Abstract
The cell surface interaction between bacterial lipopolysaccharide (LPS), Toll-like receptor 4 (TLR4) and MD-2 is central to bacterial sepsis syndromes and wound healing. We have shown that a generation (G) 3.5 polyamidoamine (PAMAM) dendrimer that was partially glycosylated with glucosamine inhibits TLR4-MD-2-LPS induced inflammation in a rabbit model of tissue scaring. However, it was a mixture of closely related chemical species because of the polydispersity of the starting PAMAM dendrimer. Generation 2 triazine dendrimers with single chemical entity material status are available at low cost and at the kilogram scale. PAMAM dendrimer can be synthetically grafted onto this triazine core dendrimer to make new triazine-PAMAM hybrid dendrimers. This led us to examine whether molecular modeling methods could be used to identify the key structural design principles for a bioactive lead molecule that could be synthesized and biologically evaluated. We describe our computer aided molecular studies of several dendrimer based constructs and the key design principles identified. Our approach should be more broadly applicable to the biologically focused, rational and accelerated design of molecules for other TLR receptors. They could be useful for treating infectious, inflammatory and malignant diseases.
Keywords: molecular modeling, dendrimer, triazine, PAMAM, glucosamine, TLR4, LPS, MD-2, inflammation, cytokines
1. Introduction
Since their introduction in 1985 and subsequent commercial availability, PAMAM dendrimers have served as the major anchoring platform for the exploration of the properties and potential of dendrimers[1]. To this end, PAMAM dendrimers have already been successfully used for diagnostic, sensing, material science and catalysis based applications[2–5].
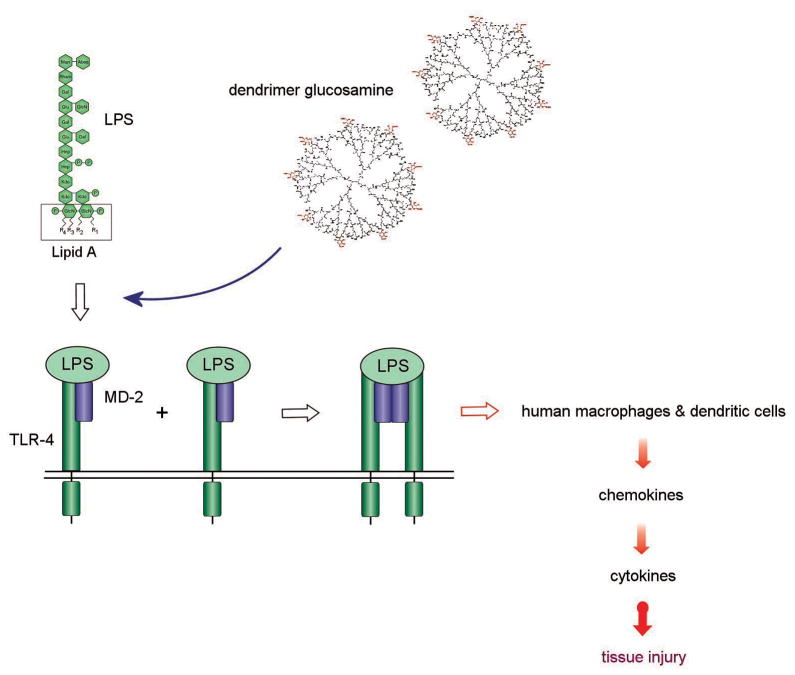
We have already shown that a partially glycosylated Generation (G) 3.5 PAMAM dendrimer blocked TLR4-MD-2-LPS mediated pro-inflammatory cytokine responses[6]. It inhibited TLR4 mediated inflammation in a rabbit model of tissue scaring (Figure 1). The biologically relevant cell surface interaction blocked was between bacterial lipopolysaccharide (LPS), host Toll-like receptor 4 (TLR4) and MD-2 [7,8]. Park et al. have recently defined the structural basis of the recognition of LPS by the TLR4-MD-2 complex[9]. In brief, the transport protein CD14 collects and delivers LPS to MD-2. The two phosphorylated glucosamines of the lipid A component of LPS bind to the charged entrance of MD-2’s hydrophobic cavity. The surface residues lining the entrance of MD-2’s pocket that have been shown to have a key role in the electrostatic binding of LPS are Arg90, Lys91, Ser118 and Lys122 [10]. This is followed by the lipid chains of LPS becoming buried in MD-2’s hydrophobic cavity. The activated TLR4-MD-2-LPS complex undergoes conformational changes and receptor dimerization, and this triggers intracellular signaling events[9]. It also initiates the pro-inflammatory chemokine and cytokine cascade responsible for host innate immune responses to pathogens and to surgical tissue injury.
Figure 1.
Illustration of the competition between LPS (agonist) and the partially glycosylated dendrimer (antagonist) for TLR4-MD-2-LPS complex induced pro-inflammatory cytokine production.
Our recent molecular docking studies have shown that these partially glycosylated dendrimers with a hydrophilic surface bind to the entrance of MD-2’s hydrophobic cavity and prevent the binding of LPS[11]. They form co-operative electrostatic interactions with residues lining the entrance to MD-2’s hydrophobic pocket (Figure 2). Crucially, dendrimer glucosamine interferes with the electrostatic binding of: (i) the 4’phosphate on the di-glucosamine of LPS to Ser118 on MD-2; (ii) LPS to Lys91 on MD-2; (iii) the subsequent binding of TLR4 to Tyr102 on MD-2. This is followed by additional co-operative interactions between several of the dendrimer glucosamine’s carboxylic acid branches and MD-2. Collectively, these interactions block the entry of the lipid chains of LPS into MD-2’s hydrophobic pocket and prevent TLR4-MD-2-LPS complex formation[11]. These partially glycosylated dendrimers did not interact with MD-2’s hydrophobic pocket, a property of other MD-2 antagonists such as eritoran.
Figure 2a & b. Frontal view of the hydrophilic and hydrophobic surfaces of MD-2.
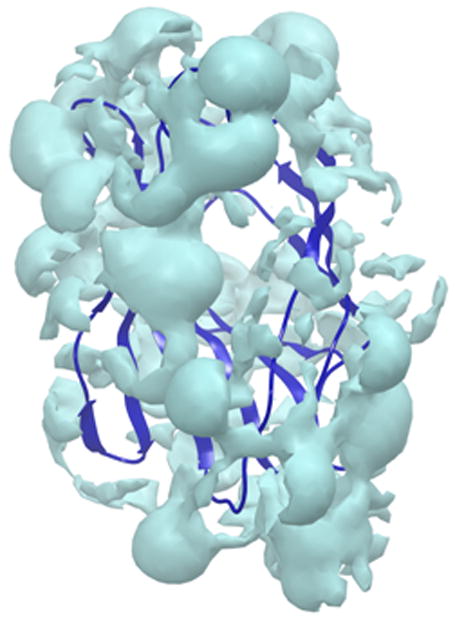
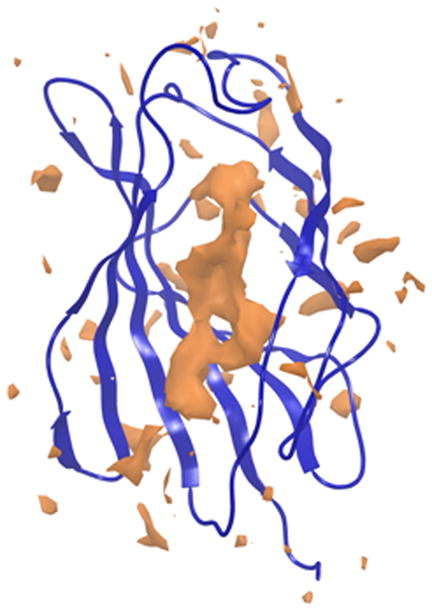
The hydrophilic (cyan) surface of MD-2 defines the need for any new dendrimer based construct to also have a hydrophilic surface. In contrast, the hydrophobic (orange) area, to which the acyl chains of Lipid A bind, lies buried deep inside MD-2’s cavity.
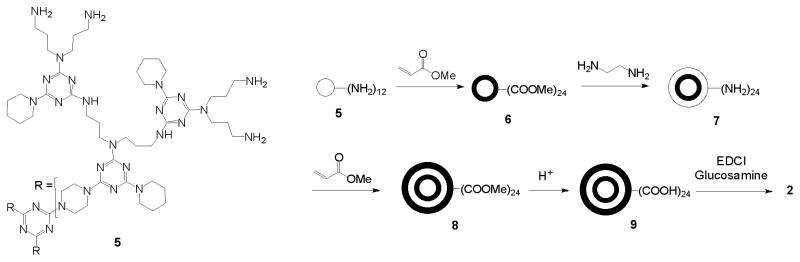
However, our active molecule 1 was a mixture of closely related chemical species. The largest contributor to this dispersity was the starting PAMAM material itself[1,12]. The availability of G2 triazine dendrimers at the kilogram scale and at low cost led us to investigate whether they could be used as an alternative starting dendrimer platform. These core materials are chemically well defined structures when compared to PAMAM core dendrimers[13]. For example, a comparative analysis with PAMAM analogues using capillary electrophoresis showed a significant difference in molecular heterogeneity; PAMAM dendrimers were mixtures of closely related molecules whereas triazine dendrimers approached single chemical entity material status[14,15].
Our initial synthetic studies of the partial glycosylation of G2 and G3 triazine dendrimers did not result in the reconstitution of the biological activity required (data not shown) despite these materials being of a size and having a surface glucosamine loading that was similar to the biologically active PAMAM dendrimer glucosamine 1. As our previous modeling studies[11,16,17] had clearly demonstrated that the surface properties of partially glycosylated dendrimers determined their bioactivity (i.e., flexibility, cluster density, surface electrostatic charge and hydrophilicity), we went on to investigate whether grafting a biologically inactive, but single entity triazine dendrimer with a minimum number of PAMAM layers could lead to molecules that replicated the flexibility, cluster density, surface electrostatic charge, hydrophilicity and biological activity of 1.
2. Materials and Methods
2.1. Molecular dynamics simulations
The minimization and molecular dynamics simulations were performed by building 2D structures of the dendrimers in ChemBioDraw Ultra 11.0 (ChemBioOffice 2008), converted to 3D ChemBio3D 11.0 (ChemBioOffice 2008), and saved as mol files. The mol files were submitted to 3D optimization in Avogadro 1.0 (http://avogadro.openmolecules.net) using MMFF force field[18] and saved in a pdb format.
The resulting structures of all of the dendrimers studied were imported into Maestro version 9 (www.schrodinger.com) and minimized with Macromodel version 9.7.211[19] for 2500 iterations using OPLS2005 force field[20]. The effects of water as solvent were taken into account by Generalized Borhn/Surface Area (GB/SA) implicit solvent model[21]. The protonation state of the nitrogen atoms at branching points and of the carboxylic groups on the surface were confirmed by pKa calculations using sparc online version 4.5 (http://sparc.chem.uga.edu/sparc/). Molecular dynamics simulations using same force field and implicit solvent methods were performed at 300k with Macromodel. The extended cut-off distances were set at their default values for implicit solvation, namely Van der Waals (8.0 Å), electrostatic (20.0 Å), and H-bond interactions (4.0 Å). A 100 ps equilibration simulation was preceded by 2500 steps of minimization. A 2 ns production run was performed as previously described[22]. A sample structure was recorded for every 10 ps of the simulation for trajectory analysis.
In addition, 10 ns molecular dynamics simulations were performed with explicit solvent. The initial structures used for these simulations were the structures resulting from the previous simulations with implicit solvation. Desmond was used to perform molecular dynamics simulations with explicit solvent[23]. The dendrimer glucosamine system was built using the SPC solvation model and the size of the box determined automatically by creating a buffer zone of 10 Å around the dendrimer. The molecular dynamics simulation, structure minimization and relaxation steps were performed for 10 ns at 300 K and 1.03 bar. Snapshot structures were recorded every 5 ps.
2.2. Molecular surface
The molecular surface of each structure was then calculated in trajectories by Maestro 9 using a 1.4 Å probe, the values exported in cvs format, and the image of the surface area of each dendrimer under study exported as jpeg. Flexibility was assessed with RMSD trajectory analysis; the first 20 structures of each trajectory were discarded and the RMSD determined in relation to the first of the remaining structures using Maestro toolkit, and then exported in cvs format. The hydrophobic and hydrophilic surfaces were calculated for a single structure of each trajectory using Maestro 9 with isovalues of 0.5 and -6 respectively. The images were exported as jpeg and the area values noted. The trajectories of each molecule were saved in mol2 format and imported into VegaZZ 2.2.0.54 [24] where the trajectory analysis tool was used for polar surface calculations along the trajectory with a probe radius of 1.4 Å. Microsoft Excel was used to plot all properties determined.
2.3. Hybrid dendrimer chemistry
A narrow polydispersity (Mw/Mn), where Mw is the weight average molar mass and Mn is the number average molar mass has been achieved for G1 and G2 PAMAM dendrimers; (a) the G1-amine had a Mn of 1,430 g/mol and a polydispersity of 1.024; (b) the G1-acid had a Mn of 2,230 g/mol and a polydispersity of 1.070; (c) the G2-amine had a Mn of 3,256 g/mol and a polydispersity of 1.054. In addition, reverse-phase HPLC has been successfully used to purify G1 and G2 PAMAM dendrimers from crude reaction products[25]. Taken together, these analytical observations encouraged us to pursue the synthesis of a partially glycosylated hybrid triazine PAMAM dendrimer.
The synthesis and analytical characterization of dendrimer 6 has been reported[14]. A solution of 6 was prepared by dissolving 8.49 g (1.692 mmol) in 50 ml methanol. The flask was immersed in a water bath at room temperature. To the dendrimer solution, 150 ml (134.7g, 2.24 mol) of ethylenediamine (EDA) corresponding to 55 mole equivalents of EDA per mole of ester was slowly added, under continuous stirring, to avoid the rapid release of significant amounts of heat. The reaction mixture was allowed to stir at room temperature. Aliquots were taken to which diethyl ether was added, resulting in the precipitation of the dendrimer. The sample was subjected to centrifugation and the collected solid analyzed by MALDI-TOF-MS. Additional EDA was added into the reaction when no change was observed between MS traces. When the reaction was complete, methanol and excess EDA were removed by adding the reaction mixture to a flask containing diethyl ether under continuous stirring. Gummy solids were produced which were allowed to settle overnight followed by removal of EDA, methanol and diethyl ether mixture by decantation. 1H NMR (CDCl3): δ 4.0-3.55 (br, m), 3.55-3.1 (br, m), 3.1-2.58 (br, m), 2.56-2.17 (br, m), 2.03-1.23 (br, m). 13C NMR (CDCl3): δ 174.41, 164.41, 72.50, 65.91, 64.39, 48.66, 44.21, 40.31, 39.72, 39.62, 32.61, 14.10.
A solution was then prepared by dissolving the oily amine terminated triazine PAMAM hybrid dendrimer 7 obtained from the previous reaction (corresponding to 9.63 g, 1.692 mmol) in 50 mL methanol. Methyl acrylate was slowly added to this solution. Initially, 7.31 ml (6.98 g, 81.22 mmol) of methyl acrylate (corresponding to 10 mole equivalents per NH2) was added. The reaction mixture was allowed to stir at room temperature. Samples were taken over time and analyzed by MALDI-TOF-MS. Additional methyl acrylate was added to the reaction until no further change was observed between the MS traces. The reaction was deemed complete after a total of 40 mole equivalents of methyl acrylate per NH2 had been added, and the reaction allowed to react for 11 days. Methanol and excess methyl acrylate were removed under reduced pressure to yield an oily substance. 1H NMR (CDCl3): δ 4.07-3.47 (br, m), 3.59 (s), 3.35(s), 3.29-3.09 (br, m), 3.03 (br, m), 2.84 (br, m), 2.80-2.60 (br, m), 2.56 (br, m), 2.52-2.10 (br, m), 1.89 (m), 1.84-1.60 (br, m), 1.60-1.26 (br, m). 13C NMR (CDCl3): δ 173.02, 165.39, 51.55, 49.06, 45.16, 44.11, 43.10, 32.35, 30.31, 25.79.
The oily dendrimer 8, obtained from the previous reaction, was dissolved in methanol and an aliquot of the solution corresponding to 1.0 g of dendrimer 8 taken. The solvent was removed under reduced pressure and the remaining material dissolved in 10 ml of 2 M NaOH. It was then allowed to react at room temperature. Progress of the hydrolysis reaction was monitored using FT-IR and MALDI-TOF-MS. The NIR spectra showed the disappearance of the signal corresponding to the carbonyl of the ester group, and the appearance of a signal corresponding to the carbonyl of the acid group; the signal corresponding to the amide carbonyl remained constant (Supplementary Figure 1). Complete hydrolysis of the esters was observed after 2 days. The mixture was acidified to pH 3 using 2 M HCl. The acidified solution was then filtered in two stages using Amicon ultrafiltration stirred cells (Amicon Bioseparations) under nitrogen pressure. The first stage required a cell with a preconditioned regenerated cellulose membrane filter with a MWt cut-off of 10 kDa. The collected filtrate was transferred into another Amicon ultrafiltration stirred cell with a similar membrane but with a MWt weight cut-off of 1 kDa. The retained solution was collected and the solvent removed under reduced pressure to yield a yellowish solid. The removal of excess acrylate was observed in the NIR spectra as the decrease in the acid carbonyl signal relative to the signal for the amide carbonyl. 1H NMR (D2O): δ 3.75-2.90 (br, m), 2.85-2.20 (br, m), 2.15-1.10 (br, m). 13C NMR (D2O): δ 177.40, 171.97, 51.43, 50.97, 50.62, 49.31, 47.70, 34.38, 30.42, 30.10.
2.4. Partially glycosylated hybrid dendrimer chemistry
The dendrimer 9 (100 mg, 10.9 μmoles) was dissolved in 1 ml of endotoxin free water and added to 100 mg (0.465 mol) of glucosamine-HCl dissolved in 2 ml of water (pH 5.0). 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDCI) (260 mg, 1.356 mmol) was dissolved in 5.2 ml water (pH 5.0) and added to 9 and glucosamine, and stirred for 3 h at room temperature at pH 5. The solution was then filtered through a Pierce Concentrator with MWt cut-off of 20 kDa and transferred into an Amicon ultrafiltration stirred cell with a MWt cut-off of 2 kDa. The dialysate was lyophilized and reconstituted in water at 50 mg/ml. 1H NMR (D2O): δ 5.28 (d), 5.06 (br, s), 3.78-2.80 (br, m), 2.78-2.1 (br, m), 2.05-1.82 (br, m), 1.81-1.65 (m), 1.60-1.10 (br, m), 1.09-0.96 (br, t), 0.95-0.75 (t). 13C NMR (D2O): δ 177.20, 171.86, 104.71, 94.86, 90.74, 89.07, 75.86, 73.75, 71.48, 70.63, 70.03, 60.64, 60.46, 55.25, 53.94, 42.67, 42.57, 36.30, 34.86, 24.92, 14.30.
2.5. Analytical chemistry
1H and 13C NMR of the samples in D2O were obtained using a Varian Mercury 300 MHz instrument with the NMR spectra of 2 acquired using a Bruker Avance 500 MHz instrument. Chemical shifts are reported in parts per million on the δ scale relative to solvent. MALDI-TOF mass spectra were obtained using an ABI Voyager-DE STR mass spectrometer operating in reflected mode using 2,4,6-trihydroxyacetophenone (THAP) as the matrix. When needed, MS and FT-IR data were reprocessed using Origin8 and Microsoft Excel software packages. Solution infrared spectra were recorded on a Bruker Tensor 27 FTIR spectrometer using 0.1 mm NaCl sealed cells. IR spectroscopy was useful for monitoring the hydrolysis of the methyl ester; the disappearance of this band confirmed our interpretation of the NMR.
2.6. TLR4-MD-2-LPS responses
Peripheral blood mononuclear cells (PBMCs) were prepared by Ficoll-Hypaque (GEHealthCare) density gradient centrifugation from human buffy coat residues. Purified PBMCs were allowed to adhere to 15 cm sterile tissue culture plates for 1 h and then washed with PBS twice to remove non-adherent cells. Adherent monocytes were removed with a cell scraper and counted using a Coulter counter.
Cytotoxicity was determined using MTT assays. Triplicate 200 μl samples of culture media (RPMI 1640 + 10% FCS) containing 105 monocytes and serial dilutions of the molecules (25 – 600 μg/ml) were incubated for 7 h at 37°C in 96 well tissue culture plates. Fifty μl of MTT (Invitrogen, 5 mg/ml in PBS) was then added and the incubation continued for 2 h. The positive control had no molecule added, and the negative control had 0.5% Triton X-100 to ensure cell death. Optical density was read at 490 nm and percentage survival calculated.
TLR4 induced inflammation was studied by culturing one million monocytes in 1 ml of culture media for 1 h in 48 well tissue culture plates to enable cell adherence. The molecules (25 – 400 μg/ml) were then added in duplicate to wells for 30 min followed by 25 ng/ml of ultra-pure LPS (Salmonella minnesota, Invivogen). Positive control wells contained LPS only. Negative control wells contained monocytes only. After a 3 h incubation, the media was removed and the cells lysed in 500 μl Tri-Reagent (Sigma).
RNA was then extracted and reverse transcription performed on 400 ng of RNA using a Qiagen kit. Triplicate aliquots of the cDNA were subjected to quantitative real time PCR for HPRT, MIP-1β [CCL4], IL-8, TNF-α, IL-6, IL-10 and IFN-β as previously described[6] using a Corbett Rotorgene 3000 PCR machine and a JumpStart Sybr Green mix (Sigma). Results were expressed as the log10 absolute copy number for each chemokine and cytokine mRNA per 105 copies of HPRT (a house keeping gene) mRNA. The results shown are from three experiments.
3. Results
3.1. Computational design parameters
We first focused on replacing the core of the PAMAM dendrimer with a hybrid triazine-PAMAM dendrimer that was derived from the extension of a reported strategy[14]. Our design criteria were shaped by three key parameters. First, the hydrophobic triazine core would have to be “hidden” with groups that conveyed water solubility and a hydrophilic surface to the hybrid dendrimer, and which also enabled the outward facing presentation of the monosaccharide via a carboxylic acid reaction site. Second, the use of a discrete, higher generation polyamine core (i.e., a triazine dendrimer) would reduce substantially the structural heterogeneity of the final hybrid dendrimer. Third, the use of PAMAM chemistry had to be reduced to a minimum because it increases structural heterogeneity[14,15].
To compare the molecular properties of biologically active 1 with those of several triazine based dendrimers that were biologically inactive, we used molecular modelling techniques and an established protocol[22] with which we have previously had considerable success for both proteins and peptides[22,26–28]. The glycosylated PAMAM, triazine, and several variations of the generations of the hybrid triazine and PAMAM dendrimers, were simulated with implicit solvation. Deriving useful data from these modelling studies required the assumption that the representative structure 1 could effectively communicate key design principles using which structures could be modelled and compared. MS and NMR analysis of 1 showed an average of 8 glucosamines per PAMAM dendrimer molecule[6]; i.e., 2 glucosamine residues on each of the 4 arms of 16 carboxylics each of the G3.5 PAMAM model of 1 (Scheme 1).
Scheme 1.
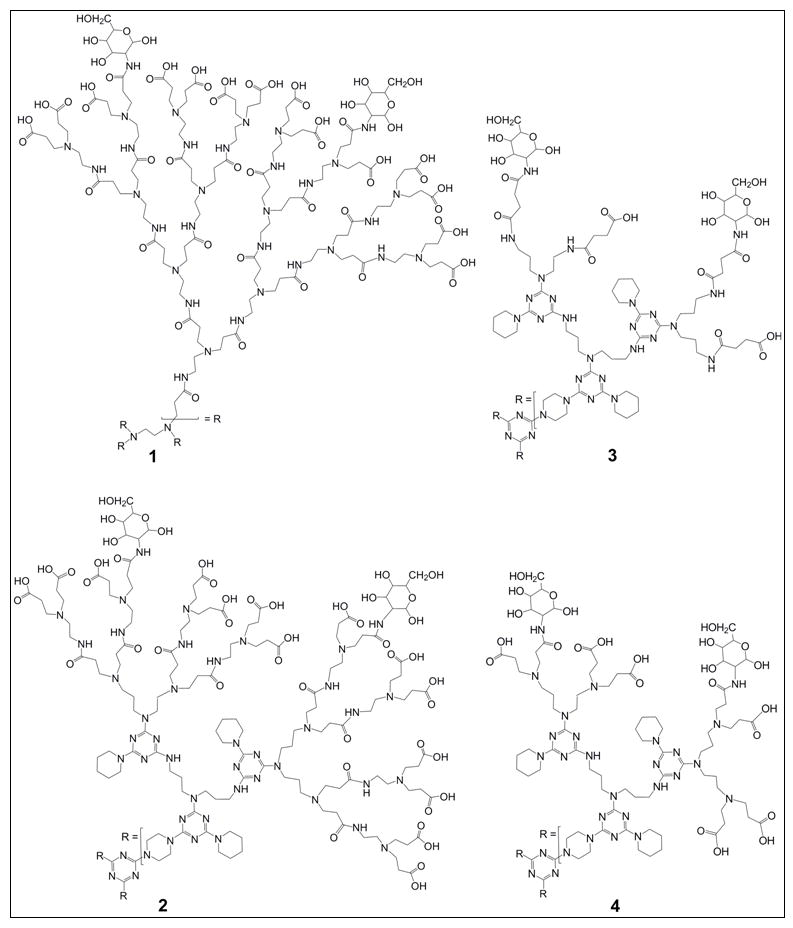
Partially glycosylated dendrimers 1 – 4.
This was confirmed by a combination of quantum chemistry calculations and molecular dynamics simulations of a series of dendrimers with different glucosamine loadings[17]. The triazine based dendrimers were then built with a total of 6 glucosamines; i.e., 2 glucosamines on each of the 3 arms of the triazine analogues because this was sterically and energetically the most favourable configuration. The position of the glucosamine molecules on each branch was randomly assigned because molecular dynamics simulation studies showed that the carboxylic acid groups were both equivalent and highly flexible[17]. In this paper, we present the detailed data for the representative hybrid dendrimers 2, 3, 4 (Scheme 1) because they were selected for chemical synthesis and biological evaluation.
3.2. Molecular dynamics simulations
A set of 2 ns molecular dynamics simulations on each of these molecules and subsequent analysis revealed molecular properties that could impact on their bioactivity. We found that their surface properties did not vary significantly once the structures had equilibrated. As expected, their surface area (SA) varied considerably. The SA’s of 3 and 4 were much lower than those of 1 and 2; they had a SA that was much closer to that of the target, 1 (Table 1). Molecular representation of the SA also showed a significant difference in the shapes of these molecules. While 3 and 4 presented a compact structure with the PAMAM branch units folding inwards, 2 presented a relaxed structure with the PAMAM branch units facing outwards; this resulted in surface properties that were similar to those of the target, 1. Measurement of polar surface area (PSA) mirrored this trend with 3 and 4 displaying a 4-fold smaller PSA than the target, 1. The PSA of 2 was closest to that of the target, 1 (Table 1). Therefore, the SA, PSA, and the more relaxed structure (i.e., with branches outstretched) of the partially glycosylated G2 triazine G1.5 PAMAM hybrid dendrimer was closest to the biologically active G3.5 PAMAM dendrimer glucosamine. This gave us the first indication of the relationship between the hydrophobic and hydrophilic surfaces of these molecules, and their impact on the design of bioactive hybrid dendrimers.
Table 1.
Dendrimer surface property analysis along the trajectory of a 2 ns molecular dynamics simulation with implicit solvent.
| 3 | 4 | 2 | 1 | |
|---|---|---|---|---|
| SA (Å2) | 3570 | 3800 | 8536 | 11212 |
| PSA (Å2) | 2035 | 2278 | 5776 | 7283 |
3.3. Hydrophobicity and hydrophilicity
Our recent studies on the interaction of partially glycosylated G3.5 PAMAM dendrimers with MD-2 showed that these molecules interact with residues lining the hydrophilic entrance of MD-2’s hydrophobic pocket without forming intermolecular contacts with the hydrophobic pocket itself[11]. It was therefore important to investigate the relative distribution of the hydrophobic and hydrophilic surfaces of these molecules because this could become a key feature of the design of bioactive hybrid dendrimers. The surface of target 1 was mainly hydrophilic (Figure 3). Dendrimer 3 had multiple exposed hydrophobic surfaces that were a consequence of the PAMAM branching units folding inwards and exposing the hydrophobic triazine core. Since the dendrimer 3 was found to be biologically inactive, this led us to conclude that surface hydrophobicity was not an important property for the molecule’s activity against the TLR4-MD-2-LPS complex. These contrasting surface properties of bioactive as compared to bioinactive dendrimers provided us with clues for the selection of candidate molecules for synthetic studies. In the case of 2, its hydrophilic surface distribution shielded the hydrophobic triazine core from solvent and led to the molecule adopting a globular shape. This meant that the surface characteristics of 2 resembled 1 much more closely than either 3 or 4 (Figure 3).
Figure 3.
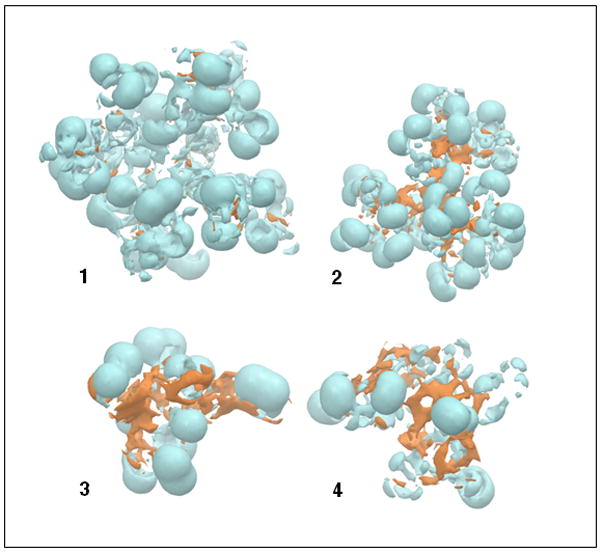
Hydrophilic (cyan) and hydrophobic (orange) surfaces of the partially glycosylated dendrimers 1 – 4.
Taken together, these modeling studies results predicted that the addition of 1.5 generations of PAMAM to the single species G2 triazine core, 5, would enable the synthesis of the simplest and most narrowly dispersed hybrid molecule that would also be bioactive. Notably, the G2 triazine G1.5 PAMAM hybrid dendrimer glucosamine also had a globular structure, a feature absent in the G2 triazine dendrimer itself.
3.4. Surface charge distribution
The surface charge distribution was also identified as an important factor for bioactivity[11]. The polar surface area values given above do not provide insights into the location of the polar surfaces. We found that the distribution of the interpolated charges on the surfaces of the partially glycosylated triazine dendrimer and the partially glycosylated G2 triazine G0.5 PAMAM hybrid dendrimer were similar; most were neutral surfaces with small areas of well separated negative charges (Figure 4). In contrast, the distribution of the interpolated charges on the surface of the partially glycosylated G2 triazine G1.5 PAMAM hybrid dendrimer were similar to those of the partially glycosylated G3.5 PAMAM dendrimer, despite differences in their internal charge distribution. Their polar surfaces were also closer to the surface for both dendrimers; this increased the probability of multiple electrostatic interactions with the target protein. These surface charge property similarities provided another explanation for why the partially glycosylated G2 triazine G0.5 PAMAM hybrid dendrimer and the partially glycosylated triazine dendrimer were not biologically active. As the partially glycosylated G2 triazine G1.5 PAMAM hybrid dendrimer had surface properties that were similar to those of the partially glycosylated G3.5 PAMAM dendrimer, we concluded, that it should be bioactive.
Figure 4. Surface charge.
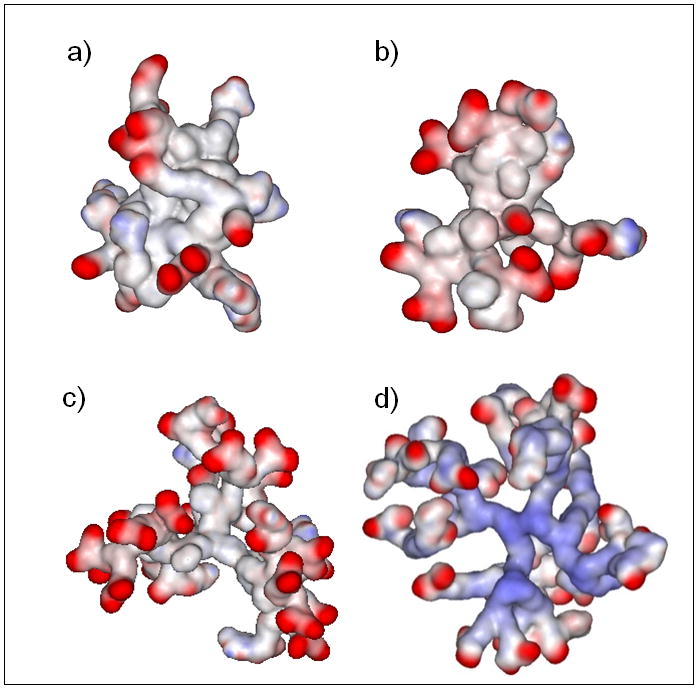
The red areas correspond to negatively charged residues and the blue areas to positively charged residues.
(A) G2 triazine dendrimer modified with 6 glucosamine molecules.
(B) G2 triazine G0.5 PAMAM hybrid dendrimer modified with 6 glucosamine molecules.
(C) G2 triazine G1.5 PAMAM hybrid dendrimer modified with 6 glucosamine molecules.
(D) G3.5 PAMAM dendrimer modified with 8 glucosamine molecules.
3.5. Other partially glycosylated hybrid dendrimers
Several other molecules were also studied including a partially glycosylated G3 triazine G0.5 PAMAM hybrid dendrimer, and a partially glycosylated G3 triazine G1.5 PAMAM hybrid dendrimer. This analysis showed that they had a similar molecular surface, hydrophilic versus hydrophobic surface distribution, and interpolated charge surfaces (Supplementary Figure 2). Taken together, these additional molecular modeling studies predicted that 1.5 layers of surface PAMAM were required to mimic the biological activity of 1. This meant that the number of PAMAM generations required for a bioactive hybrid dendrimer could be more than halved.
3.6. Hybrid dendrimer chemistry
Our synthetic efforts commenced with a single species G2 triazine dendrimer that was available at the kilogram scale[13]. It presented 12 amines at the periphery. The reaction scheme is shown in Scheme 2. It included the sequential addition of methylacrylate, ethylenediamine and methylacrylate, followed by hydrolysis to yield the penultimate intermediate in this strategy. Dendrimer 6 arises from Michael addition to triazine dendrimer, 5, and comprises two products as previously described; the major and desired material with 24 methyl acrylate groups, 6, and the minor product with 23 groups[14]. Hydrolysis of the hybrid material 8 was achieved with 2 M NaOH. Previously, 6 had been subjected to 6 M HCl for hydrolysis, but the presence of amide bonds in this material precluded that strategy. High salt concentrations and the presence of multiple carboxylate counterions (i.e., H+ or Na+) led to mass spectra that were unsuitable for the purpose of monitoring reaction progress.
Scheme 2.
Synthesis of the partially glycosylated triazine PAMAM hybrid dendrimer.
3.7. Analytical chemistry
Characterization of these materials (6 to 9, and 2) reflected both the advantages conveyed by triazine chemistry and the challenges inherent in the synthesis of larger generation dendrimers using PAMAM chemistry[14,15]. The mass spectrometry revealed a faithful elaboration of the starting material; those intermediates bearing excess graft that could arise from either residual diamine or retro-Michael reactions could be much more readily assigned than for the G3.5 PAMAM dendrimer with which we had originally started our studies. This meant that there was a better understanding of the heterogeneity that arose from the PAMAM portion of this hybrid molecule.
The unambiguous assignment of the 1H NMR spectra was precluded by the presence of several broad and overlapping signals, and was in agreement with the previously reported spectra of hybrid dendrimers[15]. We were able to define the diagnostic features of: (i) groups associated with the triazine core[13]; (ii) groups associated with the PAMAM layer[6]; (iii) the glucosamines[6]. The glucosamines provided useful lines for the H1 proton in either alpha (5.28 ppm) or beta (5.06 ppm) configurations in a region that is distinct from the dendrimer portions downfield. The rest of the glucosamine signals are found between 3 and 4 ppm and they overlapped with CH2 groups from the dendrimer (2.5 to 4 ppm) itself. The other downfield signatures observed could be subdivided into regions associated with the triazine dendrimer itself, including CH2 groups, that are adjacent to an exocyclic N-triazine (4.1-3.5 ppm), and those of the piperidine groups (2.0-1.2 ppm). Urea obscures this latter region with CH2 groups appearing from 2.05 - 1.80 ppm, and a characteristic methyl group at 1.1 - 0.9 ppm. The need for relative integrations of the CH2 groups that are associated with PAMAM derived from acrylate groups, either next to the nitrogen (3.3 - 3.1 ppm) or the carbonyl (2.8 - 2.2 ppm) was reduced, given our ability to monitor these reactions with mass spectrometry. Taken together, these results showed that 8 glucosamines were present on the surface of the desired product 2.
3.8. Biological studies
The products were evaluated using an MTT assay to determine their relative cellular toxicity. Neither 1 nor 2 was toxic to primary human blood derived monocytes at 600 μg/ml after a 7 h incubation, a time point chosen because the half-life of 1 in blood was 7 h (Supplementary Figure 3). When endotoxin free 2 was compared with 1, it was found to be an equally effective partial antagonist of TLR4-MD-2-LPS complex induced pro-inflammatory cytokine production (Figure 5). At 100 μg/ml, the mRNA copy number of IL-6 was reduced by 2.2 log10 (99.3%), Macrophage Inflammatory Protein (MIP) -1β[CCL4] by 1.7 log10 (98%), Tumor Necrosis Factor-α (TNF-α) by 1.1 log10 (96%), and Interleukin (IL) -8 by 1.6 log10 (91%) when compared to the maximal stimulation of pro-inflammatory chemokines and cytokines that was triggered with 25 ng/ml of ultra-pure LPS. The ED50 of these two molecules was similar at 75 ± 15 μg/ml. Neither 1 nor 2 affected production of the anti-inflammatory cytokines Interleukin-10 and Interferon-γ.
Figure 5. Partial antagonist activity of 2 against the TLR4-MD-2-LPS cell surface receptor complex.
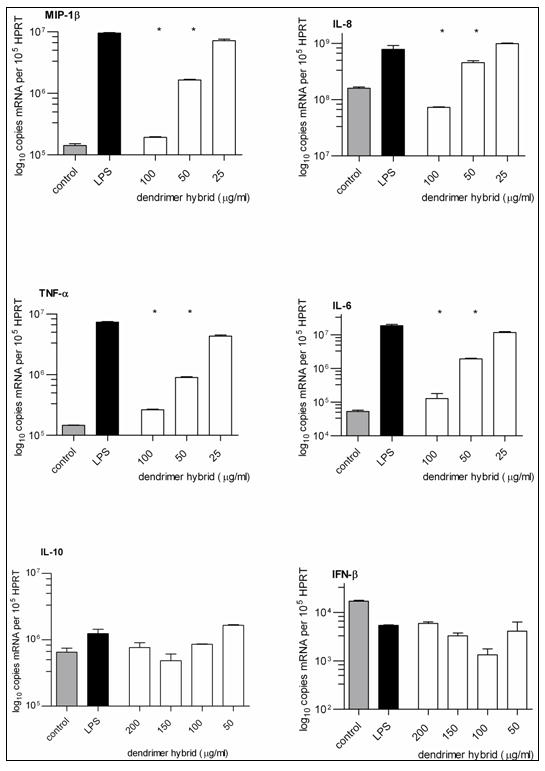
The partially glycosylated hybrid dendrimer 2 blocked LPS triggered pro-inflammatory chemokine (MIP-1β[CCL4] and IL-8) and cytokine (TNF-α and IL-6) production. It did not affect anti-inflammatory cytokine (IL-10 and IFN-β) production. The control was cultured primary human monocytes. * = p<0.001 when compared to the LPS positive control (Student’s t-test). n = 3.
3.9. Validation of computational design principles
We have previously shown, for partially glycosylated G3.5 PAMAM dendrimers, that there are no significant differences in the surface properties of the partially glycosylated molecules 7, 8 and 9 [17]. We therefore did not expect differences between the hybrid 2, as modelled with 6 surface glucosamines, and the experimentally synthesised hybrid molecule with 8 surface glucosamines. Nevertheless, to validate our molecular modelling principles, and to confirm the accuracy of the properties that we had predicted from our modelling studies, we performed an additional 2 ns molecular dynamics simulation to determine the properties for the hybrid 2 (with 8 glucosamines) as compared to the hybrid 2 (with 6 glucosamines). We found that there was no significant difference in the calculated values for either Surface Area (SA) = 8732 vs 8536 Å2 or Polar Surface Area (PSA) = 5807 vs 5776 Å2 for the hybrid 2 (with 8 glucosamines) when it was compared to the hybrid 2 (with 6 glucosamines) respectively.
In further studies, we went on to perform longer molecular dynamics simulations of these two hybrid molecules in explicit water to gain a better insight into their behaviour. Their surface properties were, once again, determined for each partially glycosylated dendrimer along the trajectory (Table 2). The surface area (SA) and polar surface area (PSA) showed an increase when comparing the triazine dendrimer with the G2 triazine G1.5 PAMAM hybrid dendrimer. In addition, these results also showed that there was no significant difference between the hybrid dendrimer with 6 surface glucosamines that was predicted to be bioactive, and the model generated of the experimentally synthesized and bioactive hybrid dendrimer with 8 surface glucosamines. The gyration radius values determined also showed the same trend; the G2 triazine G1.5 PAMAM hybrid dendrimer was a less compact structure. Notably 2(with 6 glucosamines) and 2(with 8 glucosamines) were very similar to the biologically active PAMAM dendrimer, 1.
Table 2.
Average molecular properties along the trajectory of 10 ns molecular dynamics simulation of the fully equilibrated molecules in explicit solvent.
| 3 | 4 | 2(6g) | 2(8g) | 1 | |
|---|---|---|---|---|---|
| SA (Å) | 4527.93 | 5714.03 | 10648.88 | 10555.73 | 14181.90 |
| PSA (Å) | 1793.17 | 2305.07 | 5030.59 | 4960.91 | 5782.19 |
| Gyration radius (Å) | 11.61 | 13.58 | 17.96 | 17.65 | 19.38 |
2 (6g) – with 6 glucosamines; 2(8g) – with 8 glucosamines.
3.10. Explicit versus implicit solvent
The results of longer simulations in explicit solvent were consistent with the results from implicit solvent studies for bioactive molecules. This demonstrated that the implicit solvent approach, even though it implies more approximations, can be usefully adopted as an efficient and time effective approach to aid the design of dendrimer constructs. It should be noted that our experience also suggests that, for a better understanding of the interactions of these molecules with their biological target, it is prudent to undertake longer simulations in explicit solvent[11].
4. Discussion
We describe a rational computational modelling based approach to the design of molecules with pre-defined bioactivity, and their subsequent synthesis, characterization and biological evaluation. Our studies show that bioactive molecules share several surface properties that required the addition of 1.5 generations of PAMAM to the core dendrimer. This more than halves the previously reported PAMAM requirement for a partially glycosylated dendrimer[6]. We now propose that the molecular modelling design principles described could be used to identify other structurally optimised dendrimers with interesting bioactivity. Designing and delivering these molecules with MWts of 2 – 4 kDa, which we call synthetic baby-bios (SBBs), is the next challenge in this field of biomaterials.
5. Conclusion
Computation played a central role in identifying alternative compositions of matter with the requisite hydrophilic surface areas, cluster density, flexibility and surface charge distribution required for bioactivity. The approach described should be applicable to the design and synthesis of other bioactive dendrimers for other pathogen related TLR receptors, such as TLR2 [29] and TLR3 [30], using alternative polypropyleneimine, polylysine or polypropyletherimine based chemistries. These SBBs could be useful for treating infectious, inflammatory and malignant diseases.
Supplementary Material
Acknowledgments
This research was supported by NIH grants U01 5U01AI075351-02 to SS and R01 GM64560-07 to ES, and the Hammersmith’s Biomedical Research Centre.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tomalia DA, Baker H, Dewald J, Hall M, Kallos G, Martin S, et al. A new class of polymers - Starburst-dendritic macromolecules. Polymer J. 1985;17:117–132. [Google Scholar]
- 2.Krasteva N, Besnard I, Guse B, Bauer RE, Mallen K, Yasuda A, et al. Self-assembled gold nanoparticle/dendrimer composite films for vapor sensing applications. Nano Letters. 2002;2:551–555. [Google Scholar]
- 3.Stiriba SE, Frey H, Haag R. Dendritic polymers in biomedical applications: from potential to clinical use in diagnostics and therapy. Angew Chem Int Ed Engl. 2002;41:1329–1334. doi: 10.1002/1521-3773(20020415)41:8<1329::aid-anie1329>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 4.Ribourdouille Y, Engel GD, Richard-Plouet M, Gade LH. A strongly positive dendrimer effect in asymmetric catalysis: allylic aminations with pyrphos-palladium functionalised PPI and PAMAM dendrimers. Chem Commun (Camb) 2003;11:1228–1229. doi: 10.1039/b302155f. [DOI] [PubMed] [Google Scholar]
- 5.Astruc D, Boisselier E, Ornelas C. Dendrimers designed for functions: from physical, photophysical, and supramolecular properties to applications in sensing, catalysis, molecular electronics, photonics, and nanomedicine. Chem Rev. 2010;110:1857–1959. doi: 10.1021/cr900327d. [DOI] [PubMed] [Google Scholar]
- 6.Shaunak S, Thomas S, Gianasi E, Godwin A, Jones E, Teo I, et al. Polyvalent dendrimer glucosamine conjugates prevent scar tissue formation. Nat Biotechnol. 2004;22:977–984. doi: 10.1038/nbt995. [DOI] [PubMed] [Google Scholar]
- 7.Kim JI, Lee CJ, Jin MS, Lee CH, Paik SG, Lee H, Lee JO. Crystal structure of CD14 and its implications for LPS signaling. J Biol Chem. 2005;280:11347–11351. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]
- 8.Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of LPS recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 10.Meng J, Lien E, Golenbock DT. MD-2-mediated ionic interactions between lipid A and TLR4 are essential for receptor activation. J Biol Chem. 2010;285:8695–8702. doi: 10.1074/jbc.M109.075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barata TS, Teo I, Brocchini S, Zloh M, Shaunak S. Partially glycosylated dendrimers block MD-2 and prevent TLR4-MD-2-LPS complex mediated cytokine responses. PLoS Comput Biol. 2011;7(6):e1002095. doi: 10.1371/journal.pcbi.1002095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esfand R, Tomalia DA. Poly(amidoamine) (PAMAM) dendrimers: from biomimicry to drug delivery and biomedical applications. Drug Discov Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 13.Chouai A, Simanek EE. Kilogram-scale synthesis of a second-generation dendrimer based on 1,3,5-triazine using green and industrially compatible methods with a single chromatographic step. J Org Chem. 2008;73:2357–2366. doi: 10.1021/jo702462t. [DOI] [PubMed] [Google Scholar]
- 14.Lalwani S, Chouai A, Perez LM, Santiago V, Shaunak S, Simanek EE. Mimicking PAMAM dendrimers with amphoteric, hybrid triazine dendrimers: A comparison of dispersity and stability. Macromolecules. 2009;42:6723–6732. doi: 10.1021/ma9011818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalwani S, Venditto VJ, Chouai A, Rivera GE, Shaunak S, Simanek EE. Electrophoretic behavior of anionic triazine and PAMAM dendrimers: methods for improving resolution and assessing purity using capillary electrophoresis. Macromolecules. 2009;42:3152–3161. doi: 10.1021/ma802250c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barata TS, Brocchini S, Teo I, Shaunak S, Zloh M. From sequence to 3D structure of hyperbranched molecules: application to surface modified PAMAM dendrimers. J Mol Model. doi: 10.1007/s00894-011-0966-y. [DOI] [PubMed] [Google Scholar]
- 17.Barata TS, Shaunak S, Teo I, Zloh M, Brocchini S. Structural studies of biologically active glycosylated polyamidoamine (PAMAM) dendrimers. J Mol Model. doi: 10.1007/s00894-010-0907-1. [DOI] [PubMed] [Google Scholar]
- 18.Halgren TA, Nachbar RB. MMF94: The Merck molecular force field. Bridging the gap - From small organics to proteins. Abstracts of papers of the American Chemical Society. 1996;211:70. [Google Scholar]
- 19.Fariborz M, Nigel GJR, Wayne CG, Rob L, Mark L, Craig C, et al. Macromodel - an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem. 1990;11:440–467. [Google Scholar]
- 20.Jorgensen WL, Tirado-Rives J. Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci. 2005;102:6665–6670. doi: 10.1073/pnas.0408037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J Am Chem Soc. 2002;112:6127–6129. [Google Scholar]
- 22.Zloh M, Shaunak S, Balan S, Brocchini S. Identification and insertion of 3-carbon bridges in protein disulfide bonds: a computational approach. Nat Protoc. 2007;2:1070–1083. doi: 10.1038/nprot.2007.119. [DOI] [PubMed] [Google Scholar]
- 23.Levy RM, Gallicchio E. Computer simulations with explicit solvent: Recent progress in the thermodynamic decomposition of free energies and in modeling electrostatic effects. Annu Rev Phys Chem. 2003;49:531–567. doi: 10.1146/annurev.physchem.49.1.531. [DOI] [PubMed] [Google Scholar]
- 24.Pedretti A, Villa L, Vistoli G. VEGA – An open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J Comput Aid Mol Des. 2004;18:167–173. doi: 10.1023/b:jcam.0000035186.90683.f2. [DOI] [PubMed] [Google Scholar]
- 25.Islam MT, Shi X, Balogh L, Baker JR. HPLC separation of different generations of PAMAM dendrimers modified with various terminal groups. Anal Chem. 2005;77:2063–2070. doi: 10.1021/ac048383x. [DOI] [PubMed] [Google Scholar]
- 26.Shaunak S, Godwin A, Choi JW, Balan S, Pedone E, Vijayarangam D, et al. Site-specific PEGylation of native disulfide bonds in therapeutic proteins. Nature Chemical Biology. 2006;2:312–313. doi: 10.1038/nchembio786. [DOI] [PubMed] [Google Scholar]
- 27.Brocchini S, Balan S, Godwin A, Choi JW, Zloh M, Shaunak S. PEGylation of native disulfide bonds in proteins. Nat Protoc. 2006;1:2241–2252. doi: 10.1038/nprot.2006.346. [DOI] [PubMed] [Google Scholar]
- 28.Balan S, Choi JW, Godwin A, Teo I, Laborde CM, Heidelberger S, et al. Site-specific PEGylation of protein disulfide bonds using a three-carbon bridge. Bioconjug Chem. 2007;18:61–76. doi: 10.1021/bc0601471. [DOI] [PubMed] [Google Scholar]
- 29.Choe J, Kelker MS, Wilson IA. Crystal structure of human TLR3 ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 30.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.