Abstract
Background
We recently discovered that the enzyme indoleamine 2,3-dioxygenase (IDO) is overexpressed in primary pancreatic ductal adenocarcinomas (PDA) and in lymph node metastases (J Am Coll Surg. 5:849-54: 2008). IDO2 is a recently discovered relative of IDO that has unique signaling properties (Cancer Res. 67:7082-7087: 2007). Notably, the IDO2 gene has two functional polymorphisms commonly found in human populations that abolish its enzymatic activity (R235W and Y359STOP). Both IDO and IDO2 repress the immune system and we hypothesize that expression of these enzymes in PDA may help cancer cells evade immune detection.
Methods
Based on evidence that the IDO2 may be a preferential target of D-1-methyl-tryptophan (1-MT), a clinical lead inhibitor of IDO currently being evaluated in Phase I trials, we sequenced IDO2 in 36 resected PDAs and evaluated its expression in relation to the two known genetic polymorphisms.
Results
In our patient cohort, we found that 58% (21/36) of the cases were heterozygous for the R235W polymorphism; 28% (10/36) were homozygous wild-type; and only 14% (5/36) were homozygous for the functionally inactive polymorphism. Interestingly, IDO2 had a homozygous wild-type configuration in two pancreatic cancer cell lines whereas one cell line (MiaPaCa2 cells) was homozygous for the R235W polymorphism. As for the Y359STOP polymorphism (seen in the cell line Hs766T), we found that 27% (10/36) of the cases were heterozygous, 62% (22/36) were homozygous wild-type, and only 11% (4/36) were homozygous for this functionally inactive allele. Ruling out the possibility of compound polymorphic variants, we estimated 75% of our resected patient cohort had an active IDO2 enzyme with a conservative estimate that 58% of the patients had at least one functional allele. In immunohistochemical analyses, we found that IDO2 was equally overexpressed in pancreatic cancer tissue from each genetially polymorphic subgroup. We also detected IDO2 protein expression in the genetically distinct pancreatic cancer cell lines after exposure with IFN-γ, establishing that even functionally polymorphic IDO2 sequences can generate IDO2 protein.
Conclusions
These are the first data to report IDO2 expression in PDA and indicate that IDO2 genetic polymorphisms do not negate IFN-γ-inducible protein expression. IDO2 genotyping and expression analysis of our PDA patient tissue bank and cell lines show that IDO2 is active and expressed in a majority of PDA patients. Taken together, these data strongly suggest that the clinical lead compound D-1-MT acting through IDO2 might be useful in treatment of PDA, either alone or in combination with other anti-tumor modalities.
Introduction
Ongoing efforts are unraveling the key genetic events and pathways required for the development of pancreatic cancer (1). This molecular knowledge should pave the path towards individualized treatment of this disease. However, altered genetics and signaling pathways in pancreatic cancer cells themselves may not be the only contributor to the complex process of pancreatic tumorigenesis. Further, recent attempts to target pancreatic cancer cells at the molecular level have met with little success, leaving surgery as the best therapeutic option (2, 3). Recently, several lines of research are converging to suggest that the immune system is important for tumor surveillance and in certain instances tumor killing (4). Hence, understanding how pancreatic cancer cells avoid immune detection may provide further insight and potential targets for therapy.
We previously studied an immune modulatory enzyme in pancreatic cancer, indoleamine 2,3-dioxygenase (IDO)(5), that mediates tryptophan catabolism via the kynurenine pathway (6). Notably, several studies have shown that IDO functions in facilitating the conversion of naive T lymphocytes into T regulatory cells (TRegs) (6-8). Therefore, IDO expression may foster an immuno-tolerant environment (8-12). Consistent with a role for IDO in mediating tolerance to tumors, preclinical have shown the promise of IDO inhibitors in targeting several cancers (9, 13-15). In particular, the racemic compound 1-methyl-tryptophan (1MT composed of l- and d-isomers) has been widely studied as a potent bioactive inhibitor of IDO activity (9, 10). Interestingly, recent studies found that the racemer d-1MT, which displays greater anti-tumor potency than l-1MT (9), preferentially targets an IDO-related enzyme encoded by the IDO2 gene (also termed INDOL1 or IDO like -1, Hs.122077) (10).
IDO2 resides immediately downstream of the IDO gene on chromosome 8p12 (10). Structurally, the IDO2 sequence is highly similar to IDO and an IDO2 ortholog also exists in the mouse (10). Critical catalytic residues between IDO and IDO2 are identical and thus functionally conserved (10). Biochemically, IDO2 functions like IDO in tryptophan catabolism, however, it was found that the D-1-MT but not the L-1-MT isomer selectively and potently inhibited IDO2 activity (10, 12). This difference is important because D-1-MT has been chosen as the lead compound for clinical development, based on its superior anti-tumor properties compared to L-1-MT (9).
Genetic analysis of the IDO2 gene revealed two functional polymorphisms with a high prevalence in the general population (10). Specifically, the R248W polymorphism was associated with a >90% reduction in IDO2 catalytic activity, whereas the Y359STOP polymorphism (which generates a premature stop codon) completely inactivated IDO2 activity (10). Large scale sequencing analysis revealed that these two non-functional alleles of IDO2 were commonly distributed in human populations of Asian, European, and African descent.
Taken together, these studies argued that IDO2 may be the chief target of the d-1MT, a promising immune modulating agent, and that IDO2 genotyping could predict which patients might respond to this form of chemo-immunotherapy. Thus, in the current study, we surveyed the status IDO2 in a patient cohort treated at Thomas Jefferson University in order to gain an initial assessment of this molecule as a viable target for the treatment of pancreatic cancer.
Materials and Methods
Specimens
Thirty-six cases of pancreatic ductal adenocarcinoma (PDA) were retrieved from the Thomas Jefferson University pathology archives. The Institutional Review Board approved the study. The PDA cases included 11 well-, 16 moderately- and 9 poorly-differentiated tumors. All patients signed an appropriate informed consent, authorizing the harvesting and study of their tumors for molecular studies.
Immunohistochemistry
For immunohistochemical analysis, paraffin-embedded blocks were sectioned at 5 μm. Sections were then deparaffinized first by treatment with xylene for 3 min (2x) and rehydrated by passage through a graded series of ethanol. Antigen retrieval was performed by microwaving the slides in 100 mM sodium citrate buffer for 15 min. The sections were blocked for 10 min with peroxidase blocking system and incubated at room temperature with the primary antibodies at a 1:100 dilution for 90 min, secondary biotinylated anti-rabbit IgG for 30 min and the streptavidin-biotinylated horseradish peroxidase complex for 30 min. After washing, the slides were exposed to the chromagen DABplus for 5 min and counterstained with hematoxylin (Dako, Fort Collins, CO). The percentage of IDO2 staining in tumor cells was graded by an experienced pathologist in a blinded manner. The results were divided into 4 categories: negative (0%-5% of staining cells), weakly positive (6%-25%), moderately positive (26%-50%), and strongly positive (>50%).
Immunoblot analysis
Pancreatic cancer cell lines were either treated or not treated with interferon-γ (IFN-γ) (Roche, Indianapolis, IN) and were lysed with standard SDS lysis buffer or extracted for RNA (see methods below). Equal amounts of cells were lysed and separated by electrophoresis for over 1 h at 200 volts using 10% bis-tris-polyacrylamide gels (Invitrogen, Carlsbad, CA). After electrophoretic transfer of proteins onto polyvinylidene difluoride membranes (Invitrogen), which were then blocked in Tris-buffered saline/0.5% Tween (TBS–T)/5% milk, incubated with primary IDO antibody (1:1000, Millipore, Billerica, MA) and then incubated with a horseradish peroxidase-conjugated secondary antibody (1:20,000, Santa Cruz, CA). The protein bands were visualized on X-ray film using the Immobilon Western Chemiluminescent Substrate (Millipore). Equal protein loading was confirmed using Fast Green staining (data not shown, USB, Cleveland, Ohio). At least two experiments were performed for each data point, and representative data are shown.
Immunofluorescence of IDO2 protein for detection of cellular localization
CAPAN1 cell lines were treated with 750 U/mL of IFN-γ (Roche Applied Science) over 48 hours. Cells were plated in chamber slides overnight for immunofluorescence. Cells were fixed in 3% paraformaldehyde (Sigma Aldrich) for 20 minutes, followed by cell permeabilization in 0.5% Triton X-100 /1% normal goat serum in PBS for 15 minutes. Cells were incubated for 1 hour in polyclonal rabbit IDO2 primary antibody (1:50). Cells were fluorescently labeled using goat anti-rabbit secondary antibody (1:400, Alexa Fluor 488, Molecular Probes, Eugene, OR) and incubated in the dark for one hour. Cell nuclei were stained with DAPI and cells were analyzed under a Zeiss LSM-510 Confocal Laser Microscope.
DNA sequencing of IDO2 polymorphisms
Genomic DNA was isolated from surgically resected pancreatic tissue extracted using the DNeasy Blood and Tissue Kit (Qiagen, Valencia CA). PCR amplification of the relevant IDO2 exons were carried out using specific primer sets used previously (10). For sequencing of the R235W polymorphism we used (forward 5’ GAA CAT TCT ATC CCC CGT TGC 3’ and reverse 5’TTA CCT GAG AGT GGA TCC CTA GCA 3’). For sequencing of the premature stop codon (Y359STOP) we used (forward 5’ TCT TGT GCT CCC TCC AAA ACA 3’ and reverse 5’TGG TTT GGC TTC CCA TGC TT 3’). PCR reactions were done in 25 μL reactions using 2 μL of DNA, 0.5 un/μL of Taq polymerase (USB, Cleveland OH), 2.5 μL of 10X PCR buffer (USB, Cleveland OH), and 0.5 μL 10 mM dNTP Mix (Invitrogen, Carlsbad, CA). Conditions were set for 35 cycles at (95°C for 2 min, 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, followed by an extension of 2 min at 72°C). Sequencing reactions included PCR purified products using DNA purification columns (Qiagen) and the above forward primers for each PCR reaction. Each PCR reaction was checked by electrophoretic separation on a DNA agarose gel (16). Sequencing was then performed by capillary electrophoresis in the Kimmel Cancer Center DNA core facility at Thomas Jefferson University.
Results
Genotyping of IDO2 functional polymorphisms in patient PDA specimens
Genotyping of R235W
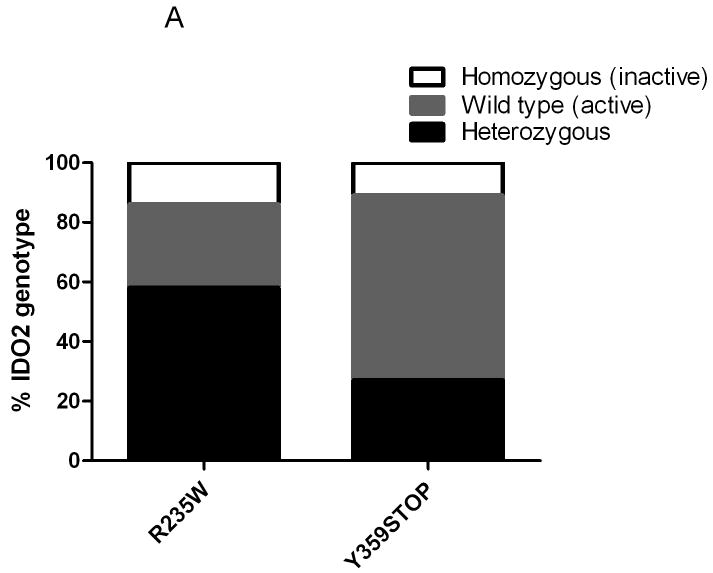
Through direct sequencing of specific exons using conditions we previously reported (10), we found 58% of our patient cohort were heterozygous for the R235W polymorphism; 28% were homozygous wild-type; and only 14% were homozygous for this functionally inactive polymorphism of the IDO2 gene (10) (Figures 1 and 2A).
Figure 1.
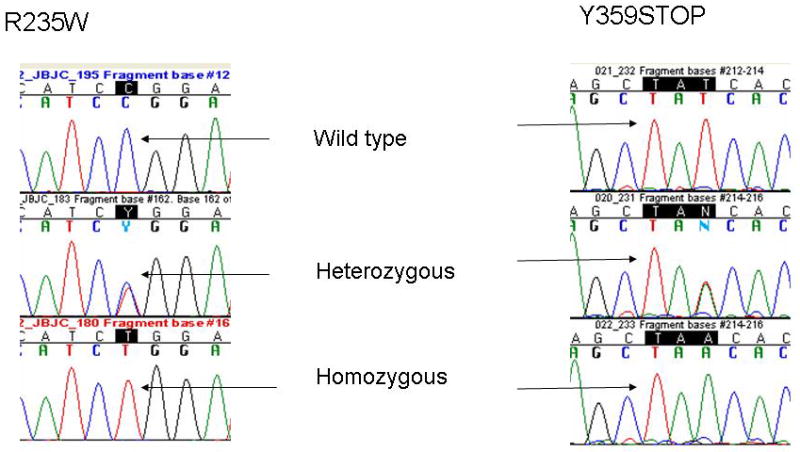
Representative chromatograms of direct sequencing of patient constitutional genomic DNA showing the three possible sequences of homozygous, heterozygous, or wild-type sequence: R235W polymorphism (left) and Y359STOP (right).
Figure 2.
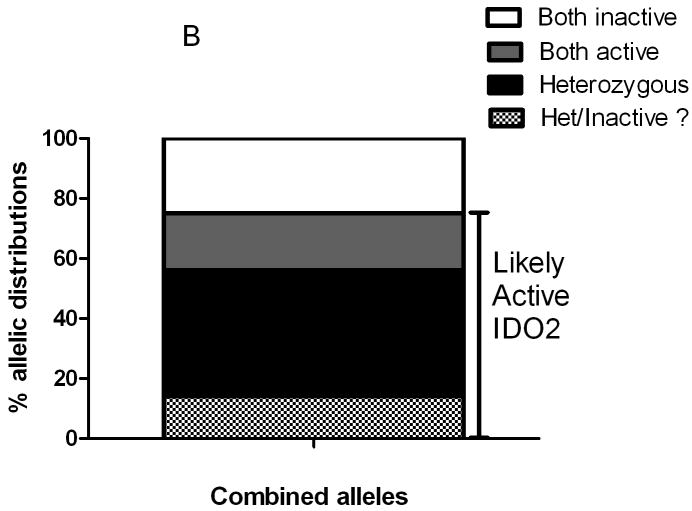
Functionally active IDO2 alleles are frequently found in PDA patients. A) Percent distribution of alleles for the R235W and the Y359STOP polymorphisms in our patient cohort. B) Combined distribution of amino acid changes when surveying both polymorphisms in our patient cohort. In the corresponding legend: both active is the presence of wild type sequence present on both alleles (R/R - Y / Y); both inactive is the presence of a functional polymorphism on both alleles (i.e., R /W -stop / stop, R /R– stop/stop, W / W - Y / Y, W/W - Y / stop); heterozygous is the presence of at least one functional allele; het/Inactive ? indicates that direct sequencing could not determine if the polymorphism was located on the same alleles. The bar on the right indicates that the majority of the alleles likely translate into active functional IDO2 enzyme.
Genotyping of Y359STOP
We found that only 11% of the genomic DNA from patient samples were homozygous for the premature stop codon polymorphism (Y359STOP). The Hs766T cell line harbored this homozygous stop codon as well. While 27% of our patient samples were heterozygous for the Y359STOP polymorphism, the majority (62%) were homozygous wild-type (Figure 2A). When analyzing the combined frequency of the two polymorphic variants (Figure 2B), the majority (75%) of our resected patient cohort had a genetic profile predicting an active IDO2 enzyme (i.e., a functional IDO2 allele). When including the possibility that compound polymorphic sequences existed, this number was conservatively corrected to 58% (Figure 2B). However, a previous report suggested that both inactivating polymorphisms rarely segregate to the same allele (10), so the 75% figure for active IDO2 is likely more representative in our patient cohort.
Expression of IDO2 in PDA
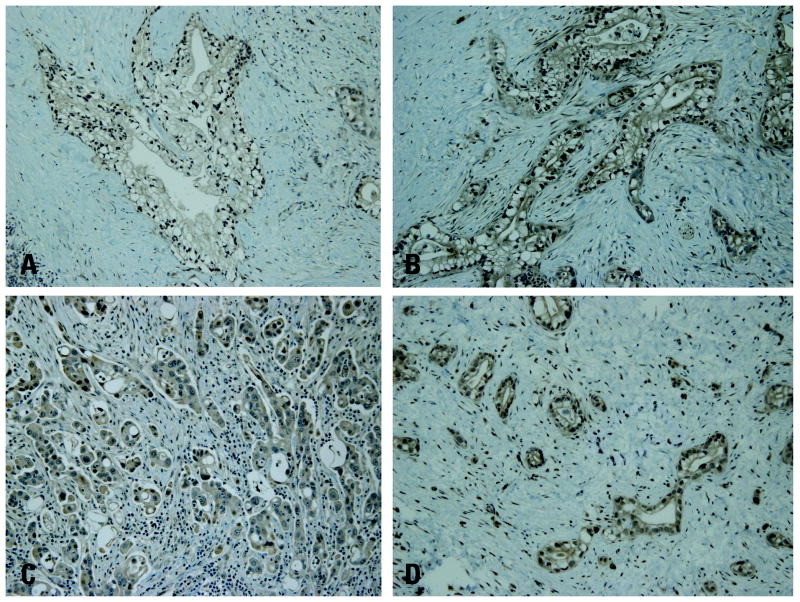
Immunohistochemical analysis of IDO2 protein was performed in three cases each for different genetic subgroups (Y359STOP homozygous, Y346STOP heterozygous, R235W homozygous and homozygous wild type at 235) (Figure 3). Five of 12 cases were moderately positive whereas 7 were weakly positive for IDO2. Of the five moderately IDO2 positive cases, all displayed lymph node metastasis, whereas only 2 of the 7 weakly IDO2 positive tumors were associated with nodal disease. Intensity of IDO2 and percentage of tumor cell staining did not correlate with any IDO2 polymorphic sub-group. There was also no correlation between IDO2 immunolabeling and tumor grade. In contrast to labeling of tumor tissues, we found that normal pancreatic tissue showed no IDO2 staining (data not shown).
Figure 3.
IDO2 protein expression in pancreatic ductal adenocarcinoma ex vivo. A) Weak cytoplasmic IDO2 staining in well differentiated pancreatic ductal adenocarcinoma heterozygous for the R235W polymorphism. B) Moderate cytoplasmic IDO2 staining in well differentiated pancreatic adenocarcinoma homozygous for the R235W polymorphism. Both panels A and B were wild type at Y359. C) Moderate cytoplasmic staining in poorly differentiated pancreatic ductal adenocarcinoma heterozygous for the STOP polymorphism (Y359STOP). D) Cytoplasmic IDO2 staining in moderately differentiated PDA homozygous for the STOP polymorphism (Y359STOP). Both panels C and D were wild type at R235.
Analysis of IDO protein expression in pancreatic cancer cell lines
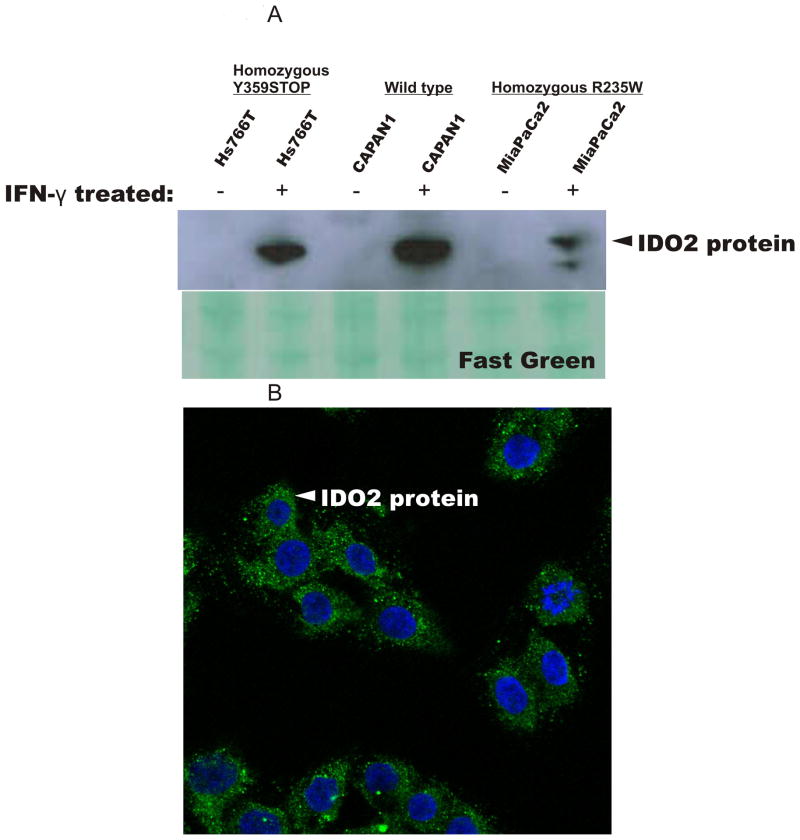
We also included five pancreatic cancer cell lines in our study (see Figure legends for sequencing data on cell lines). In previous studies (5, 17-19), we and others detected IDO protein expression induced by IFN-γ in pancreatic cancer cells and human cells. Protein lysates from primary pancreatic cancer cell lines, Hs766T, CAPAN1, and MiaPaCa2 cells showed IDO2 protein expression when treated with IFN-γ as detected by immunoblot analysis (Figure 4A). Interestingly, we observed no molecular weight alterations of the IDO2 protein even in cell lines harboring a pre-mature stop codon (Figure 4A), possibly due to a similar gel mobility of the truncated protein. To further validate our findings, we were able to detect cytoplasmic expression of IDO2 in CAPAN1 cells (Figure 4B) after IFN-γ treatment.
Figure 4.
IDO2 expression in pancreatic cancer cell lines (Hs766T, CAPAN1, and MiaPaCa2 cells) is expressed only after IFN-γ exposure (750U/ml) for 48 h. A) IDO protein expression assessed by immunoblot analysis. Equal protein loading was confirmed by Fast Green staining of the membrane. Amino acid polymorphic sequences of the cell lines are: Hs766T (wild type at codon R235, harbored a homozygous premature stop codon at 346), CAPAN1 (wild type at codon R235, wild type at codon 359), and MiaPaCa2 (homozygous at codon R235W, wild type at codon 359). Two additional pancreatic cancer cell lines were sequenced (PL-5 and BxPC3) and both were heterozygous at codon R235W and wild type at codon 359. B) CAPAN1 cells exposed to IFN-γ (750U/ml) for 48 h. IDO2 protein detected in the cytoplasm via immunoflouresence (Alexa Fluor 488 staining, green). DAPI, blue stain, represents nuclear staining.
Discussion
PDA is one of the deadliest of all cancers, accounting for over 35,000 deaths in the U.S. annually. Unfortunately, novel and conventional therapeutic strategies employed to date have yielded disappointing results. The many factors underlying the aggressiveness of pancreatic cancer must include the ability of pancreatic tumor cells to avoid immune detection. In this study and in previous work (5), we have shown that IDO and its related protein IDO2 are upregulated in pancreatic cancer cells. These enzymes can locally regulate the immune environment in order to provide an immuno-tolerant milieu which allows for the tumor to thrive (13). We explored IDO (5) and IDO2 because pancreatic cancer is a good candidate for immunotherapeutic strategies and pre-clinical studies show IDO inhibitors (e.g. 1-MT) to be selective targeting agents (10, 13).
The majority of our patient cohort had at least one wild-type or functioning IDO2 allele (Figure 2B). These data combined with our expression work in ex vivo tissue and cell culture experiments show that IDO2 is an active target in pancreatic cancer cells. While the size of our patient cohort was small, the frequencies of the polymorphisms we saw in the cohort coincided with those seen in a larger population study (10), offering an initial argument against any link between IDO2 genotype and pancreatic cancer susceptibility. Since these polymorphisms inactivate IDO2 as a target, it is nonetheless reasonable to imagine that IDO2 genotyping of patient tissue or blood could help select the most appropriate patients for treatment with an IDO inhibitor such as D-1-MT that preferentially targets IDO2. We believe the use of constitutional genomic DNA in our studies offers an accurate read of the IDO2 sequence, because the chromosome 8q arm where both IDO and IDO2 reside have been shown to be intact in large scale allelotyping of pancreatic cancer cell lines and xenografted tumors (20, 21).
In summary, IDO inhibitors are highly specific and have shown great promise in pre-clinical studies (13). So far in vivo use of these compounds in various models have proven them to be effective anti-tumor agents (13). The current study and our previous work show that both IDO and IDO2 can be rationalized as good targets for eradication of pancreatic cancer cells. Future studies will dictate whether D-1-MT or other IDO inhibitors will target one or both of these enzymes and how much functional redundancy and cross-talk may exist between IDO and IDO2. Also, future work will explore the biologic significance of IDO and IDO2 enzymatic activity (e.g. recruitment of Tregs) in circulating pancreatic tumor cells (5). Clinically, the most promising concept is to combine current vaccine strategies (22) or chemotherapies (23) with D-1-MT or other IDO inhibitors for the benefit for pancreatic cancer patients. Such a concept could take advantage of the multiple pathways involved in the complex process of pancreatic tumorigenesis and offer new promise for improvements in survival yet to be realized in this deadly disease.
Footnotes
To be presented at the 120th annual meeting of the Southern Surgical Association, December 2008
References
- 1.Jones S, Zhang X, Parsons DW, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008 doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kennedy EP, Yeo CJ. The case for routine use of adjuvant therapy in pancreatic cancer. J Surg Oncol. 2007;95:597–603. doi: 10.1002/jso.20719. [DOI] [PubMed] [Google Scholar]
- 3.Winter JM, Cameron JL, Campbell KA, et al. 1423 pancreaticoduodenectomies for pancreatic cancer: A single-institution experience. J Gastrointest Surg. 2006;10:1199–210. doi: 10.1016/j.gassur.2006.08.018. discussion 210-1. [DOI] [PubMed] [Google Scholar]
- 4.Zitvogel L, Tesniere A, Kroemer G. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nat Rev Immunol. 2006;6:715–27. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 5.Witkiewicz A, Williams TK, Cozzitorto J, et al. Expression of indoleamine 2,3-dioxygenase in metastatic pancreatic ductal adenocarcinoma recruits regulatory T cells to avoid immune detection. J Am Coll Surg. 2008;206:849–54. doi: 10.1016/j.jamcollsurg.2007.12.014. discussion 54-6. [DOI] [PubMed] [Google Scholar]
- 6.Uyttenhove C, Pilotte L, Theate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–74. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 7.Colombo MP, Piconese S. Regulatory-T-cell inhibition versus depletion: the right choice in cancer immunotherapy. Nat Rev Cancer. 2007;7:880–7. doi: 10.1038/nrc2250. [DOI] [PubMed] [Google Scholar]
- 8.Fallarino F, Grohmann U, You S, et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–61. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 9.Hou DY, Muller AJ, Sharma MD, et al. Inhibition of indoleamine 2,3-dioxygenase in dendritic cells by stereoisomers of 1-methyl-tryptophan correlates with antitumor responses. Cancer Res. 2007;67:792–801. doi: 10.1158/0008-5472.CAN-06-2925. [DOI] [PubMed] [Google Scholar]
- 10.Metz R, Duhadaway JB, Kamasani U, Laury-Kleintop L, Muller AJ, Prendergast GC. Novel tryptophan catabolic enzyme IDO2 is the preferred biochemical target of the antitumor indoleamine 2,3-dioxygenase inhibitory compound D-1-methyl-tryptophan. Cancer Res. 2007;67:7082–7. doi: 10.1158/0008-5472.CAN-07-1872. [DOI] [PubMed] [Google Scholar]
- 11.Muller AJ, DuHadaway JB, Donover PS, Sutanto-Ward E, Prendergast GC. Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat Med. 2005;11:312–9. doi: 10.1038/nm1196. [DOI] [PubMed] [Google Scholar]
- 12.Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117:1147–54. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prendergast GC. Immune escape as a fundamental trait of cancer: focus on IDO. Oncogene. 2008;27:3889–900. doi: 10.1038/onc.2008.35. [DOI] [PubMed] [Google Scholar]
- 14.Muller AJ, Prendergast GC. Indoleamine 2,3-dioxygenase in immune suppression and cancer. Curr Cancer Drug Targets. 2007;7:31–40. doi: 10.2174/156800907780006896. [DOI] [PubMed] [Google Scholar]
- 15.Muller AJ, Malachowski WP, Prendergast GC. Indoleamine 2,3-dioxygenase in cancer: targeting pathological immune tolerance with small-molecule inhibitors. Expert Opin Ther Targets. 2005;9:831–49. doi: 10.1517/14728222.9.4.831. [DOI] [PubMed] [Google Scholar]
- 16.Brody JR, Calhoun ES, Gallmeier E, Creavalle TD, Kern SE. Ultra-fast high-resolution agarose electrophoresis of DNA and RNA using low-molarity conductive media. Biotechniques. 2004;37:598–602. doi: 10.2144/04374ST04. [DOI] [PubMed] [Google Scholar]
- 17.Brandacher G, Perathoner A, Ladurner R, et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: effect on tumor-infiltrating T cells. Clin Cancer Res. 2006;12:1144–51. doi: 10.1158/1078-0432.CCR-05-1966. [DOI] [PubMed] [Google Scholar]
- 18.Pfefferkorn ER. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci U S A. 1984;81:908–12. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasui H, Takai K, Yoshida R, Hayaishi O. Interferon enhances tryptophan metabolism by inducing pulmonary indoleamine 2,3-dioxygenase: its possible occurrence in cancer patients. Proc Natl Acad Sci U S A. 1986;83:6622–6. doi: 10.1073/pnas.83.17.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calhoun ES, Hucl T, Gallmeier E, et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66:7920–8. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 21.Iacobuzio-Donahue CA, van der Heijden MS, Baumgartner MR, et al. Large-scale allelotype of pancreaticobiliary carcinoma provides quantitative estimates of genome-wide allelic loss. Cancer Res. 2004;64:871–5. doi: 10.1158/0008-5472.can-03-2756. [DOI] [PubMed] [Google Scholar]
- 22.Laheru D, Jaffee EM. Immunotherapy for pancreatic cancer - science driving clinical progress. Nat Rev Cancer. 2005;5:459–67. doi: 10.1038/nrc1630. [DOI] [PubMed] [Google Scholar]
- 23.Muller AJ, Prendergast GC. Marrying immunotherapy with chemotherapy: why say IDO? Cancer Res. 2005;65:8065–8. doi: 10.1158/0008-5472.CAN-05-2213. [DOI] [PubMed] [Google Scholar]