Abstract
We introduce a novel method for annotating protein function that combines Naïve Bayes and association rules, and takes advantage of the underlying topology in protein interaction networks and the structure of graphs in the Gene Ontology. We apply our method to proteins from the Human Protein Reference Database (HPRD) and show that, in comparison with other approaches, it predicts protein functions with significantly higher recall with no loss of precision. Specifically, it achieves 51% precision and 60% recall versus 45% and 26% for Majority and 24% and 61% for χ2–Statistics, respectively.
Keywords: protein function, protein interaction networks, Naïve Bayes, association rules, Gene Ontology
1. Introduction
Understanding protein function is one of the most challenging problems in biology. While many genome sequences have been generated, a large fraction of the newly discovered genes lack functional characterization. This is particularly true for higher eukaryotes. While many experimental approaches, including both individual protein or gene-specific efforts and large scale, whole-genome projects, are used successfully, these are time consuming and expensive. Large scale, computational methods to predict protein function, therefore, can potentially play important roles.
Early computational methods inferred functions of novel proteins from their amino acid sequence similarity to proteins of known function [1] or from observations of pairs of interacting proteins that had orthologs in another organism fused into a single protein chain [2]. Correlated evolution, correlated RNA expression patterns, plus patterns of domain fusion, have also been used to predict similarities in protein functions [3,4]. Several other approaches have annotated proteins based on phylogenetic profiles of orthologous proteins [5–9]. Bayesian reasoning was used to combine large-scale yeast two-hybrid (Y2H) screens and multiple microarray analyses [10] and Support Vector Machines were used to combine protein sequence and structure data [11] to produce functional predictions. Other methods related features extracted from protein 3D structures to function [12,13]. Recently, a consensus method, GOPred [14] predicted protein function by combining three different classifiers, namely, BLAST k-nearest neighbor, Subsequence Profile Map and Peptide statistics combined with support vector machine. While each of these approaches has had some success, generally they produce high false positive rates because their underlying principles/assumptions are valid for only a small number of proteins [15,19]. In addition, many methods were appropriate largely for prokaryotic sequences [15].
Protein-protein interaction (PPI) data have proven valuable for inferring protein function from functions of interaction partners. Facilitating this work, whole genome interaction data have been and/or are being generated for E. coli, yeast, worm, fly and human [16–24]. The curated databases consolidate these datasets [25–29] that have been used by several methods. The Majority method annotated yeast proteins based on the most frequent functional properties of nearest neighbors [19]. However, because the whole network was not considered, a function that occurred at a very high frequency was not annotated when it did not occur in the nearest neighbor set. In an approach that extended the Majority method, functions were annotated by exploiting indirect neighbors and using a topological weight [30], and χ2–statistics were used to look at all proteins within a particular radius, although it did not consider any aspect of the underlying topology of the PPI network [31]. FunctionalFlow considered each protein as a source of functional flow for its associated function, which spread through the neighborhoods of the source [35]. Proteins receiving the highest amount of flow of a function were assigned that function. This algorithm did not take into account the indirect flow of functions to other proteins after labeling them. Markov random fields (MRF) and belief propagation in PPI networks were combined to assign protein functions based on a probabilistic analysis of graph neighborhoods [32–34]. This assumed that the probability distribution for the annotation of any node was conditionally independent of all other nodes, given its neighbors. These methods were sensitive to the neighborhood size and the parameters of the prior distribution. The MRF methods were later extended by combining PPI data, with gene expression data, protein motif information, mutant phenotype data, and protein localization data to specify which proteins might be active in a given biological process [36,37]. Other global approaches integrated PPI network with more heterogeneous data sources (such as large-scale two-hybrid screens and multiple microarray analyses) [10,38]. Our algorithm ClusFCM [39] assigned biological homology scores to interacting proteins and performed agglomerative clustering on the weighted network to cluster the proteins by known functions and cellular location; functions then were assigned to proteins by a Fuzzy Cognitive Map. PRODISTIN formulated a distance function (the Czekanowski-Dice distance) that uses information on shared interactome to mirror a functional distance between proteins [15]. Other approaches predicted protein functions via the patterns found among neighbors of proteins within a network [40,41]. Recently, a network-based method combined the likelihood scores of local classifiers with a relaxation labeling technique [42]. Several approaches applied clustering algorithms to PPI networks to predict functional modules, protein complexes and protein functions, however, the performance of these algorithms differs substantially when run on the same network which leads to uncertainties regarding the reliability of their results [43].
Here, we extend our previous work [44] by exploring the hierarchical structure of the Gene Ontology database. For each of protein, a predicted function will be considered as a true positive if the function is a parent of any function in the annotated set of the protein. Thus, this work is a less conservative approach than our previous work. We use Naïve Bayes combined with association rules and take into account the underlying topology of a PPI network. Predicted functions are analyzed by association rules to discover relationships among the assigned functions, i.e., when one set of functions occurs in a protein then the protein may be annotated with an additional set of other specific functions at some confidence level. We test our method on human protein data and compare its performance with the Majority [19] and χ2 statistics [31] methods.
2. Materials
Gene Ontology (GO) database
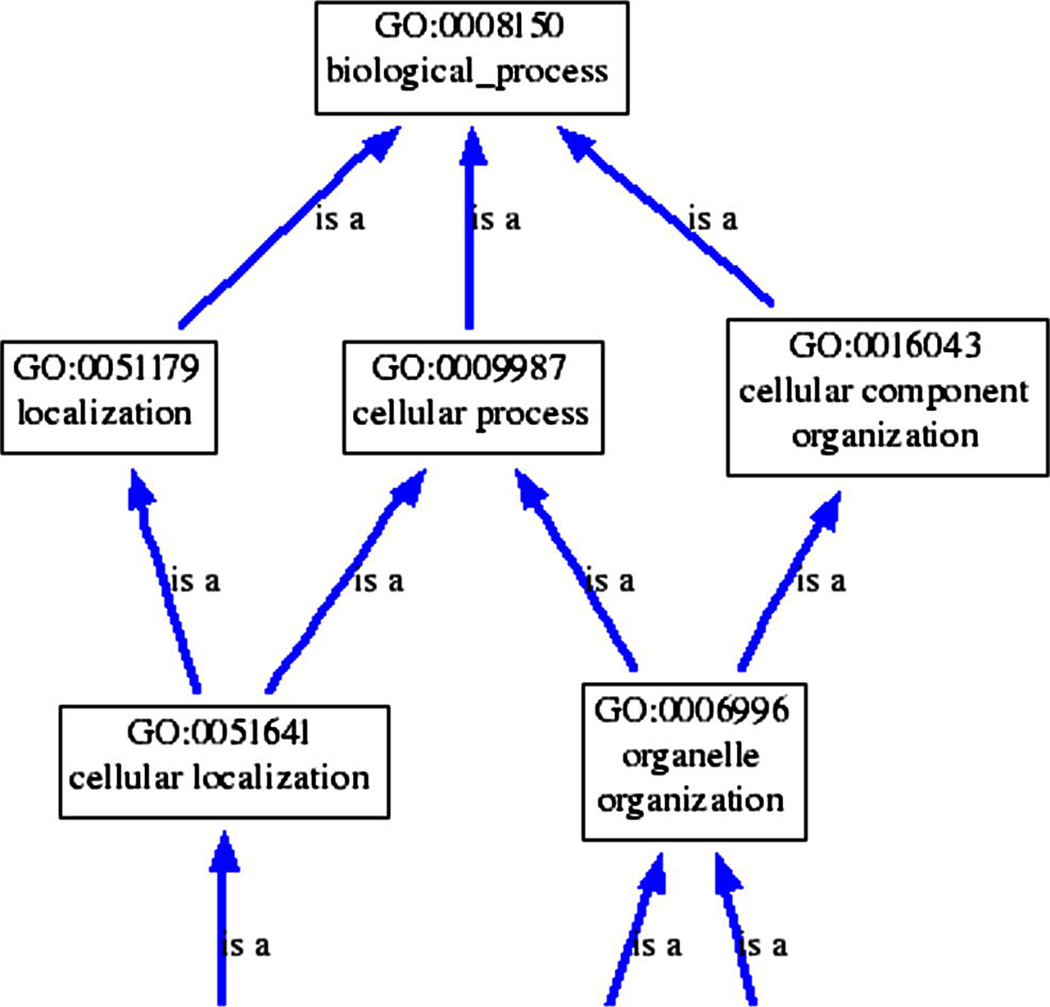
The Gene Ontology (GO) [45] was established to provide a common language to describe aspects of the biology of a gene product. The use of a consistent vocabulary allows genes from different species to be compared based on their GO annotations. GO terms are composed of the three structured controlled vocabularies (ontologies): the molecular function of gene products, their associated biological processes, and their physical location as cellular components. Each ontology is constructed as a directed acyclic graph through a parent-child “is-a” relationship (see Figure 1). We used the GO database (version 1.1.940 released 1/2010).
Fig. 1.
A hierarchical structure of GO terms: many-to-many parent-child relationships are allowed in the ontologies. A gene may be annotated to any level of ontology, and to more than one term within an ontology [45].
Human interaction dataset
The human interaction data were retrieved from HPRD [29] (release 7/2009). The entire dataset contains 38,788 direct molecular interactions from three types of experiments (in vivo, in vitro, and in Y2H). There are 9,630 distinct proteins annotated with 433 GO functions in the three categories. Because some estimates suggest that more than half of all current Y2H data are spurious [46,47], we first excluded interactions supported only by the Y2H experiments, leaving 29,557 interactions from in vivo and in vitro experiments, and 422 GO functions annotating the 7,953 unique proteins. A more recent study showed that Y2H data for human proteins were actually more accurate than literature-curated interactions supported by a single publication. Therefore, we also separately analyzed the complete HPRD PPI dataset.
Note that we use the “is-a” relationships to eliminate all parent GO terms annotated for a protein, i.e. suppose there are two GO terms A and B annotated for a protein, and if A “is-a” B, then B is removed from the annotated GO terms of the protein.
3. Methods
3.1 Notation
G=(V,E): an undirected graph to define the PPI network, where V is a set of proteins and E is a set of edges connecting proteins u and v if the corresponding proteins interact physically;
K: the total number proteins in the PPI network
F: the whole GO function collection set and |F|: the cardinality of the set F
fi: a function in the set F (i=1‥|F|)
O(fi) is the parent ontology set of fi, that is ∀g∈O, fi “is-a” g
Cu: the cluster coefficient of protein u
Nu: the neighbor set of protein u (proteins interacting directly with protein u)
: the number of proteins annotated with function fi in Nu and : the number of proteins un-annotated with function fi in Nu where .
3.2 Posing the Problem: From Annotation to Classification
For a function of interest fi, we want to annotate the function fi to the proteins in a PPI network. We pose the functional annotation problem as a classification problem. The training data are in the form of observations d ∈ Rk (k dimensions) and their corresponding class information. For each protein u in the network, a function of interest fi is considered as a class label 1 if the protein u is annotated with fi, and otherwise as 0. The features to deduce class information are selected as follows. Exploiting the fact that proteins of known functions tend to cluster together [19], the first feature we take into account, A1, is the number of proteins annotated with the function fi in the neighborhood set of protein u (i.e. ). The second feature (A2) is the number of proteins not annotated with the function fi in the neighborhood set of the protein u (i.e. ). Several studies indicate that other features can be useful to predict functions and drug targets for a protein, such as the number of functions annotated in proteins in the neighborhood set at level 2 of the protein [31], the connectivity (the total number of incoming and outgoing arcs of a protein, which is equal to ), the betweenness (the number of times a node appears in the shortest path between two other nodes) and the clustering coefficient Cu (the ratio of the actual number of direct connections between the neighbors of protein u to the maximum possible number of such direct arcs between its neighbors) [48]. To select the best features for a robust learning method, we use a feature selection method. First, we form a sub set, S, containing two features: . Second, we perform a heuristic search by iteratively adding one feature at a time to the set S (without using class information) to form a new subset, S’. Next, we classify the HPRD data with the selected S’ features by the Radial Basis Function, Support Vector Machine, Logistic Regression and Naïve Bayes. The feature to be added to S’ is the feature that achieves the maximum average value of the harmonic mean of the four classification methods. The heuristic search terminates when the average of the harmonic mean of the four methods does not increase. At the end of this process we came up with three selected features, namely, and A3=Cu. We use Weka [49] to implement the four classifiers with default parameters. Performance in terms of recall, precision, and harmonic mean of the four classifiers on the HPRD data using the three selected features using 10-fold cross validation is shown in Table 1. Because Naïve Bayes performs the best in terms of the harmonic mean, we use it in our predictive modeling.
Table 1.
Performance of the Radial Basic Function, Support Vector Machine, Logistic Regression and Naïve Bayes methods on the HPRD data set using 10-fold cross validation.
| Precision | Recall | Harmonic Mean | |
|---|---|---|---|
| Radial Basis Function | 0.63 | 0.11 | 0.19 |
| Support Vector Machine | 0.81 | 0.04 | 0.09 |
| Logistic Regression | 0.68 | 0.15 | 0.24 |
| Naïve Bayes | 0.53 | 0.27 | 0.36 |
3.3 Predictive modeling
Phase I: Naïve Bayes
If d=<A1, A2, A3> is an observation for a protein u, we decide a class membership for the observation d (corresponding to a function of interest fi) by assigning d to the class with the maximal probability computed as follows:
| (1) |
Note that P(d | fi) can be ignored because it is the same for all classes:
| (2) |
The likelihood P(d | c, fi) is the probability of obtaining the observation d for a protein u in class c and is calculated as:
| (3) |
Thus equation (2) becomes:
| (4) |
Because the factorials in equation (4) are constant, we can rewrite the maximum a posteriori class c as follows:
| (5) |
Two key issues arise here. First, the problem of zero counts can occur when given class and feature values never appear together in the training data. This can be problematic because the resulting zero probabilities will eliminate the information from all other probabilities. We use the Laplace correction to avoid this [50]. Second, in equation (5), the conditional probabilities are multiplied and this can result in a floating point underflow. Therefore, it is better to perform the computations using logarithms of the probabilities. Equation (5) then becomes:
| (6) |
The parameters of the model, in our case, P̂(A1 | c, fi), P̂ (A2 | c, fi), P̂ (A3 | c, fi) and P̂(c | fi) can be estimated as follows:
| (7) |
| (8) |
| (9) |
| (10) |
where lc1=1 and lc2=2 are the Laplace corrections and the attributes A2 and A3 can be similarly estimated.
Phase II: Association Rules
Association rules are statements of the form {fX} => {fY}, meaning that if we find all of {fX} in a protein, we have a good chance of finding {fY} with some user-specified confidence (derived as an estimate of the probability P({fY} | {fX})) and support (the fraction of proteins that contain both functions {fX} (in the antecedent) and {fY} (in the consequent) of the rule in the entire network). With 0.1% support and 75% confidence thresholds, we found 900 association rules in the HPRD, and 1,154 rules in the HPRD without Y2H. For example, the rule GO:0004894 → GO:0006955 was found with 100% confidence. Next, we derive new functions from the predicted functions in Phase I by using the mined rules and the following axioms [52]:
if X ⊇ Y then X → Y,
if X → Y then XZ → YZ for any Z, and
if X → Y and Y → Z then X → Z.
Below we briefly describe the Majority and χ2 statistics methods used in comparisons.
Majority
For each protein u in a PPI network, we count the number of times each function fi ∈ F occurs in neighbors of the protein u. The functions with the highest frequencies are assigned to the query protein u.
χ2 statistics
For each function of interest fi we derive the fraction πfi (number of proteins annotated with function fi / K). Then, we calculate efi as the expected number for a query protein u annotated with fi: efi = Nu πfi. The query protein u is annotated with the function with the highest χ2 value among the functions of all proteins in its neighbors, where .
The assessment of the proposed method, Majority and χ2 statistics, which takes into account the many-to-many relationships of GO terms, is performed as follows. For a protein, a predicted function will be i) a true positive if it exists in the annotated functions or the set O(annotated functions), and ii) a false positive if it does not exists in the annotated set. A function existing in the annotated set but not existing in the predicted set will be considered as a false negative while functions in the entire GO set not existing in both annotated and predicted sets will be true negatives.
4. Results and Discussions
We implement our method in Java as a combination of Naïve Bayes and the association rule algorithm. In addition, we implement the Majority and χ2 statistics methods and test all on the HPRD data (with and without interactions identified by Y2H). To compare the performance of our method we use implicit thresholds, τ. We normalize the posterior probability of a query protein u annotated with the function fi: P(c=1| d, fi) and decide the protein u to be annotated with the function fi if the normalized P(c=1| d,fi) > τ, where τ assumes a value between 0 and 1, in increments of 0.1.
Our method assumes that a newly annotated protein propagates its newly acquired function(s) to its direct neighbors. Thus, the method is repeated in two iterations. In the second iteration, to calculate the value for a protein u, we count both the number of proteins in its neighborhood annotated with fi and predicted with fi in the first iteration. In the Majority and χ2 statistics methods top k functions having the highest scores (k ranges from 0, 1, … 20) are selected and those functions are assigned to the query protein.
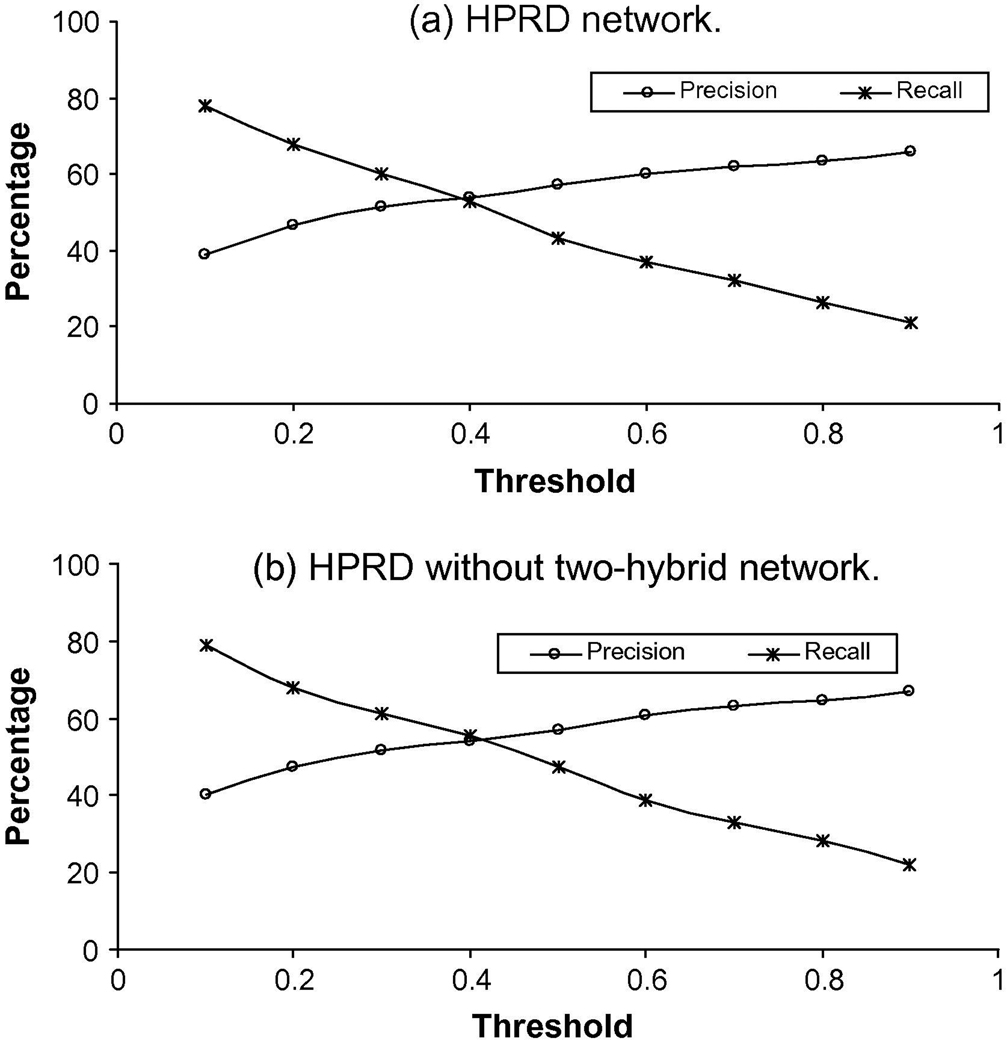
We use the leave-one-out method to evaluate the predictions. For each query protein u in a PPI network we assume that it is not annotated. Then, we the methods described above to predict protein functions for protein u. For each method, we choose the threshold which yields the highest Matthews Correlation Coefficient (MCC) values. Figure 2 shows the relationship between precision and recall using different thresholds for the normalized probabilities of query proteins on the HPRD data sets. The threshold resulting in the highest MCC measures for the HPRD and HPRD without Y2H data sets is 0.3. Because functional annotations for proteins are incomplete at present, a protein may have a function that has not yet been experimentally detected. Our goal is to decrease the number of annotated functions that are not predicted and increase the number of predicted functions that are actually annotated. The fact that the values of recall are always higher than the values of precision in all datasets increases confidence in our method.
Fig. 2.
Precision and recall results of our method on a) the HPRD network data and b) HPRD without two-hybrid network data.
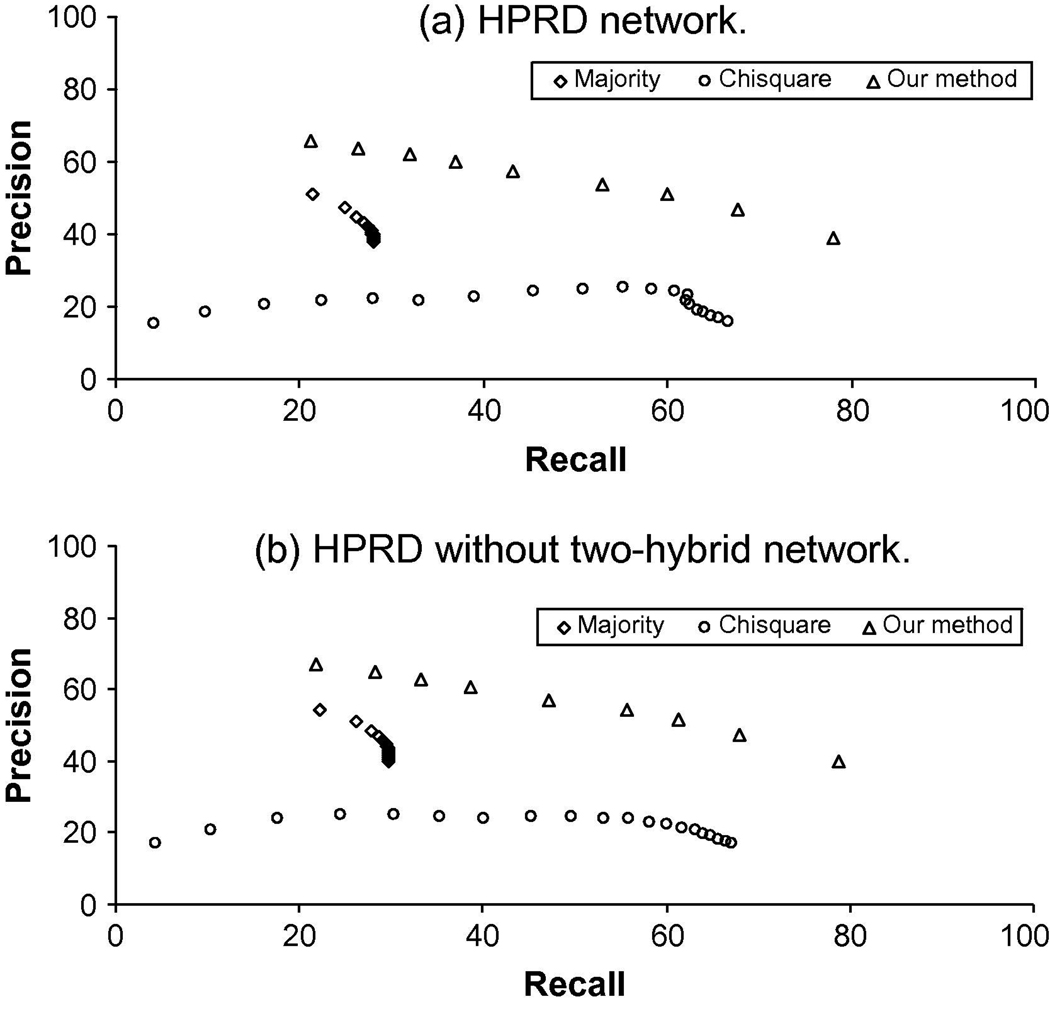
The Figure 3 shows that, for any precision, the recall of our method outperforms Majority and χ2 statistics. Performance measures of the three methods are shown in Table 2. Our method performs equally well in both data sets. To gauge the robustness of the algorithms’ performance (the MCC value), we use the ANOVA one-way-test for statistical significance from the leave one out cross validation. The ANOVA statistics shown in Table 3 confirm that our method indeed performed better than the compared methods.
Fig. 3.
Precision and recall of the three methods on a) the HPRD network data and b) HPRD without two-hybrid network data.
Table 2.
Performance of the three methods on two datasets using leave-one-out validation*; (1): Our method (2): Majority (3): χ2 statistics.
| HPRD | HPRD without Y2H | |||||
|---|---|---|---|---|---|---|
| (1) | (2) | (3) | (1) | (2) | (3) | |
| Precision | 0.51 | 0.45 | 0.24 | 0.52 | 0.49 | 0.23 |
| Recall | 0.60 | 0.26 | 0.61 | 0.61 | 0.28 | 0.58 |
| MCC | 0.55 | 0.34 | 0.37 | 0.56 | 0.36 | 0.36 |
The selected implicit thresholds for our method, Majority, χ2 statistics are .3, 3, and 12, respectively.
Table 3.
Results from one way ANOVA test for MCC value on the two datasets; (1): Our method (2): Majority (3): χ2 statistics.
| HPRD | HPRD without Y2H | |||||
|---|---|---|---|---|---|---|
| (1) | (2) | (3) | (1) | (2) | (3) | |
| Average | 0.55 | 0.34 | 0.37 | 0.56 | 0.36 | 0.36 |
| Variance | 0.07 | 0.08 | 0.03 | 0.07 | 0.07 | 0.03 |
| F | 1841.2 | 1581.16 | ||||
| P | 0 | 0 | ||||
In addition to providing statistics tests to cement our method, it is worth noting that this work takes into account the hierarchical structure of GO database for predicting functions of a protein. If a new function of a protein exists in the set O(annotated functions) of the protein, it will be a true positive. Thus we expect to sacrifice the cost of recall (sensitivity) to increase precision. However, the performance (i.e. recall and precision) of our method was not significantly changed in comparison with our previous work. The reason is probably that in comparison with an average of 1.56 (1.77) shared functions per each interactome, there is only an average of 0.19 (0.15) functions in the O(annotated functions) per each interactome in the HPRD (HPRD without Y2H) dataset. Thus, those functions account for a minor proportion of the performance measures.
Because protein functional annotation is incomplete, it is possible that novel predicted functions that are at present false positives may eventually be discovered to be true positives. We list in Table 4 some proteins from the HPRD without Y2H dataset that are annotated with novel functions at very high probabilities (>.9). The full list of predicted functions for human proteins and Java source code are shown at http://chr21.egr.vcu.edu/bayesian
Table 4.
Novel protein functions predicted by the Naive Bayes.
| HPRD ID |
Symbol | Protein Name | GO Term ID | GO Name | Predicted Probability |
|---|---|---|---|---|---|
| 00010 | ACHE | Acetylcholinesterase | GO:0005201 | extracellular matrix structural constituent | 0.99 |
| 00010 | ACHE | Acetylcholinesterase | GO:0004872 | receptor activity | 0.92 |
| 00015 | ACTC1 | Actin alpha, cardiac muscle | GO:0008092 | cytoskeletal protein binding | 1.00 |
| 00017 | ACTG1 | Actin gamma 1 | GO:0008092 | cytoskeletal protein binding | 1.00 |
| 00019 | ACTN2 | Actinin alpha 2 | GO:0005194 | cell adhesion molecule activity | 0.99 |
| 00019 | ACTN2 | Actinin alpha 2 | GO:0005554 | molecular_function | 0.96 |
| 00021 | ACVR1 | Activin A receptor, type I | GO:0003924 | GTPase activity | 1.00 |
| 00023 | PML | PML | GO:0003700 | transcription factor activity | 1.00 |
| 00025 | ACVR2A | Activin A receptor, type II | GO:0005102 | receptor binding | 1.00 |
| 00030 | ACTA1 | Actin alpha, skeletal muscle 1 | GO:0008092 | cytoskeletal protein binding | 1.00 |
| 00030 | ACTA1 | Actin alpha, skeletal muscle 1 | GO:0005200 | structural constituent of cytoskeleton | 1.00 |
| 00032 | ACTB | Actin beta | GO:0008092 | cytoskeletal protein binding | 1.00 |
| 00038 | ADA | Adenosine deaminase | GO:0004930 | G-protein coupled receptor activity | 0.99 |
| 00043 | ADORA2A | Adenosine A2 receptor | GO:0008092 | cytoskeletal protein binding | 0.92 |
| 00054 | ARF1 | ADP ribosylation factor 1 | GO:0015457 | auxiliary transport protein activity | 1.00 |
| 00059 | FDX1 | Adrenodoxin | GO:0003824 | catalytic activity | 1.00 |
| 00061 | ADM | Adrenomedullin | GO:0005179 | hormone activity | 0.96 |
| 00070 | ALDOA | Aldolase 1 | GO:0003824 | catalytic activity | 0.98 |
| 00072 | A2M | Macroglobulin, alpha 2 | GO:0008236 | serine-type peptidase activity | 0.97 |
| 00082 | LRPAP1 | RAP | GO:0004872 | receptor activity | 1.00 |
| 00097 | SAA1 | Serum Amyloid A1 | GO:0005201 | extracellular matrix structural constituent | 1.00 |
| 00098 | SAA2 | Serum amyloid A2 | GO:0005201 | extracellular matrix structural constituent | 0.97 |
| 00101 | APCS | Serum amyloid P | GO:0004872 | receptor activity | 0.96 |
| 00106 | AGT | Angiotensin I | GO:0008236 | serine-type peptidase activity | 1.00 |
| 00106 | AGT | Angiotensin I | GO:0004177 | aminopeptidase activity | 0.96 |
| 00106 | AGT | Angiotensin I | GO:0004180 | carboxypeptidase activity | 0.92 |
| 00113 | CD19 | CD19 | GO:0004713 | protein-tyrosine kinase activity | 1.00 |
| 00113 | CD19 | CD19 | GO:0030159 | receptor signaling complex scaffold activity | 0.98 |
| 00114 | CD22 | SIGLEC2 | GO:0030159 | receptor signaling complex scaffold activity | 0.99 |
| 00114 | CD22 | SIGLEC2 | GO:0004713 | protein-tyrosine kinase activity | 0.95 |
| 00116 | CD38 | Cyclic ADP ribose hydrolase | GO:0004872 | receptor activity | 0.97 |
| 00120 | SERPINA3 | Alpha 1 antichymotrypsin | GO:0008236 | serine-type peptidase activity | 1.00 |
| 00121 | SLPI | Secretory leukocyte protease inhibitor | GO:0008236 | serine-type peptidase activity | 0.98 |
| 00123 | SLC9A1 | Solute carrier family 9, isoform A1 | GO:0005509 | calcium ion binding | 0.93 |
| 00125 | CD3EAP | Antisense ERCC1 | GO:0003899 | DNA-directed RNA polymerase activity | 0.91 |
| 00126 | IFNAR1 | Interferon, alpha receptor | GO:0005125 | cytokine activity | 1.00 |
| 00133 | APOB | Apolipoprotein B | GO:0003754 | chaperone activity | 1.00 |
| 00135 | APOE | Apolipoprotein E | GO:0004872 | receptor activity | 0.98 |
| 00138 | LRP1 | Low density lipoprotein receptor-related protein 1 | GO:0008236 | serine-type peptidase activity | 0.98 |
| 00139 | NR2F2 | Nuclear receptor subfamily 2, group F, member 2 | GO:0003700 | transcription factor activity | 1.00 |
| 00139 | NR2F2 | Nuclear receptor subfamily 2, group F, member 2 | GO:0030528 | transcription regulator activity | 1.00 |
| 00146 | ARRB1 | Beta arrestin 1 | GO:0004930 | G-protein coupled receptor activity | 1.00 |
| 00147 | ARRB2 | Beta arrestin 2 | GO:0004930 | G-protein coupled receptor activity | 1.00 |
| 00120 | SERPINA3 | Alpha 1 antichymotrypsin | GO:0008236 | serine-type peptidase activity | 1.00 |
| 00121 | SLPI | Secretory leukocyte protease inhibitor | GO:0008236 | serine-type peptidase activity | 0.98 |
| 00123 | SLC9A1 | Solute carrier family 9, isoform A1 | GO:0005509 | calcium ion binding | 0.93 |
| 00125 | CD3EAP | Antisense ERCC1 | GO:0003899 | DNA-directed RNA polymerase activity | 0.91 |
| 00126 | IFNAR1 | Interferon, alpha receptor | GO:0005125 | cytokine activity | 1.00 |
| 00133 | APOB | Apolipoprotein B | GO:0003754 | chaperone activity | 1.00 |
5. Conclusions
We have described a novel method for protein functional annotation that combines Naïve Bayes and association rules. The method used global optimization that took into account the following features of interaction networks: direct and indirect interactions, the underlying topology (cluster coefficients), and functional protein clustering, as well as the many-to-many relationships of the GO terms in the GO database. We have shown the robustness of our method by testing it on two interaction data sets using the leave-one-out cross-validation. The results showed that our method consistently outperformed the Majority and the χ2–statistics methods in predicting protein functions. In addition, our method predicts new relationships among the predicted functions that can provide new experimental directions. Finally, in comparison with previous work, the results empirically showed that our approach does not depend on the GO’s hierarchy.
Acknowledgments
The authors acknowledge support from the National Institutes of Health (1R01HD05235-01A1).
References
- 1.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marcotte E, Pellegrini M, Ng H, Rice D, Yeates T, Eisenberg D. Detecting protein function and protein–protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 3.Marcotte E, Pellegrini M, Thompson M, Yeates T, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X, Kao M, Wong W. Transitive functional annotation by shortest-path analysis of gene expression data. Proc Natl. Acad. Sci. USA. 2002;99:12783–12788. doi: 10.1073/pnas.192159399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellegrini M, Marcotte E, Thompson M, Eisenberg D, Yeates T. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowers P, Cokus S, Eisenberg D, Yeates T. Use of logic relationships to decipher protein network organisation. Science. 2004;306:2246–2259. doi: 10.1126/science.1103330. [DOI] [PubMed] [Google Scholar]
- 7.Pagel P, Wong P, Frishman D. A domain interaction map based on phylogenetic profiling. J Mol Biol. 2004;344:1331–1346. doi: 10.1016/j.jmb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Xu J, Liu Z, Liu Q, Zhao A, Shi T, Li Y. Refined phylogenetic profiles method for predicting protein–protein interactions. Bioinformatics. 2005;21:3409–3415. doi: 10.1093/bioinformatics/bti532. [DOI] [PubMed] [Google Scholar]
- 9.Ranea J, Yeats C, Grant A, Orengo C. Predicting Protein Function with Hierarchical Phylogenetic Profiles: The Gene3D Phylo-Tuner Method Applied to Eukaryotic Genomes. PLoS Comput Biol. 2007;3(11):e237. doi: 10.1371/journal.pcbi.0030237. doi:10.1371/journal.pcbi.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troyanskaya O, Dolinski K, Owen A, Altman R, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc Natl. Acad. Sci. USA. 2003;100:8348–8353. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis D, Jebara T, Noble W. Support vector machine learning from heterogeneous data: an empirical analysis using protein sequence and structure. Bioinformatics. 2006;22:2753–2760. doi: 10.1093/bioinformatics/btl475. [DOI] [PubMed] [Google Scholar]
- 12.Pal D, Eisenberg D. Inference of protein function from protein structure. Structure. 2005;13:121–130. doi: 10.1016/j.str.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Petrey D, Fischer M, Honig B. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proc Natl Acad Sci USA. 2009;106(41):17377–17382. doi: 10.1073/pnas.0907971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saraç ÖS, Atalay V, Cetin-Atalay R. GOPred: GO Molecular Function Prediction by Combined Classifiers. PLoS ONE. 2010;5(8):e12382. doi: 10.1371/journal.pone.0012382. doi:10.1371/journal.pone.0012382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brun C, Chevenet F, Martin D, Wojcik J, Guénoche A, Jacq B. Functional classification of proteins for the prediction of cellular function from a protein–protein interaction network. Genome Biol. 2003;5(1):R6. doi: 10.1186/gb-2003-5-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, et al. A map of the interactome network of the metazoan C.elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 18.Fromont-Racine M, et al. Toward a functional analysis of the yeast genome through exhaustive Y2H screens. Nat. Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 19.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nature Biotechnology. 2000;18:1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 20.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 21.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 22.Yu H. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–104. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7(4):e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rual JF, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 25.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32(Database issue):D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pagel P, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21(6):832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 28.Aranda B, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Research. 2009;38(Database issue):D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peri S, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Research. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chua H, Sung W, Wong L. Exploiting indirect neighbours and topological weight to predict protein function from protein–protein interactions. Bioinformatics. 2006;22:1623–1630. doi: 10.1093/bioinformatics/btl145. [DOI] [PubMed] [Google Scholar]
- 31.Hishigaki H, Nakai K, Ono T, Tanigami A, Takagi T. Assessment of prediction accuracy of protein function from protein-protein interaction data. Yeast. 2001;18:523–531. doi: 10.1002/yea.706. [DOI] [PubMed] [Google Scholar]
- 32.Deng M, Zhang K, Mehta S, Chen T, Sun F. Prediction of protein function using protein-protein interaction data. Journal of Computational Biology. 2003;10:947–960. doi: 10.1089/106652703322756168. [DOI] [PubMed] [Google Scholar]
- 33.Letovsky S, Kasif S. Predicting protein function from protein/protein interaction data: a probabilistic approach. Bioinformatics. 2003;19:197–204. doi: 10.1093/bioinformatics/btg1026. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez A, Flammi A, Maritan A, Vespignani A. Global protein function prediction from protein-protein interaction networks. Nature Biotechnology. 2003;21(6):697–670. doi: 10.1038/nbt825. [DOI] [PubMed] [Google Scholar]
- 35.Nabieva E, Jim K, Agarwal A, Chazelle B, Singh M. Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics. 2005;21:302–310. doi: 10.1093/bioinformatics/bti1054. [DOI] [PubMed] [Google Scholar]
- 36.Deng M, Chen T, Sun F. An integrated probabilistic model for functional prediction of proteins. Journal of Computational Biology. 2004;11:463–475. doi: 10.1089/1066527041410346. [DOI] [PubMed] [Google Scholar]
- 37.Nariai N, Kolaczyk ED, Kasif S. Probabilistic Protein Function Prediction from Heterogeneous Genome-Wide Data. PLoS ONE. 2007;2(3):e337. doi: 10.1371/journal.pone.0000337. doi:10.1371/journal.pone.0000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin CH, Chen SH, Ho CW, Ko MT, Lin CY. A hub-attachment based method to detect functional modules from confidence-scored protein interactions and expression profiles. BMC Bioinformatics. 2010 Jan 18;11 Suppl 1:S25. doi: 10.1186/1471-2105-11-S1-S25. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen C, Mannino M, Gardiner K, Cios K. ClusFCM: An algorithm for predicting protein functions using homologies and protein interactions. J Bioinform Comput Biol. 2008;6(1):203–222. doi: 10.1142/s0219720008003333. [DOI] [PubMed] [Google Scholar]
- 40.Kirac M, Ozsoyoglu G. Protein function prediction based on patterns in biological networks; Proceedings of 12th International Conference on Research in Computational Molecular Biology (RECOMB); 2008. pp. 197–213. [Google Scholar]
- 41.Cho YR, Zhang A. Predicting protein function by frequent functional association pattern mining in protein interaction networks. IEEE Trans Inf Technol Biomed. 2010;14(1):30–36. doi: 10.1109/TITB.2009.2028234. [DOI] [PubMed] [Google Scholar]
- 42.Hu P, Jiang H, Emili A. Predicting protein functions by relaxation labelling protein interaction network. BMC Bioinformatics. 2010 Jan 18;11 Suppl 1:S64. doi: 10.1186/1471-2105-11-S1-S64. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song J, Singh M. How and when should interactome-derived clusters be used to predict functional modules and protein function? Bioinformatics. 2009;1, 25(23):3143–3150. doi: 10.1093/bioinformatics/btp551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen CD, Gardiner KJ, Nguyen D, Cios KJ. PRICAI 2008. Vol. 5351. LNAI; 2008. Prediction of Protein Functions from Protein Interaction Networks: A Naïve Bayes Approach; pp. 788–798. [Google Scholar]
- 45.Ashburner M, et al. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sprinzak E, Sattath S, Margalit H. How reliable are experimental protein–protein interaction data? Journal of Molecular Biology. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 47.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 48.Yao L, Rzhetsky A. Quantitative systems-level determinants of human genes targeted by successful drugs. Genome Res. 2008;18(2):206–213. doi: 10.1101/gr.6888208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witten IH, Frank E. Data Mining: Practical machine learning tools and techniques. 2nd Edition. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 50.Niblett T. Proceedings of the Second European Working Session on Learning. Bled, Yugoslavia: Sigma; 1987. Constructing decision trees in noisy domains; pp. 67–78. [Google Scholar]
- 51.Agrawal R, Imielinski T, Swami A. Mining Association Rules Between Sets of Items in Large Databases; SIGMOD Conference; 1993. pp. 207–216. [Google Scholar]
- 52.Armstrong W. Information Processing. Vol. 74. North Holland: 1974. Dependency Structures of Data Base Relationships. [Google Scholar]