Abstract
Alternative splicing (AS) has the potential to greatly expand the functional repertoire of mammalian transcriptomes. However, few variant transcripts have been characterized functionally, making it difficult to assess the contribution of AS to the generation of phenotypic complexity and to study the evolution of splicing patterns. We have compared the AS of 309 protein-coding genes in the human ENCODE pilot regions against their mouse orthologs in unprecedented detail, utilizing traditional transcriptomic and RNAseq data. The conservation status of every transcript has been investigated, and each functionally categorized as coding (separated into coding sequence [CDS] or nonsense-mediated decay [NMD] linked) or noncoding. In total, 36.7% of human and 19.3% of mouse coding transcripts are species specific, and we observe a 3.6 times excess of human NMD transcripts compared with mouse; in contrast to previous studies, the majority of species-specific AS is unlinked to transposable elements. We observe one conserved CDS variant and one conserved NMD variant per 2.3 and 11.4 genes, respectively. Subsequently, we identify and characterize equivalent AS patterns for 22.9% of these CDS or NMD-linked events in nonmammalian vertebrate genomes, and our data indicate that functional NMD-linked AS is more widespread and ancient than previously thought. Furthermore, although we observe an association between conserved AS and elevated sequence conservation, as previously reported, we emphasize that 30% of conserved AS exons display sequence conservation below the average score for constitutive exons. In conclusion, we demonstrate the value of detailed comparative annotation in generating a comprehensive set of AS transcripts, increasing our understanding of AS evolution in vertebrates. Our data supports a model whereby the acquisition of functional AS has occurred throughout vertebrate evolution and is considered alongside amino acid change as a key mechanism in gene evolution.
Keywords: alternative splicing, nonsense-mediated decay, vertebrate evolution, RBM39
Introduction
The majority of human genes are subject to alternative splicing (AS) (Harrow et al. 2006; Tress et al. 2007; Kim et al. 2008; Wang et al. 2008; Chen and Manley 2009). AS occurs in invertebrate, plant, and fungal genomes (Cheah et al. 2007; McGuire et al. 2008; Hansen et al. 2009; Simpson et al. 2010), although its higher frequency in vertebrates suggests a link with phenotypic complexity (Kim et al. 2007). However, the vast majority of AS transcripts identified in the human genome lack described functions, and it is now clear that AS does not simply act to generate protein isoforms. Notably, the introduction of a premature termination codon (PTC) can induce the nonsense-mediated decay (NMD) pathway, causing the degradation of the transcript (Mendell et al. 2004). It has been suggested that the majority of AS in human may not generate protein isoforms (Sorek et al. 2004; Skandalis et al. 2010) and that AS may play a significant role in gene regulation (Lewis et al. 2003; Skandalis et al. 2010). In fact, 60% of AS transcripts characterized in the ENCODE project pilot phase lacked annotated CDS (Tress et al. 2007).
Because the demonstration of AS functionality in the laboratory is a low-throughput endeavor, attempts to study genome-wide AS must rely on in silico methods using proxies for functionality. Recently, several such analyses have uncovered trends in AS by studying de novo alignments of transcriptional data or existing computational genebuilds. Notably, human splicing patterns are frequently seen to correlate with tissue-specific expression profiles (Wang et al. 2008), and both tissue-specific AS exons and those conserved between human and mouse typically display elevated sequence conservation (Sorek and Ast 2003; Koren et al. 2007; Wang et al. 2008). Tissue specificity and sequence conservation are thus markers of AS functionality, although the mechanistic relationship between sequence conservation and AS remains unclear. Nevertheless, the major portion of human AS appears to be species specific, and it is clear that transposable elements (TEs) have played a significant role in the creation of new exons (Sorek et al. 2002; Amit et al. 2007; Sela et al. 2007; Gal-Mark et al. 2008; Ram et al. 2008; Schwartz et al. 2009; Sela et al. 2010). However, in this context, the definition of “functionality” becomes looser because new transcripts are likely to remain selectively neutral, while they are “tested” for functionality (Modrek and Lee 2003). Furthermore, the link between transcriptional and phenotypic complexity is unfortunately obscured by practical considerations; nucleotide entries in databases such as GenBank contain sequencing errors (Benson et al. 2010), whereas transcript libraries contain “noise,” for example, objects resulting from in vivo splicing errors. All these may cause false interpretations (Graveley 2001; Sorek et al. 2004; Pickrell et al. 2010).
In this study, we compare the AS repertoire of 309 human coding genes within the 44 ENCODE pilot regions against their mouse orthologs in a systematic manner, using conservation as an indicator of functionality; essentially, we describe the AS complement of a smaller genomic region (approximately 30 Mb) to an unprecedented level of detail. We demonstrate that the description of AS based on intensive manual annotation results in a significant gain of information, increasing AS coverage while providing greater insights into the functionality of each transcript. Our manual annotation is recognized as more accurate than computational methods in creating gene models; we provided the reference gene annotation for the ENCODE pilot regions and are now annotating the whole human genome as part of the GENCODE consortium (Guigo et al. 2006; Harrow et al. 2006; Wilming et al. 2008). This methodology is particularly suited to the identification of spurious transcripts and judges the coding potential of each valid spliceform on a case-by-case basis. By annotating human and mouse in tandem, we distinguish transcripts that are conserved in both species from those that are species specific. In addition, we utilize RNAseq data sets to validate human and mouse models that could initially be constructed based on transcriptional data from the other species due to spice site conservation, whereas a subset of such human models are also confirmed using reverse transcriptase–polymerase chain reaction (RT-PCR). We judge the age of our conserved splicing events by a comparison with nonmammalian genomes, primarily the manually annotated zebrafish genome. We also examine the sequence-level conservation of our AS events and the extent to which these exons overlap with TEs. Overall, our data set provides a highly informative assessment of the evolution of AS patterns in vertebrates, in particular regarding the potential role of NMD in functional splicing.
Materials and Methods
Genome Annotation
We had previously annotated the ENCODE pilot phase regions of the human reference genome, which approximates to 1% of the assembly (Harrow et al. 2006). These regions were originally selected to help develop methods for the identification of functional genome elements in a community-wide manner (Birney et al. 2007). Approximately 15 Mb of this sequence was selected manually, targeting genomic regions of known biological interest, whereas the other 15 Mb was selected using a stratified random approach; further information is provided in supplementary file 1 (Supplementary Material online). For this study, the existing gene models in GRCh37/hg19 were reexamined and updated as appropriate. Mouse regions orthologous to the 44 pilot sections were available from the ENCODE consortium, and genes were annotated in tandem with their human counterparts using the annotation criteria of the Vega project (Wilming et al. 2008). These detailed criteria are discussed in supplementary file 1 (Supplementary Material online). Briefly, this process uses in-house software and is based on the genomic alignment of multispecies transcriptional (expressed sequence tags [ESTs], mRNAs, and cDNAs) and protein evidence in GenBank and UniProt. Canonical splice sites were required for the construction of splice junctions. Transcript models were annotated as noncoding (including nonpolyadenylated retained introns, which cannot be distinguished from immature transcripts), CDS linked, or NMD linked. The putative annotation of NMD required the presence of an in-frame termination codon located 50 bp or more upstream of a splice junction (this is standard in NMD classification [Lewis et al. 2003]). Interspersed repeats overlapping with gene models were identified firstly as part of a standard RepeatMasker analysis included in the Vega annotation pipeline (Smit et al. 1996–2010) and secondly by querying the RepBase data set (Jurka et al. 2005). Zebrafish loci orthologous to human and mouse genes were identified using the orthology resources at ZFIN (Sprague et al. 2006); these assignments were manually queried using the Vertebrate Multiz Alignment and Conservation resources at the UCSC genome browser (Siepel et al. 2005; Pollard et al. 2010). Zebrafish annotation was performed using the same essential methodology as for human and mouse (Jekosch 2004), making additional use of RNAseq gene models constructed by S. White based on in house paired end data generated on the Illumina Genome Analyzer platform (see Ensembl release 59).
Analysis of Alternative Splicing Conservation
Exonic and intronic sequences from human and mouse were extracted, aligned, and compared using in-house software based on the Ensembl Compara API (a multispecies database containing the results of genome-wide species comparisons [Hubbard et al. 2009]). Alignments between human and zebrafish exons were performed individually using ClustalW (Larkin et al. 2007), with exonic orthology being assigned during the annotation process. Conservation in non-Vega annotated genomes was examined using the Vertebrate Multiz Alignment and Conservation resources at the UCSC genome browser (Siepel et al. 2005; Pollard et al. 2010), in conjunction with independent Blast analyses (Altschul et al. 1990) against the relevant genome using the human AS exon and flanking exons. Transcriptional support for AS events in such genomes was established using the UCSC genome browser. Two-tailed z-tests were used for statistical testing.
RNAseq Analysis
Complications remain in performing the de novo alignment of RNAseq reads to genomic sequence (Morin et al. 2008; Mortazavi et al. 2008; Wang et al. 2009); hence, the annotation process began by extracting the maximum information from the traditional transcriptomics and proteomics databanks. Subsequently, each object based entirely on evidence from another species was used to query RNAseq libraries in an attempt to provide species-specific support. For human, this was the library generated by Wang et al. (2008); for mouse, the library generated by Mortazavi et al. (2008). The RNAseq reads were mapped using GEM (http://gemlibrary.sourceforge.net), allowing up to two mismatches against a combined index including the corresponding genome sequence and annotated splice junctions. In each case, the alignments of the supporting reads were manually reanalyzed, and ambiguous matches were rejected (e.g., matches with more than one equivalent genome location). Human models based on mouse evidence were also validated against human RNAseq models generated by S. White at Ensembl, using an Exonerate-based methodology (Slater and Birney 2005) for the RNAseq Genome Annotation Assessment Project 1; information on the libraries used is presented at http://gencodegenes.org/rgasp.
RT-PCR Analysis
Each human transcript based on mouse evidence was subjected to RT-PCR validation. Double-stranded cDNA from eight human tissues (brain, heart, kidney, testis, liver, spleen, lung, skeletal muscle) was generated with the Marathon cDNA amplification kit (Clontech). Flanking primer pairs were designed to specifically target in human the AS form identified in mouse. Primer sequences are included in supplementary file 2 (Supplementary Material online). The PCRs were performed with JumpStart REDTaq ReadyMix (Sigma). For each tissue, 2 μl of each RT-PCR reaction were pooled together (a total of 760 reactions, the majority of which were not related to the present study) and purified with the QIAquick PCR purification Kit (QIAGEN). This purified DNA was used to generate a sequencing library with the “Genomic DNA sample prep kit” (Illumina) according to the manufacturer's recommendations, minus the fragmentation step. This library was subsequently sequenced on an Illumina Genome Analyzer 2 platform. The 75-bp reads were mapped onto both the reference human genome (hg19) and the predicted spliced amplicons using bowtie 0.12.5 (Langmead et al. 2009). Uniquely mapping reads with no mismatch were used to validate splice sites. Splice junctions were validated if spanned by a minimum of 10 reads with a minimum of 8 nucleotides flanking the breakpoints.
Results and Discussion
Generation of the AS Data Set
Human and Mouse Gene Annotation
We defined 309 pairs of orthologous protein-coding genes in the human and mouse genomes; these are listed in supplementary file 2 (Supplementary Material online). Table 1 categorizes each individual transcript constructed for these gene pairs based on its conservation status and functional annotation. Transcripts are divided into: “conserved” transcripts supported by equivalent transcriptional evidence in both species, human and mouse specific transcripts (collectively referred to as “species specific”), and transcripts built in one species using mRNAs or ESTs from the other on the basis of splice site and reading frame conservation (“cross-species transcripts,” separated into “human to mouse” and “mouse to human”). Finally, the “status change” category includes a small number of transcripts that were not added cross-species in spite of splice site conservation because their coding category would have been changed. Further analysis is limited to coding transcripts (i.e., CDS and NMD), and redundancy due to untranslated region (UTR) variation was eliminated; analysis of conserved AS in UTRs is complicated by the decoupling of functional and positional conservation in vertebrate promoter regions (Birney et al. 2007; Schmidt et al. 2010).
Table 1.
Total Transcripts Built within 309 Pairs of Orthologous Human and Mouse Protein Coding Loci
| Human | Coding | Noncoding | NMD | CDS | Conserved | Hs specific | Cross-species Hs | Status change | Cross-species Mm |
| Total | 1,675 | 1,049 | 324 | 1,351 | |||||
| Nonredundant | 1,264 | 424 | 464 | 260 | 15 | 101 | |||
| NMD | 288 | 22 | 146 | 89 | 4 | 27 | |||
| CDS | 976 | 402 | 318 | 171 | 11 | 74 | |||
| Mouse | Coding | Noncoding | NMD | CDS | Conserved | Mm specific | Cross-species Mm | Status change | Cross-species Hs |
| Total | 1,207 | 748 | 193 | 1,014 | |||||
| Nonredundant | 979 | 424 | 189 | 101 | 5 | 260 | |||
| NMD | 180 | 22 | 41 | 27 | 1 | 89 | |||
| CDS | 799 | 402 | 148 | 74 | 4 | 171 |
NOTE.—Hs, human; Mm, mouse; cross-species Hs, added to mouse based on human evidence; cross-species Mm, added to human based on mouse evidence; status change, cross-species transcripts with conserved splice sites though nonconserved CDS/NMD annotation. Gray shading highlights transcripts supported by evidence in that species.
Classification of Human and Mouse AS
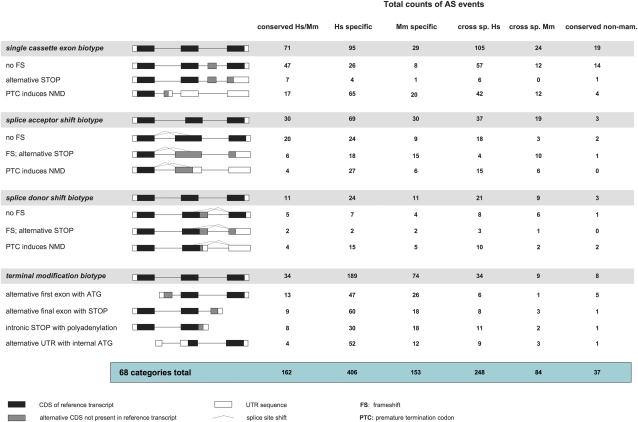
For each locus we classified, a “reference” transcript against which AS events could be compared (essentially the conserved CDS transcript with the most transcriptional support; see supplementary file 1, Supplementary Material online). Supplementary file 3 (Supplementary Material online) divides the AS transcripts from table 1 into 68 categories based on their structure and coding annotation (henceforth referred to as “biotypes”), considering only the nature of the AS event of interest and not the whole transcript (this creates the small differences in total counts seen between the two data sets; see supplementary file 1, Supplementary Material online). Subsequent analysis is limited to the two major biotype categories presented in figure 1: interior modifications, representing splice site shifts and single cassette exons (cassettes) that are flanked by coding exons present in the reference transcript, and terminal modifications, where a new or modified terminal coding exon is linked to a variant CDS. Multi-cassettes are thus ignored in subsequent calculations unless stated; these cassettes combine to generate the final coding category and are low in number.
FIG. 1.
Summary of interior and terminal modification alternative splicing biotypes. This figure summarizes the most common biotype categories in the data set (interior and terminal modifications); a complete set of 68 biotypes is represented in supplementary file 3 (Supplementary Material online). The total numbers highlighted in blue relate to the full 68 biotypes, whereas the total numbers of AS event within each biotype category are highlighted within the gray bars. Each entry represents the categorization of a particular AS event described within the context of the reference transcript structure and not the complete transcript itself; for this reason, the total counts differ slightly from those in table 1 (see supplementary file 1, Supplementary Material online, for further information). From left to right, “conserved Hs/Mm” lists events supported in human and mouse, “Hs-specific” and “Mm-specific” events that are species specific, “cross sp. Hs” and “cross sp. Mm” events that could be aligned cross-species, and “conserved non-mam.” events for which AS is also supported in nonmammals (this is thus a subset of “conserved Hs/Mm”; see supplementary file 2, Supplementary Material online). Although cassette events are pictorially represented by the insertion of an additional exon, these categories also include the “skipping” of exons from the reference transcript. Cassette counts do not include multiple exon events or mutually exclusive exon pairs. For splice site shifts, the counts for shifts in both the 5′ and 3′ directions have been combined. Furthermore, these counts do not distinguish between alternative STOPs and PTCs found within the AS region and those that appear downstream out of frame with the reference CDS. In all cases, these biotypes are distinguished in supplementary file 3 (Supplementary Material online).
Validation of Cross-Species Transcripts
We identified 134 conserved AS events based on our initial use of traditional transcriptomics data. Table 1 and figure 1 also incorporate RNAseq and RT-PCR data. First, our RNAseq analysis using existing libraries provided support for 9 human transcripts initially based on mouse mRNAs or ESTs and 12 mouse transcripts initially based on human evidence (see Materials and Methods). Second, we provided support for three additional human transcripts by comparison to RNAseq gene models produced by the Ensembl group (S. White, unpublished data). Finally, we validated a further four mouse-supported AS events in human via RT-PCR (see Materials and Methods). Overall, we reclassified 28 AS events as supported in both species, giving 162 events in total. This 20.9% increase demonstrates the value of comparative annotation in predicting AS events for validation and confirms that existing EST and mRNA databases for human and mouse are nonsaturated.
Analysis of Species-Specific Splicing
The Human Data Set Contains More AS Transcripts than Found in Mouse
Considering all data in table 1, we have annotated 769 more transcripts in human than in mouse; human contains an extra 1.0 noncoding transcripts and 1.5 coding transcripts per gene. However, the Expressed Sequence Tags database contains approximately 1.7 times as many human ESTs as mouse (Boguski et al. 1993), and the detection of AS levels is related to EST coverage (Kan et al. 2002; Kim et al. 2004, 2007). Although the total transcript counts cannot be interpreted with confidence, a comparison of the CDS to NMD ratios in each species may be informative. Supplementary file 4 (Supplementary Material online) summarizes the CDS and NMD transcripts highlighted in gray in table 1 as a Venn diagram, thus considering for both species only transcripts supported by evidence from that species. We observe a higher proportion of NMD transcripts in human: 22.4% of the 1,163 human supported coding transcripts are NMD, compared with 12.7% of the 719 mouse supported. These values suggest an increase in NMD-linked transcription in human, although we cannot eliminate the possibility that they are affected by inconsistencies in the creation of the various human and mouse EST libraries. Figure 1 indicates that the excess of human NMD transcripts is largely explained by a similar excess of cassettes; 61.5% of the 174 NMD events in the human-specific and added cross-species to mouse categories are linked to single cassettes, whereas there are 3.8 times as many cassette events in these categories as in their mouse equivalents. Here, we define cassette events as whole exon exclusions (skips) or insertions; cassettes that generate alternative CDS are referred to as “CDS cassettes” and those predicted to induce the NMD pathway as “NMD cassettes.” Exon skipping has been reported as the prevalent form of AS in metazoans (Kim et al. 2007). However, human cassette events do not induce NMD at a higher frequency compared with mouse; 68.4% and 69.0% of the respective species-specific cassettes are NMD cassettes.
AS Exons Are Formed from Transposable Elements and Noncoding Sequence in Human and Mouse
AS in human has been linked to the expansion of Alu elements in the genome, and such elements are absent in mouse (Sorek et al. 2002; Amit et al. 2007; Sela et al. 2007; Gal-Mark et al. 2008; Ram et al. 2008; Schwartz et al. 2009; Sela et al. 2010). Furthermore, the exonization of TEs has been reported as more widespread in the human genome compared with mouse (Sela et al. 2007). We thus examined the association between species-specific AS events and TEs in our data set; these data are presented in supplementary file 2 (Supplementary Material online). If the excess of human cassettes is linked to TEs, we would expect a similar excess in the proportion of cassettes overlapping TE sequences. Each species-specific cassette event represents an insertion; a human skip will inevitably be added to mouse provided the flanking exons are present in both species. We observe that 49.2% of human-specific NMD cassettes overlap with TEs (typically Alus), compared with 45.0% in mouse. The respective values for CDS cassettes are 40.0% and 33.3%. The two species also possess comparable rates of TE association in the terminal modification biotypes. For example, 21.3% of alternative first coding exons and 46.7% of alternative final coding exons overlap with TEs in human, compared with 23.1% and 55.6%, respectively, in mouse. In contrast, we observe a lower association between TEs and splice site shifts; of the total 75 human and mouse-specific shifts that extend into intronic sequence, only 6 overlap with TEs. In summary, the exonization of TEs apparently does not explain any human excesses in the major biotypes categories; human possesses similar excesses in the proportions of events that are not linked to TEs.
However, although the majority of species-specific AS events appear unlinked to TEs, some may have formed from TEs whose sequences have degraded past the point of recognition. Alternately, a species-specific AS event may actually signify a loss of function in the reciprocal species, for example, the degradation of splicing capability. However, we do not detect convincing genomic or transcriptomic evidence for the existence of any human or mouse-specific AS events prior to the formation of the rodent/primate clade (data not shown). Finally, new AS events may have been created by localized exonic duplication; again, we see no evidence for this within our data set (data not shown). Instead, we infer on the basis of parsimony that AS creation unlinked to TEs is largely due to the exonization of intronic or nongenic sequence, presumably where splicing motifs are created by point mutation. In summary, our analysis indicates that TEs and non-transposon sequence play similarly important roles in the creation of new AS events in the human and mouse genomes.
Analysis of AS Conserved between Human and Mouse
Gene Regulation by NMD Is Widespread in the Human and Mouse Genomes
Our 162 conserved AS events represent one CDS transcript per 2.3 genes, and one NMD transcript per 11.4 genes; these events are contained within 95 out of 309 genes (supplementary files 2 and 3, Supplementary Material online). Considering all 68 AS biotypes in supplementary file 3 (Supplementary Material online), the cassette is the most common AS form conserved between human and mouse (48.8%). Furthermore, although 68.4% of human-specific single cassettes are NMD cassettes, 76.1% of conserved single cassette events are CDS cassettes (fig. 1), supporting an assumption that functional AS chiefly acts to generate protein isoforms. Nonetheless, our identification of conserved NMD is interesting, although not unprecedented. It has been reported that most mammalian NMD transcripts are not under selective pressure and therefore likely to be noise (Pan et al. 2006). However, members of the SR (serine–arginine) family of splicing factors use NMD isoforms to downregulate their own translation in a homeostatic manner, and it is speculated that this strategy (regulated unproductive splicing and translation or “RUST”) may be common to splicing factors generally (Sureau et al. 2001; Lareau et al. 2007). We have identified 27 NMD-inducing AS events that are conserved between human and mouse. Of these 27, RBM39, SF1, SON, and LUC7L have demonstrated or putative roles as splicing factors. Although these four genes are not categorized as SR proteins, we predict that they may be subject to RUST. Although functional information is not available for all the remaining 23 genes, most do not appear to function in splicing; PISD, for example, is involved in the formation of phosphatidylethanolamine. Our analysis therefore suggests that RUST may be a widespread mechanism, not limited to particular gene families.
Conserved AS Events Can Be Formed from Transposon Sequence
Two of the 162 conserved AS events are linked to TEs, indicating that exons created by this mechanism may acquire genuine functionality (see supplementary file 5, Supplementary Material online). First, the splice acceptor site and CDS portion of an alternative final exon of the OBSL1 gene are contained within a CHARLIE8 element, a mammal-specific TE (Jurka et al. 2005). The splice site and intact CDS are found in all placental mammal genomes for which sequence is available, and the absence of the STOP codon in the TE consensus sequence suggests a gain of function mutation. The most divergent alignment identified is to the opossum genome, where sequence conservation is weaker and the CDS disrupted. Second, a 69-bp cassette of the ST7 gene is embedded within an L3 element. This exon is 100% identical between human and mouse at the sequence level; it is in fact 99–100% identical among all placental mammals, although apparently absent outside eutherians. This suggests the exon may have been rapidly subjected to purifying selection following its incorporation into the ST7 gene.
A Subset of AS Exons Display an Increase in Sequence Conservation
It is well established that AS exons in general display elevated sequence conservation, often extending into intronic regions (Sorek and Ast 2003; Sorek et al. 2004; Koren et al. 2007). We sought to reappraise this association in the context of our wider range of AS biotypes and conservation categories. We have calculated the percentage sequence similarity between human and mouse of each conserved AS exon (exonic similarity scores), as well as the scores for the 1,118 constitutive exons within these same genes for comparison (ignoring terminal CDS exons). Similarly, we have calculated the percentage identity of the flanking intronic sequences in 40-bp and 100-bp windows from all splice donor and acceptor sites for both exon sets (intronic similarity scores). We define 95% sequence identity as our marker for significance in the 40-bp window because below 1.5% of the total 5′ and 3′ flanks in the control set exceed this value. All scores calculated for AS exons are presented in supplementary file 2 (Supplementary Material online).
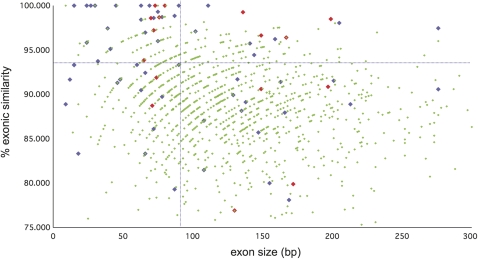
In total, 74.2% of conserved AS events represent interior modifications; one conserved cassette event per 3.9 genes and one conserved splice site shift per 7.5 genes (supplementary file 3, Supplementary Material online). Figure 2 plots the exonic similarity scores of constitutive exons, conserved single CDS cassettes, and conserved single NMD cassettes. The tendency of cassettes to be shorter than constitutive exons has been reported (Sorek et al. 2004). Here, 62.3% of conserved cassettes are shorter than the constitutive lower quartile of 90 bp, whereas 25.9% of CDS cassettes are under 30 bp in size, compared with 1.0% of constitutive cassettes. The average exonic similarity score is 88.1 ± 5.5% for constitutive exons, and we observe significantly elevated (z-test, P < 0.001) similarities of 90.7 ± 11.1% and 91.7 ± 10.4% for CDS and NMD cassettes, respectively. In fact, 14 of these 71 exons are 100% conserved between human and mouse, compared with 5 out of 1,118 constitutive cassettes; a 44-fold excess. In addition, 30 cassettes have 95% or above intronic similarity scores over one or both 40-bp flanks, a 27.8-fold excess over the control set. However, although our data confirm that exonic conservation, short size, and intronic conservation are general attributes of cassette-based AS, it is clear from figure 2 that none of these attributes are universal; 29.9% of AS cassettes are larger than the constitutive mean and 22.1% have exon similarity scores below the constitutive mean.
FIG. 2.
The percent identity of human and mouse AS cassette exons plotted against exon size. Only AS single cassette events that are transcriptionally supported in both species are included. “% exonic similarity” represents the percentage similarity of each orthologous AS cassette pair at the base pair level. Cassettes linked to CDS are blue, whereas those linked to NMD are red. For comparison, the 1,118 constitutive internal exons for those genes displaying conserved AS are plotted in green. A cut-off of 300 bp has been used for exon size. The vertical dotted line marks the first quartile value of the median exon size, whereas the horizontal dotted line marks the upper standard deviation limit of the average exon percentage identity. AS cassettes linked to both CDS and NMD tend to be shorter and more similar at the sequence level than the control exons. However, this does not hold true for all AS cassettes; for example, although 14 have exon scores of 100%, 17 fall have scores falling below the average score for the control set exons.
Because splice site shifts affect exon edges, whole exon conservation and exon size are not informative metrics; the largest and smallest exons in this category are 21-bp exon and 1,491 bp, respectively. Instead, we note that 15 out of 41 shifts associate with elevated 40-bp intronic conservation at the relevant flank (8 acceptors and 7 donors), a similar enrichment over the constitutive set as seen for cassette exons. Again, however, elevated sequence conservation is not a universal attribute of splice site shifts. The most striking examples of conservation are from the ENPP1 and GCFC1 genes, both of which induce NMD. For ENPP1, the 40-bp intronic flank of the shifted splice site is 100% identical between human and mouse. A degree of conservation would be expected because the splice site is shifted into the interior of the reference transcript exon; however, this 40-bp region is fully conserved in the chicken, lizard, and Xenopus genomes, that is, over synonymous and nonsynonymous codon positions. This conserved region—like the majority of those identified here—does not appear to have been reported by any previous whole-genome investigation, likely due to its short size coupled with the fact that it overlaps exonic sequence. In the case of GCFC1, both alternative splice sites are found within a 31-bp region that is 100% identical in 43 out of 46 vertebrate genomes, including four fish species (data not shown).
The average exonic similarity score of the 13 alternative first exons (minus the 5′ UTR) is 81.9 ± 8.1%, compared with 90.4 ± 7.2% for the constitutive first exons. All alternative first exon scores fall under 95%, and we detect no evidence for elevated intronic conservation scores. In contrast, six of the nine alternative final exons have exonic similarity scores exceeding the upper standard deviation of the average score for constitutive final exons (87.6 ± 8.4%). In addition, six are associated with elevated intronic conservation over the splice acceptor 40-bp window, compared with 2 out of 86 constitutive final exons. Finally, we detect no signatures of sequence conservation for final coding exons that utilize intronic STOPs; the average exonic similarity score is 81.1 ± 9.6%, with all these values falling below 95%. Although these low counts prevent a robust statistical analysis, our preliminary findings generally support a model whereby elevated sequence conservation at the splice acceptor site is linked to the control of AS. In short, the incorporation of alternative terminal exons requires the selection of the splice acceptor site prior to the usage of a polyadenylation site, whereas alternative promoters likely control the selection of alternative first exons. Similarly, the usage of a retained intron STOP would seem to depend on the rejection rather than selection of a splice site, followed again by polyadenylation. However, we emphasize the need to study terminal modifications as part of a larger sample size.
Analysis of AS in Nonmammalian Genomes
A Quarter of Conserved Mammalian AS Events Occur in Nonmammalian Genomes
We next examined the relationship between sequence conservation at the AS event and the age of the splicing event. AS is not restricted to mammalian genomes; its activity is particularly well studied in fish, Drosophila, and nematodes (Stolc et al. 2004; Hansen et al. 2009; Graveley et al. 2010; Lu et al. 2010; Nilsen and Graveley 2010; Ramani et al. 2011). Initially, we focused our analysis on the zebrafish genome and manually annotated the 89 out of 95 genes for which orthology could be established (see Materials and Methods). We searched for further AS support in all other nonmammalian vertebrate genomes, using a simpler database mining approach (see Materials and Methods). In total, 31 out of 95 genes (32.6%) were seen to display conserved AS patterns beyond mammals (i.e., there is transcriptional support for both the reference and AS events in the same genome). This represents 37 out of 162 AS events (22.9%), including 19 described in zebrafish and 14 in Xenopus; these data are presented in supplementary file 2 (Supplementary Material online) and integrated into figure 1. This work may underestimate the true extent of splicing conservation because transcriptional coverage in nonmammalian genomes is less complete than in human or mouse.
AS Exons Present in Nonmammals Display Elevated Sequence Conservation
The average exonic score for the 37 AS events supported in human, mouse, and at least one nonmammal vertebrate (henceforth “nonmammalian AS”) is 93.5 ± 5.8% between human and mouse, compared with 88.5 ± 10.1% for those 125 events where AS is supported in mouse and human though not outside mammals (henceforth “mammal-only AS”). We thus detect an association between elevated conservation at the sequence level and nonmammalian conservation at the splicing level (z-test P < 0.001). Of these 37 events, 29 are interior modifications; 10 of these AS exons are 100% identical between human and mouse, compared with 6 of the 92 mammal-only interior modification AS exons (a 5.5 times enrichment). Notably, 22.2% of conserved NMD events occur in nonmammalian genomes, often linked to striking sequence conservation; the GCFC1 splice shift event noted above occurs in human and zebrafish alongside 100% regional sequence conservation between the two genomes. Five other genes undergo NMD-linked AS in nonmammals, including all four splicing factors discussed above: SON, SF1, RBM39, and LUC7L. We therefore demonstrate that the conservation of NMD-linked AS in splicing factors can extend far beyond the mammalian order. However, GCFC1 has a postulated role in transcriptional regulation via DNA binding, whereas the remaining gene CTTNBP2 associates with the actin cytoskeleton in rat (Ohoka and Takai 1998). Unlike for GCFC1, we do not observe any obvious elevated conservation surrounding the alternative CTTNBP2 splice donor site.
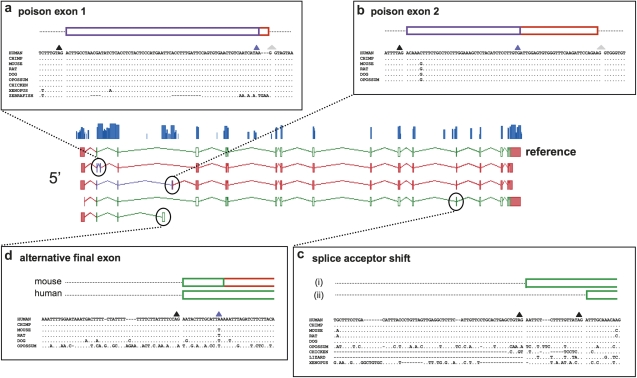
RBM39: The Evolution of AS within a Single Gene
Our data indicate that conserved human and mouse AS events were not created at a single point in vertebrate evolution, and this statement holds true even within a single gene. RBM39 contains significant splice variation in human and mouse; 42 transcripts have been annotated in the former and 24 in the latter, whereas 6 AS events are supported in both. RBM39 is a potential splicing factor, linked to both AS and constitutive exons (Cazalla et al. 2005); it may also function in transcriptional regulation (Dutta et al. 2008). However, to our knowledge the differential functionality of individual RBM39 transcripts remains unstudied. Four conserved AS events of particular interest are depicted in figure 3. This includes two NMD cassettes containing PTCs (poison exons), the first of which is 98.6% identical between human and Xenopus (fig. 3a), and 69.9% identical between human and zebrafish (where it also displays AS). However, although the second NMD cassette is 99–100% identical among all mammalian genomes, it cannot be aligned to nonmammalian genomes (fig.3b); it appears to represent a more recent evolutionary innovation. Although we can predict that RBM39 is subject to RUST, the fact that this gene utilizes two distinct poison exons in mammals raises the possibility that the two transcripts are generated under the control of distinct regulatory processes. Further downstream, an RBM39 exon possesses a pair of alternative splice acceptor sites within an 80-bp region that is 100% identical between all but three available placental mammal genomes, although only 68.5% identical to opossum (fig.3c). Although the downstream site is conserved in all genomes back to Xenopus (where AS is supported), the upstream site in only found in placental mammals, where AS is supported in 11 species (data not shown). This AS event is thus apparently younger than either poison exon event. Finally, RMB39 contains an alternative final exon in human and mouse (fig.3d), the splice acceptor site of which is found in all placental mammal genomes and not beyond. Because human and mouse use nonequivalent termination codons, this exon may represent a genuine functional difference between the two species. However, at this point, we reach the limits of what can be extrapolated from an annotation-based analysis.
FIG. 3.
The conservation of four alternative splicing events within RBM39. The RMB39 gene for RNA binding motif protein 39 contains six AS events conserved between human and mouse, four of which are highlighted here. The central panel shows the structure of these four splice variants in human (5′ and 3′ UTRs shown in red; CDS in green; NMD region in purple) and the Phastcons conservation plot. The peripheral panels contain genomic alignments taken initially from the conservation track resources at the UCSC genome browser (Siepel et al. 2005; Pollard et al. 2010). Splice acceptor sites of the form [N/GT] are represented by black triangles, splice donor sites of the form [NAG] by gray triangles, and termination codons by blue triangles. Two of the AS events are “poison” exons, introducing PTCs predicted to induce NMD. (a) Poison exon 1 is 98.6% identical at the base pair level between human and Xenopus and has transcriptional support in zebrafish. The UCSC alignment of the zebrafish splice donor site has been corrected based on manual analysis, whereas the PTC used in this genome is found in the subsequent exon. (b) In contrast, poison exon 2 cannot be aligned in any genomes beyond that of opossum. (c) Two AS acceptor sites are found for this exon in human and mouse; the first (i) is limited to mammalian genomes, whereas the second (ii) exists (often with a “wobble” on the first base pair) in all genomes back to Xenopus (where there is transcriptional support). (d) The alternative final exon uses a splice site found in mammals genomes only, although the STOP codon seen in mouse and other mammalian genomes is absent in apes; the human and mouse CDS are thus significantly different.
Conclusions
We have demonstrated the value of intensive manual annotation for systematically describing and comparing the AS content of vertebrate transcriptomes. We have analyzed in detail the relationships between predicted AS transcript functionality, TE association, and AS conservation at the transcript and sequence levels. Although each of these phenomena had been previously described to some degree, ours is the first investigation to combine each of these strands of analysis into a single picture of gene evolution. Our analyses have focused on 1% of the human genome, and we do not “scale up” our values here due to sampling biases inherent in the creation of the ENCODE pilot regions. However, we believe that we have reported the most accurate values yet obtained for these attributes at the gene level, achieving a level of detail that approaches exhaustive within the limits of current technology.
First, our comparison of human and mouse AS levels indicates that although just 13.5% of human and 19.1% of mouse transcripts are supported in both genomes, one AS event is conserved per 1.9 multiexon genes. Although conserved CDS transcripts outnumber NMD transcripts 5-fold, we infer that gene regulation by NMD may be more common than previously thought (conserved in 9% of human/mouse genes) and not particular to splicing factors. In contrast, although our conservation-based approach alone cannot confirm the functionality of several hundred species-specific CDS, their annotation will be an essential first step in the mining of incipient large-scale proteomics data sets. Second, our results indicate that novel human and mouse exons are created at similar rates from TE and non-TE sequence. Third, we have shown that the patterns of AS biotype association are different between conserved and nonconserved AS events and between TE-linked and nonlinked exons. Following this, our results indicate that elevated sequence conservation is an attribute of specific conserved AS events and more common to certain biotypes, whereas the ST7 cassette exon indicates that elevated conservation can also be coupled to TE-derived AS events. Finally, we have demonstrated that at least a quarter of conserved human/mouse AS events predate the mammalian order and have shown for the first time that this includes NMD-linked events. In combination, these final two observations indicate that while AS events have arisen throughout vertebrate evolution, elevated conservation (where observed) often appears at a significantly later time. We speculate that AS events may undergo increased purifying selection when they become coupled to new regulatory processes (perhaps linked to tissue or developmental specificity), and that this may occur at any point in evolution after the creation of the AS event.
In conclusion, the original paradigm for gene evolution centers on the adaptive potential of amino acid changes in a single protein molecule. In 2003, however, Modrek and Lee presented a model in which AS represents an alternate pathway for gene evolution, whereby novel functionality can evolve without disruption to the main protein isoform (Modrek and Lee 2003). Almost a decade later, we believe that the contribution of AS to gene evolution remains largely unexplored and still unappreciated; AS is routinely ignored in studies considering the evolution of single genes. In fact, we believe that AS may prove to be the more significant form of ongoing gene evolution, particularly for genes where the main protein product is under strong selective constraint. For example, the 324 aa reference CDS of human RBM39 differs from the mouse CDS at a single residue, whereas the chicken CDS has just nine differences. When we change our focus to the AS complement of RBM39, however, a gene that appeared inert in terms of evolution is seen to show dramatic change between species. Overall, our work demonstrates that we cannot truly understand the genome of a species until we have characterized its AS complement and that our view of evolution will remain incomplete until we understand how these patterns have changed between species.
Supplementary Material
Supplementary files 1–5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
Wellcome Trust (grant number WT077198/Z/05/Z to J.M.M., A.F., J.F.-B., T.H., and J.H.); National Institute of Health (grant number 5U54HG004555 to J.M.M., A.F., J.F.-B., T.H., and J.H.); Ministerio de Ciencia e Innovación/Instituto Nacional de Bioinformatica (ISCIII GNC-1 to R.G., T.A., and T.D.); the Ministerio de Educación y Ciencia (BIO2006-03380 to R.G., T.A., and T.D.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amit M, Sela N, Keren H, Melamed Z, Muler I, Shomron N, Izraeli S, Ast G. Biased exonization of transposed elements in duplicated genes: a lesson from the TIF-IA gene. BMC Mol Biol. 2007;8:109. doi: 10.1186/1471-2199-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. 420 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST–database for "expressed sequence tags. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Cazalla D, Newton K, Caceres JF. A novel SR-related protein is required for the second step of Pre-mRNA splicing. Mol Cell Biol. 2005;25:2969–2980. doi: 10.1128/MCB.25.8.2969-2980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta J, Fan G, Gelinas C. CAPERalpha is a novel Rel-TAD-interacting factor that inhibits lymphocyte transformation by the potent Rel/NF-kappaB oncoprotein v-Rel. J Virol. 2008;82:10792–10802. doi: 10.1128/JVI.00903-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mark N, Schwartz S, Ast G. Alternative splicing of Alu exons–two arms are better than one. Nucleic Acids Res. 2008;36:2012–2023. doi: 10.1093/nar/gkn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, et al. (11 co-authors) The developmental transcriptome of Drosophila melanogaster. Nature. 2010 doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigo R, Flicek P, Abril JF, et al. (18 co-authors) EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006;7(Suppl. 1):S2.1–S31. doi: 10.1186/gb-2006-7-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Lareau LF, Blanchette M, et al. (11 co-authors) Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, et al. (15 co-authors) GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S4.1–S9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, et al. (58 co-authors) Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jekosch K. The zebrafish genome project: sequence analysis and annotation. Methods Cell Biol. 2004;77:225–239. doi: 10.1016/s0091-679x(04)77012-0. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kan Z, States D, Gish W. Selecting for functional alternative splices in ESTs. Genome Res. 2002;12:1837–1845. doi: 10.1101/gr.764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G. Alternative splicing: current perspectives. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Klein R, Majewski J, Ott J. Estimating rates of alternative splicing in mammals and invertebrates. Nat Genet. 2004;36:915–916. doi: 10.1038/ng0904-915. ; author reply: 916–917. [DOI] [PubMed] [Google Scholar]
- Koren E, Lev-Maor G, Ast G. The emergence of alternative 3' and 5' splice site exons from constitutive exons. PLoS Comput Biol. 2007;3:e95. doi: 10.1371/journal.pcbi.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. (13 co-authors) Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Peatman E, Wang W, Yang Q, Abernathy J, Wang S, Kucuktas H, Liu Z. Alternative splicing in teleost fish genomes: same-species and cross-species analysis and comparisons. Mol Genet Genomics. 2010;283:531–539. doi: 10.1007/s00438-010-0538-3. [DOI] [PubMed] [Google Scholar]
- McGuire AM, Pearson MD, Neafsey DE, Galagan JE. Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol. 2008;9:R50. doi: 10.1186/gb-2008-9-3-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, McDonald H, Varhol R, Jones S, Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoka Y, Takai Y. Isolation and characterization of cortactin isoforms and a novel cortactin-binding protein, CBP90. Genes Cells. 1998;3:603–612. doi: 10.1046/j.1365-2443.1998.00216.x. [DOI] [PubMed] [Google Scholar]
- Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet. 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Schwartz S, Ast G. Multifactorial interplay controls the splicing profile of Alu-derived exons. Mol Cell Biol. 2008;28:3513–3525. doi: 10.1128/MCB.02279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, et al. (11 co-authors) Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 2011;21:342–348. doi: 10.1101/gr.114645.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, et al. (13 co-authors) Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Gal-Mark N, Kfir N, Oren R, Kim E, Ast G. Alu exonization events reveal features required for precise recognition of exons by the splicing machinery. PLoS Comput Biol. 2009;5:e1000300. doi: 10.1371/journal.pcbi.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela N, Mersch B, Hotz-Wagenblatt A, Ast G. Characteristics of transposable element exonization within human and mouse. PLoS One. 2010;5:e10907. doi: 10.1371/journal.pone.0010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, et al. (16 co-authors) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CG, Manthri S, Raczynska KD, et al. (11 co-authors) Regulation of plant gene expression by alternative splicing. Biochem Soc Trans. 2010;38:667–671. doi: 10.1042/BST0380667. [DOI] [PubMed] [Google Scholar]
- Skandalis A, Frampton M, Seger J, Richards MH. The adaptive significance of unproductive alternative splicing in primates. RNA. 2010;16:2014–2022. doi: 10.1261/rna.2127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2010. http://www.repeatmasker.org. [Google Scholar]
- Sorek R, Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Clements D, et al. (17 co-authors) The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, et al. (12 co-authors) A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress ML, Martelli PL, Frankish A, et al. (46 co-authors). The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci U S A. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.