry (Fragaria vesca). Nat Genet. 2011;43:109–116. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KS, Burd S, et al. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiol. 2010;154:1929–1956. doi: 10.1104/pp.110.160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development. 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, Ha D, Xie Z, Wang C, Wang H, et al. Ectopic expression of soybean GmKNT1 in Arabidopsis results in altered leaf morphology and flower identity. J Genet Genomics. 2008;35:441–449. doi: 10.1016/S1673-8527(08)60061-2. [DOI] [PubMed] [Google Scholar]
- 49.Sato Y, Sentoku N, Nagato Y, Matsuoka M. Isolation and characterization of a rice homebox gene, OSH15. Plant Mol Biol. 1998;38:983–998. doi: 10.1023/a:1006065622251. [DOI] [PubMed] [Google Scholar]
- 50.Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- 51.Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–979. doi: 10.1046/j.1365-313x.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- 52.Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- 53.Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. [Google Scholar]
- 54.Muller KJ, He X, Fischer R, Prufer D. Constitutive knox1 gene expression in dandelion (Taraxacum officinale, Web.) changes leaf morphology from simple to compound. Planta. 2006;224:1023–1027. doi: 10.1007/s00425-006-0288-y. [DOI] [PubMed] [Google Scholar]
- 55.Chang S, Puryear J, Cairney J. Simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rept. 1993;11:113–116. [Google Scholar]
- 56.Dhingra A, Bies DH, Lehner KR, Folta KM. Green light adjusts the plastid transcriptome during early photomorphogenic development. Plant Physiol. 2006;142:1256–1266. doi: 10.1104/pp.106.088351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson D. In situ hybridisation in plants; In: Bowles DJ, Gurr SJ, McPhereson M, editors. U.K.: Oxford University Press; 1991. [Google Scholar]
- 58.Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 59.El Mansouri I MJ, Valpuesta V, López-Aranda JM, Pliego F, Quesada MA. Shoot regeneration and Agrobacterium-mediated transformation of Fragaria vesca L. Plant Cell Reports. 1996;15:642–646. doi: 10.1007/BF00232469. [DOI] [PubMed] [Google Scholar]
- 60.Bent A. Arabidopsis thaliana floral dip transformation method. Methods Mol Biol. 2006;343:87–103. doi: 10.1385/1-59745-130-4:87. [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 62.Felsenstein J. Phylogenies and the comparative method. The American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR analysis for detection of full-length FaKNOX1 transcript from four independent lines of FaKNOX1 RNAi transgenic plants. Flower and runner tip mRNA were prepared as tissue samples as FaKNOX1 is expressed in high abundance in these tissue. Based on availability of samples some lines were tested with flower and some with runner tip sample. ACTIN was used as an internal control.
(TIF)
Real-time PCR analysis for quantitation of transcript of different members of KNOX family members in WT and FaKNOX1 RNAi lines. Runner tip was used as tissue samples as KNOX is expressed in high abundance in this tissue. Data from four FaKNOX1 RNAi lines are presented here, three lines from M1 class and one from M2 class.
(TIF)
RNA-gel-blot analysis for quantitation of FaKNOX1 transcript from four independent lines of FaKNOX1 OX transgenic plants. RNA was prepared from mature leaves for overexpression lines.
(TIF)
A relative real-time PCR analysis to show tissue-specific expression of FvKNOX2 - FvKNOX5 transcripts. The flower sample was used as reference tissue. Error bars represent standard error of the mean derived from three replicates.
(TIF)
Accession numbers and their corresponding genes used for phylogenetic clustering.
(DOC)