Abstract
Development of a computational decision aid for a new medical imaging modality typically is a long and complicated process. It consists of collecting data in the form of images and annotations, development of image processing and pattern recognition algorithms for analysis of the new images and finally testing of the resulting system. Since new imaging modalities are developed more rapidly than ever before, any effort for decreasing the time and cost of this development process could result in maximizing the benefit of the new imaging modality to patients by making the computer aids quickly available to radiologists that interpret the images. In this paper, we make a step in this direction and investigate the possibility of translating the knowledge about the detection problem from one imaging modality to another. Specifically, we present a computer-aided detection (CAD) system for mammographic masses that uses a mutual information-based template matching scheme with intelligently selected templates. We presented principles of template matching with mutual information for mammography before. In this paper, we present an implementation of those principles in a complete computer-aided detection system. The proposed system, through an automatic optimization process, chooses the most useful templates (mammographic regions of interest) using a large database of previously collected and annotated mammograms. Through this process, the knowledge about the task of detecting masses in mammograms is incorporated in the system. Then we evaluate whether our system developed for screen-film mammograms can be successfully applied not only to other mammograms but also to digital breast tomosynthesis (DBT) reconstructed slices without adding any DBT cases for training. Our rationale is that since mutual information is known to be a robust intermodality image similarity measure, it has high potential of transferring knowledge between modalities in the context of the mass detection task. Experimental evaluation of the system on mammograms showed competitive performance compared to other mammography CAD systems recently published in the literature. When the system was applied “as-is” to DBT, its performance was notably worse than that for mammograms. However, with a simple additional preprocessing step, the performance of the system reached levels similar to that obtained for mammograms. In conclusion, the presented CAD system not only performed competitively on screen-film mammograms but it also performed robustly on DBT showing that direct transfer of knowledge across breast imaging modalities for mass detection is in fact possible.
Keywords: image informatics, pattern recognition, mammography, digital breast tomosynthesis
1. Introduction
Breast cancer is the most common non-skin cancer in women in the United States. The current screening standard for breast cancer is mammography, with some new modalities such as digital breast tomosynthesis (DBT) recently approved for clinical use. One of the ways to improve the overall benefit of breast cancer screening is with the support of computer-aided detection (CAD) systems designed to offer second opinions to the radiologists interpreting breast images. In the last two decades there has been great progress in the investigation, development and clinical deployment of such systems in screening mammography. However, there is still room for improvement, particularly with respect to the detection of breast masses, further reduction of false positives, and ideally direct translation across imaging platforms (from screen-film to digital mammography to DBT).
Multiple approaches have been proposed for the computer-aided detection of breast masses involving a variety of image processing and machine learning algorithms [1]. A recently published review paper [2] provides a good overview of the available approaches with some discussion on their advantages and disadvantages. One such approach for breast masses is template matching [3, 4, 5, 6, 7, 8, 9] where the similarity of a region of interest to a template or a set of templates is assessed. Then, based on the similarity score, the suspicion level of each region in the entire image is evaluated.
CAD systems that are based on the template matching approach vary in the measures used to assess image similarity and the generation or selection of appropriate templates. With respect to similarity criteria, the two most popular ones are correlation and mutual information. With respect to appropriate mass templates, researchers have applied artificial templates, probabilistic templates, as well as libraries of actual masses as templates. As pointed out in [2], the template matching approach appears to be a competitive alternative to other approaches but its success depends on the generation of effective templates that cover the diverse appearance range of breast masses.
Our group has been developing a template matching approach for mass detection in screening mammograms [7, 8, 10, 11, 12, 13] that capitalizes on a library of real mass and normal (i.e. not containing abnormalities) templates with mutual information as the similarity measure. Although the approach has been shown to be quite effective for region-based analysis [7], its implementation on full mammograms becomes computationally expensive as more templates are deposited in the library. Therefore, in subsequent studies, we proposed several improvements to our technique including case selection [10, 12] and ensemble classification [13], all applied for specific chosen locations, not entire mammograms. In a recent study [14] we showed preliminary results regarding the extension of our system to full mammograms as a base of a context-sensitive mammography CAD system. However, the focus of that study was on the context-sensitive (eye tracking-based) aspect and only limited details of the system and limited system evaluation were presented. The current study focuses on the possibility of translation of the results between mammography and digital breast tomosynthesis. It also contains the full description and performance evaluation of the system.
Despite the computational challenge, the general mutual information-based template matching approach has several advantages. The leading one is its simplicity. As it is based on template matching, it does not require segmentation of structures in the breast and extraction of features. Even though feature-based CAD systems have been shown to be quite efficient, image features can be sensitive to image acquisition parameters including digitizer characteristics for screen-film mammograms or vendor-specific pre-processing and post-processing algorithms for digital mammography. Consequently, feature-based CAD systems developed for certain acquisition parameters require at the very minimum careful recalibration in order to be successfully applied across platforms. In contrast, our mutual information-based template matching approach has the potential for direct translation across platforms. It does not depend on the numerical values of computer-extracted features but on the notion of mutual information that captures the statistical relationship of two pixel intensity probability distributions. Mutual information gained interest in the related task of image registration [15] where it has been shown that it is particularly effective for assessing similarity between images from different modalities depicting the same anatomical structure (multi-modality image matching). This property can be particularly useful when translating a CAD system developed using a set of images coming from one imaging device to images coming from other devices or even other imaging modalities. Our pilot investigation of this potential for location specific mass detection was presented in [16].
Developing a fully automated CAD system that can easily adapt across imaging platforms could be a very efficient decision support strategy as emerging breast imaging modalities continue to rapidly evolve and advance. It could significantly facilitate the development of CAD systems as the new modalities appear and make the decision aids available on time for the radiologists interpreting the images.
The purpose of this paper is (1) to present in detail a computationally efficient implementation of a computer-aided system for detection of masses in mammograms that is based on previously proposed principles of mutual information-based template matching as well as (2) to evaluate whether the proposed detection scheme has the desired flexibility for robust translation across modalities, from screen-film mammography to DBT.
2. Methods
In this section we describe the methodology used in our experiments. The databases used in our study are described in Section 2.1. Section 2.2 describes the framework of the CAD system, including pre-processing (Section 2.2.1), template matching (Section 2.2.2) and post-processing (Section 2.2.3). Training of the system is described in Section 2.3. Training, in this context means incorporating the domain knowledge (about mass detection in mammography) into the framework described in Section 2.2. Finally, section 2.4 describes how the proposed system was tested on mammograms and on digital breast tomosynthesis reconstructed slices. Therefore, in Section 2.4 we describe how we evaluated whether the domain knowledge incorporated in our CAD system using mammography transfers to another modality.
2.1. Databases
In our experiments we used a publicly available database of screening mammograms as well as a digital breast tomosynthesis database collected at Duke University Medical Center.
2.1.1. Database 1: Screening mammography
Database 1 was based entirely on the Digital Database of Screening Mammography [17] (DDSM). We used all cases digitized with the Lumisys scanner (there are 944 such cases) with a few exceptions. Specifically, we excluded 141 cases classified as benign without call-back (i.e., BENIGN_WITHOUT_CALLBACK folder in DDSM) since these cases are usually considered not challenging. To minimize the evaluation bias, we excluded 100 cases that were used as an internal validation set to optimize some of the system parameters during the initial system development stages. We also excluded 201 cases that contained only calcifications as well as 39 cases for which the masses were not included in our breast segmentation mask (e.g., masses within the pectoral muscle region of a mammogram). This resulted in 463 cases which we call Database 1. Since each case contains 4 images (two views of each breast), Database 1 consisted of 1852 mammographic images that contained 401 masses in total (239 malignant and 162 benign). This database was used to train and test the CAD system.
2.1.2. Database 2: Digital breast tomosynthesis
We collected the DBT data at Duke University Medical Center using a prototype DBT system Mammomat NovationTOMO (Siemens Healthcare, Erlangen Germany)1. System parameters including the acquisition and reconstruction process have been reported previously[18]. The study was approved by the institutional review board and it was compliant with the Health Insurance Portability and Accountability Act. We used a total number of 134 cases. Each case contained up to four volumes, each corresponding to one view of one breast. The total number of volumes used in this study was 346. Out of all cases 15 contained lesions that were classified as masses or architectural distortions based on their radiological presentation. Three cases contained biopsy-proven malignant masses. In the remaining 12 cases, the lesions were considered benign based on further examination or their radiological appearance. Given that for many cases we had two views of a breast available (CC and MLO), there were 27 occurrences of a lesion in these cases (the same lesion visible in two views is counted as two appearances). Five of them corresponded to malignant masses. The remaining cases (119) were considered normal.
Since the focus is on the applicability of our CAD system developed using the mammography dataset to DBT reconstructed slices, we extracted single (1 mm) slices from each volume. For abnormal volumes, we extracted the central slice for each lesion. For normal volumes, we extracted one, randomly selected slice. These slices (total of 348 out of which 28 were abnormal) constituted Database 2.
2.2. Description of the CAD system
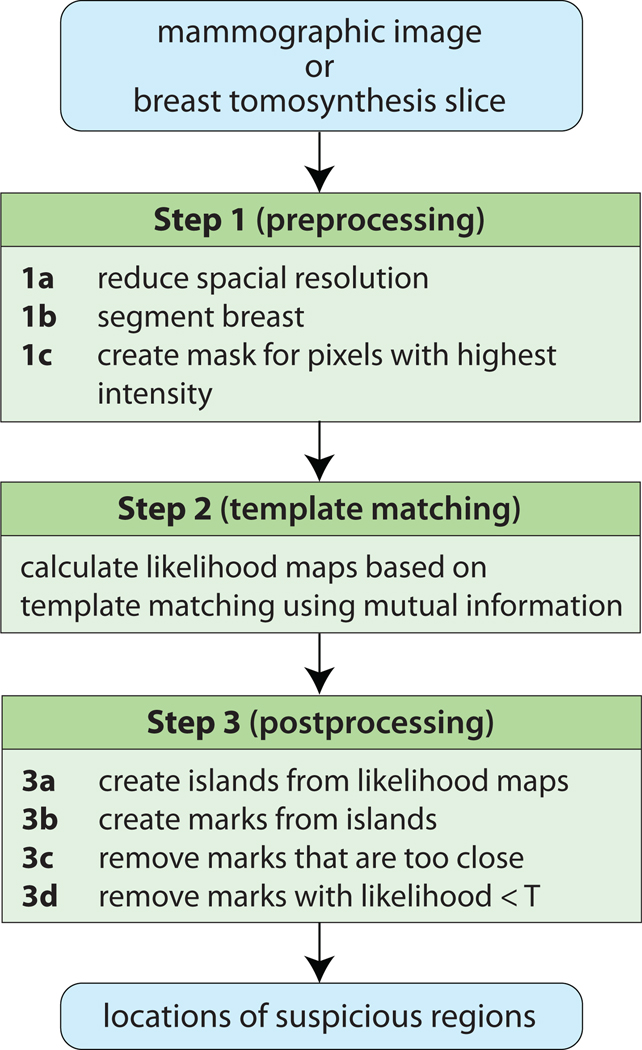
A diagram of the proposed CAD system is presented in Fig. 1. The system proceeds in three steps. These steps are identical when the system is tested on mammograms and when it is tested on DBT reconstructed slices except for the breast segmentation algorithm (Step 1b).
Figure 1.
Diagram of the proposed CAD system
2.2.1. Step 1: Preprocessing
This step consists of three parts. It starts with reducing the spatial resolution of the original images (mammograms and DBT slices) to 0.4 mm per pixel. This reduced resolution was found sufficient in our preliminary experiments with mammograms. Please note that the original resolution of mammograms and DBT slices are different (50 µm per pixel and 85 µm per pixel respectively) and therefore the scaling factor differed as well: it was 8 for mammograms and approximately 4.7 for DBT slices. It is important that the images presented to the CAD system have the same resolution so that a similarity between structures (e.g., masses) of approximately the same size in two different modalities can be successfully captured. The spatial resolution reduction was performed simply by sampling values of pixels in the original image distanced by the scaling factor (i.e., no averaging was done). In addition to ensuring the same resolution of images in the two modalities, this step drastically increases the computational efficiency of the system.
The next part (Step 1b) differs when the system is tested on mammograms and DBT reconstructed slices. When the system is tested on mammograms, a simple breast segmentation algorithm is utilized based on global thresholding of the images. The global threshold is established separately for each mammographic image via peak detection in the image histogram. Additionally for mammograms, a pectoral muscle segmentation was applied (a simple straight line-fitting approach was used). When the system is tested on DBT reconstructed slices, the segmentation algorithm was applied directly to each of the reconstructed slices and consisted of several steps. First, the noise was removed from the images through median filtering (only for the purpose of segmentation). Then, for each line of pixels in the image a point of transition from the background to the breast tissue was identified by detecting the point when the smoothed gradient reached an empirically determined threshold. Then, after removing outliers among the detected transition points, a spline was fitted to the points. Finally, the mask was slightly smoothed and trimmed. This algorithm can result in visually aggressive segmentation in cases when only very low gradients are observed in some parts of the breast. However, this is particularly the case in the areas of fatty tissue (low pixel values) in the breast which are of less interest when looking for masses (usually higher pixel values). Such aggressive segmentation was in fact often observed in our database. However, none of the abnormalities in DBT slices was excluded due to the segmentation error.
Please note, that different segmentation algorithms could be used in our experiments and a breast CAD researcher that will decide to use our template matching approach can simply apply the algorithm that he/she has at hand. Since the focus of our paper is on the template matching and inter-modality translation of the CAD system rather than the preprocessing issues, we used the basic segmentation algorithms described above.
Finally, in the last part of this step (Step 1c) a mask is created that includes the 30% highest intensity pixels within the breast. Only pixels within the mask are considered as suspicious enough in the further steps. The underlying assumption of this step is that masses correspond to high intensity regions of the breast. In our experiments through varying this threshold from 20% to 100% we observed that this step not only allows for substantial improvement in the time complexity of our algorithm but also for slight improvement in its overall performance. The threshold of 30% was chosen as providing a good balance between sensitivity and specificity for out test set of 100 cases. Before identifying the 30% highest intensity pixels, a 3×3 median filter was applied to the original images to reduce the noise in the image that could result in creating small (even single pixel) islands throughout the image. Even though we believe that such preprocessing assures better quality masks, according to our preliminary experiments it had only minimal impact on the overall system performance. The median filter was applied for the purpose of this step only. In the further steps, the original images were used.
2.2.2. Step 2: Template matching
This is the main step of the proposed CAD system. For each pixel within the mask generated in step 1, a likelihood score is calculated of that pixel corresponding to an abnormality. This likelihood calculation is based on the template matching scheme using mutual information originally presented in [7]. It proceeds as follows: For each pixel p, a region of interest (ROI) is extracted centered on p. The ROI is 64×64 pixels in size (i.e. 25.6×26.5 mm). This size of ROI has been previously determined optimal for our application [19]. Then, the mutual information (I) between the query ROI and each template ROI stored in the database of the CAD system is calculated as:
| (1) |
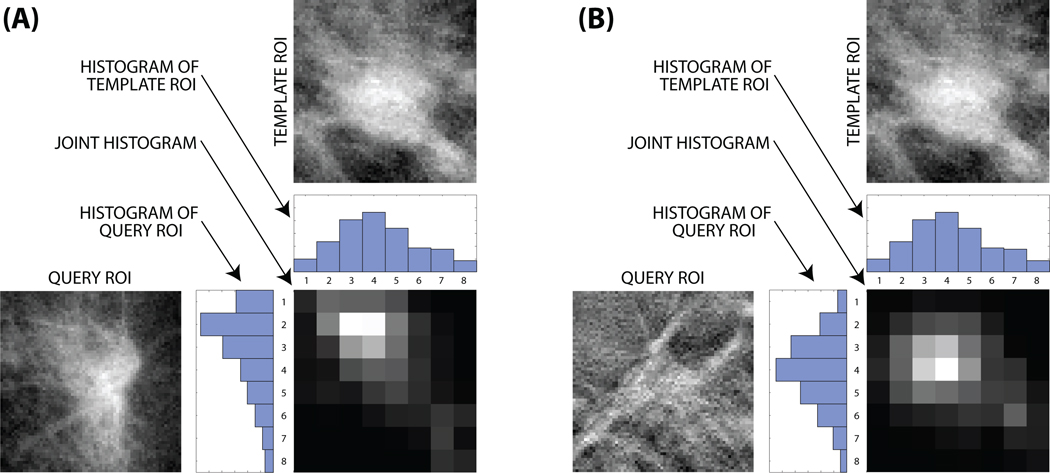
The joint probability density function pAB (a, b) and the marginal probability density functions pA (a) and pB (b) are estimated using the histogram approach [20] with 8 bins [7]. The bins are uniformly distributed between the minimum and maximum pixel values in the image with 0.5% of the pixels with the highest intensity and 0.5% pixels with the lowest intensity being ignored in calculating such minimum and maximum values to reduce the impact of noise. The ignored pixels are then included in the marginal (first and last) bins. Calculating mutual information between two images from the same modality and two images from different modalities are illustrated in Fig. 2.
Figure 2.
Calculating mutual information: (A) between two mammography ROIs and (B) between a tomosynthesis and a mammography ROI
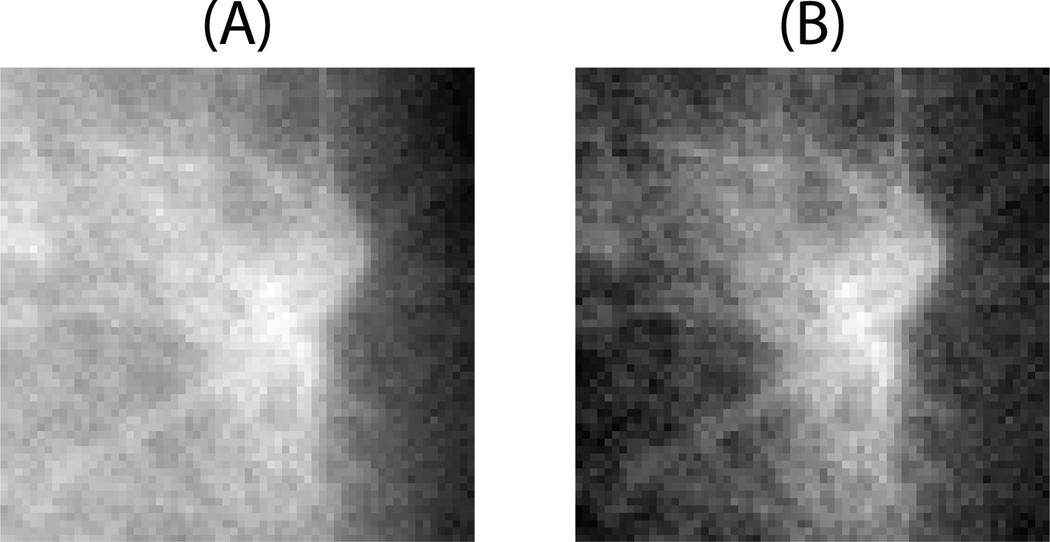
Background correction is applied to each query ROI and each template ROI before calculating their mutual information. It proceeds as follows. Given the image in the form f(x, y), where x and y are pixel coordinates, initially, a first order polynomial g(x, y) = c1x + c2y is fitted into the image. Then the background corrected image is obtained by subtracting the fitted plane from the original image: f(x, y) − g(x, y). The background correction for an example ROIs is presented in Fig. 3. Similar background correction has been previously adapted to mammograms [21] and chest radiographs [22]. Our preliminary experiments showed that background correction is an important step that contributes to better overall CAD performance.
Figure 3.
An example ROI before (A) and after (B) background correction
After the mutual information is calculated between the query ROI and each template ROI, the likelihood score is calculated as
| (2) |
where Q is a query ROI, Mi are mass templates, Nj are normal templates, m is the number of mass templates and n is the number of normal templates. This entire step results in a likelihood map with each candidate pixel assigned a likelihood value L of belonging to a mass.
2.2.3. Step 3: Postprocessing
To extract CAD marks from likelihood maps calculated in Step 2, an iterative multi-level thresholding algorithm is applied similar to the one in [23, 24]. Specifically, at each threshold regions of pixels are created that have been assigned likelihood values higher than the threshold LT. The threshold LT is varied from the maximum value of likelihood present in the likelihood map (Lmax) to the value that is established as Lmin+0.4(Lmax − Lmin) where Lmin is the minimum value of likelihood present in the entire likelihood map. Decreasing LT results in growing regions with high likelihood. When two such regions are about to merge they are saved separately indicating two separate suspicious areas. If a suspicious region does not merge with another by the end of all iterations, it is also saved. After the entire process, a mark is created corresponding to each saved region placed at the pixel with the highest likelihood score within the region. The value of the maximum likelihood score in the region is assigned to the mark to indicate the predicted overall likelihood that the marked location corresponds to a true mass (Li).
Then, threshold T is applied such that all marks in the image with likelihood Li less than T are removed. The value of T determines the operating point of the system and sets a balance between its sensitivity and specificity. Finally, for any marks within a distance of 20 pixels (8 mm) or less from each other, the mark with the lower Li value was removed. This final step is repeated until there were no such pairs left. The resulting marks were the final output of the CAD system.
2.3. System training
The only part of our CAD system that requires machine learning-based optimization is the selection of the ROI templates stored in the systems database. Note that this step is different from classifier training used in typical CAD systems (e.g. [25]) that involve classifiers such as linear discriminant analysis, artificial neural networks, support vector machines etc. However, we still use the term “training” as it involves machine learning-based system optimization using previously collected data.
We include this template selection step since we previously discovered [12] that it allows for dramatic reduction of time complexity of the template matching algorithm while improving its overall performance. The time complexity is crucial in our CAD system since calculation of the mutual information measure is time consuming when template matching is performed scrolling across the full mammogram.
The selection of the template ROIs proceeds as follows. First, two sets of ROIs (64×64 pixels in size), S and V, are generated from the available mammograms. These two sets are extracted in the following way. For all mammograms available for training, ROIs are extracted around each mass present in those mammograms. Five ROIs are extracted for each mass: one ROI centered on the radiologist annotation and 4 additional ROIs centered 5 pixels (2mm) from the center of the radiologist annotation in each direction (up, down, left and right). Then, from each mammogram, 6 normal ROIs are extracted. An ROI is considered normal if it is at least 40 pixels (16mm) from the centroid of the closest DDSM mass annotation. Then, from all extracted ROIs, 500 normal and 500 mass ROIs are selected randomly to constitute the set S. This set is used as the pool of ROIs from which the template ROIs are selected. Set V consists of all remaining mass ROIs and a number of normal ROIs so that the total number of ROIs in V is 10000. Set V is used to evaluate the performance of the system that is based on a subset of ROIs selected from S. Specifically, each ROI from V is assigned a decision value by the system using the subset of S as the database and the decision rule from equation (2). Then the area under the receiver operator characteristic for all ROIs in V is calculated as the performance measure.
Given the two sets, the random mutation hill climbing (RMHC) [26] algorithm is applied to select ROIs to store in the system's database of templates. In our previous studies [12], we determined RMHC to be a superior selection algorithm among multiple available alternative choices. In short, RMHC is an iterative algorithm that proceeds as follows. First a number k of ROIs is randomly drawn from the set S. Then, at each iteration, one randomly drawn ROI from the selected set is swapped with one randomly drawn ROI from the remainder of S. If the swapping increases system performance (evaluated using V) then the change is kept. Otherwise it is reversed. This operation is repeated for 2000 iterations. This number was previously found [12] to provide a good balance between system performance and computational complexity. At every iteration, the system performance is measured using the area under the ROC curve on the validation set V. In our experiments the number of selected ROIs was 30. This number was optimized with use of the 100 cases that were excluded for the final validation experiments. The 30 ROIs selected in this step constitute the knowledge database of the CAD system to be used as templates for the calculation of likelihood maps (i.e., in step 2 described Section 2.2.2).
2.4. Experimental design
We conducted two experiments. In Experiment 1, we developed and tested our CAD system for screening mammograms using 10-fold crossvalidation with Database 1. In Experiment 2 the performance of mammography CAD system was further tested on DBT reconstructed slices from Database 2. When evaluating performance, we took the following measures into account:
Image-based sensitivity: the ratio of detected occurrences of lesions to the total number of occurrences of lesions (lesion that appears in two views counts as two occurrences)
Case-based sensitivity: the ratio of detected lesions (in at least one of two views of the breast) to the total number of lesions (lesion that appears in two views counts as one)
Number of false positives per image (FPs/I: the average number of false positive marks (FP) in an image (i.e. marks that do not correspond to an annotated lesion).
To evaluate the overall performance of the systems, we used free response operating characteristic (FROC) analysis [27]. FROC curves show how sensitivity (image-based or case-based) and the number of false positives per image change as the threshold T (see Section 2.2.3) change. FROC curves are a common choice when evaluating the performance of computer-aided detection systems.
Furthermore, we provide a detailed analysis for specific sub-groups of cases that have higher clinical significance: malignant masses and architectural distortions. Malignant masses are masses that represent cancer and as such are of special interest. Even though our system aims at detecting masses in general, it is important to know whether malignant masses are detected as accurately as all suspicious lesions. Architectural distortions are distortions of the breast parenchyma that do not have a focal abnormality. They are known to present a challenging detection task for both mammographers as well as computer-aided detection systems.
2.4.1. Experiment 1: evaluating performance for mammography
Following the 10-fold crossvalidation data handling scheme, we randomly divided Database 1 into 10 approximately equal parts (folds). Each fold contained 46 or 47 cases. Each case was included entirely in one fold to ensure that mammographic views of the same case do not appear in both training and testing at the same time. Then, we used 9 folds for system training and the remaining fold for testing. We repeated this process 10 times, each time with a different fold used for testing. We obtained ten free receiver operator characteristic (FROC) [27] curves which relate the average number of false positive marks (FP) in an image to which we combined by averaging the true positive fraction (TPF) for a given number of false positive marks (FP) per image. The FROC curves were obtained empirically by varying the threshold T in Step 3 of our system.
For each threshold T, the average number of FP marks per image (FPs/I) was calculated only on images that did not contain any masses. To calculate TPF (sensitivity), we assumed that a mass is detected if there was a CAD mark within the DDSM mass annotation.
2.4.2. Experiment 2: evaluating performance for digital breast tomosynthesis
In Experiment 2 we evaluated whether the system trained using mammographic cases only (and therefore having knowledge of only such cases) can be successfully applied to DBT reconstructed slices. Specifically, we used the entire Database 1 for system training and Database 2 for testing. A lesion was considered detected if there was a CAD mark within the rectangle outlining the lesion. The number of false positives per image was assessed on slices from normal volumes. The performance was evaluated using FROC analysis.
This experiment consisted of two parts. In the first part (Experiment 2a), we applied the system trained on mammographic cases (Database 1) “blindly” to DBT slices (Database 2). In the second part (Experiment 2b), we applied the same system with an addition in the preprocessing step. Specifically, upon completion of step 1c during preprocessing, the pixels that were within 64 pixels (size of a single ROI) from the breast edge (based on the segmentation mask) were automatically assigned a likelihood value of 0. If a lesion was not detected because of this modification, it was counted as missed and it was reflected in the overall sensitivity of the system. The rationale for this step is based on our discoveries in Experiment 2a and is discussed below.
3. Results
3.1. Experiment 1
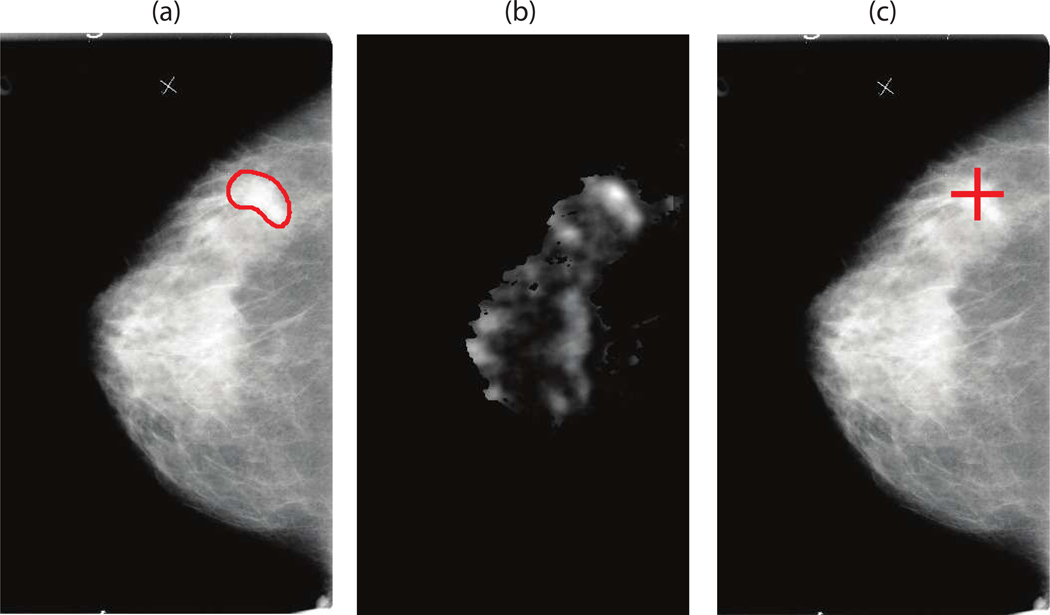
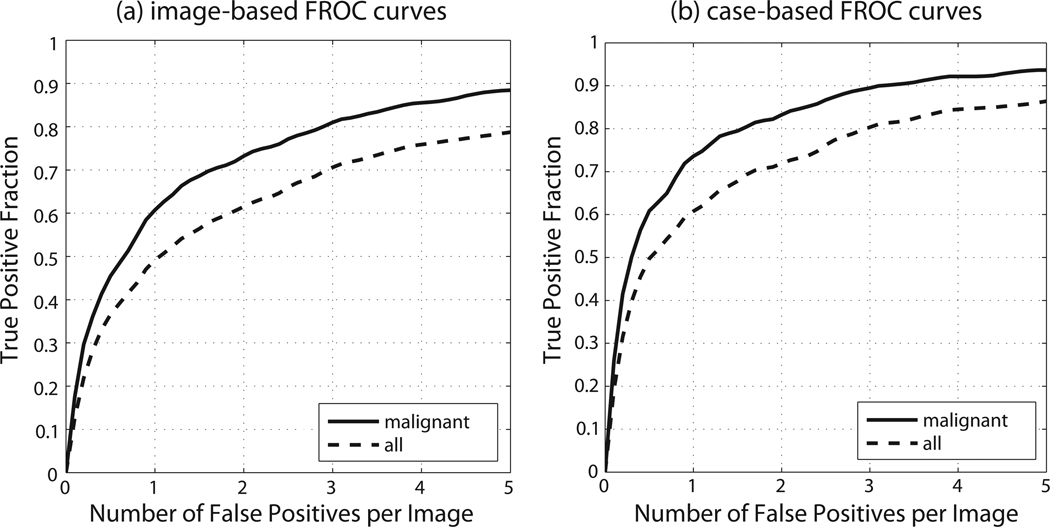
Performance of our system for an example case is shown in Fig. 4. The complete FROC curves for our system, when tested on mammograms are presented in Fig. 5. For malignant masses, the average case-based sensitivity was 0.736±0.084 and 0.832±0.044 for 1 and 2 FPsI respectively. The image-based sensitivity for malignant masses was 0.607±0.077 and 0.732±0.052 for 1 and 2 FPs/I respectively. For all masses, the average case-based sensitivity was 0.608±0.065 and 0.719±0.047 for 1 and 2 FPs/I respectively. The image-based sensitivity for all masses was 0.492±0.064 and 0.615±0.063 for 1 and 2 FPs/I respectively.
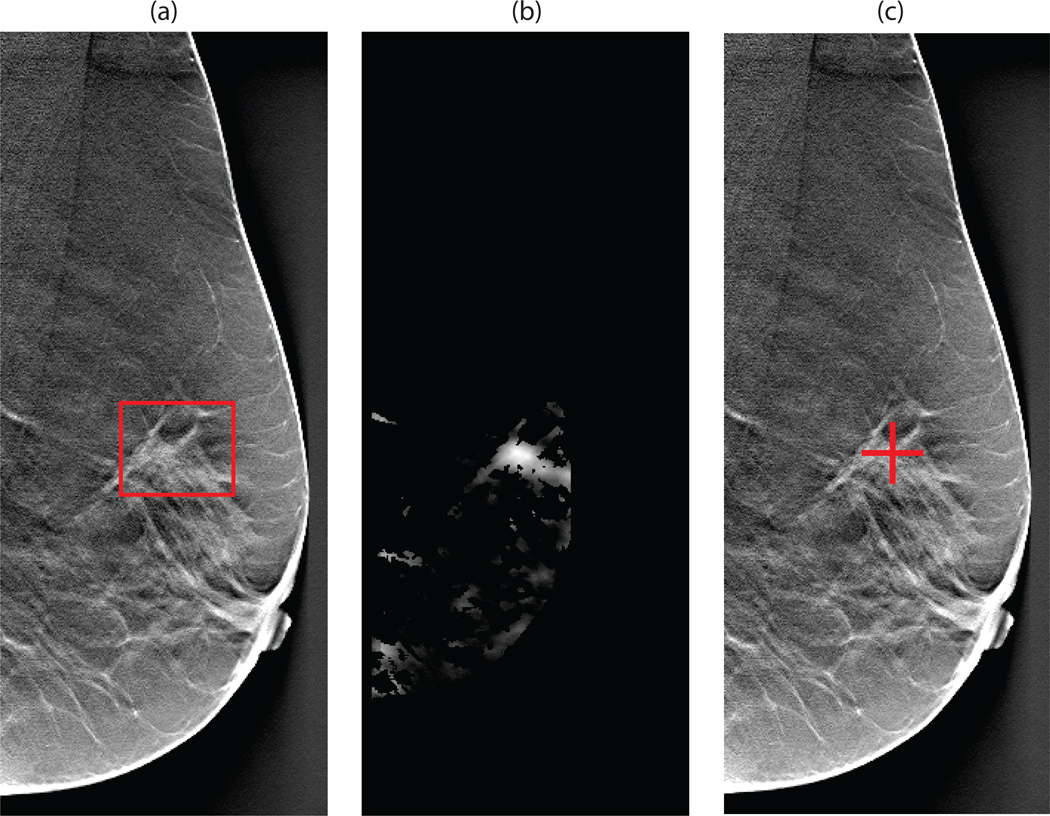
Figure 4.
Illustration of the CAD steps in mammography for the system operating at approximately 1 FP/image: (a) original DDSM mammogram with the DDSM-provided annotation of a mass present (b) the likelihood map output of the template matching CAD step (c) CAD mark generated during the final step
Figure 5.
Mammography: crossvalidation FROC performance
We also examined the system performance separately on architectural distortions (i.e. lesions that in DDSM were described as masses with shape “ARCHITECTURAL DISTORTION”). The average image-based sensitivity was 0.355±0.203 and 0.503±0.204 for 1 and 2 FPs/I respectively. Only slightly lower sensitivity for architectural distortions as compared to sensitivity for all masses is an additional indicator of the robustness of our system.
3.2. Experiment 2
When the system trained using mammographic cases was applied to DBT reconstructed slices “as is”, the system performance was poor (e.g. case-based sensitivity of 0.625 for 5 FPs/I). However, upon further investigation we observed that the poor system performance was caused by highly a suboptimal system response for regions close to the breast edge. The appearance of the breast edge region differs between mammograms and DBT reconstructed slices. Specifically, artifacts are quite possible in the DBT reconstructed slices due to the acquisition as well as the reconstruction process. Furthermore, DBT slices do not demonstrate the fading effect present along the breast edge in mammograms. This fading effect causes areas close to the breast edge to be often eliminated during step 1c. Such effect does not take place in DBT. To account for the breast edge differences between the two modalities, in the second part of Experiment 2 we simply excluded a 64 pixel wide stripe (i.e., the size of an ROI) along the breast edge. This ad-hoc step allows us to evaluate whether the CAD system performs robustly on the remaining breast region.
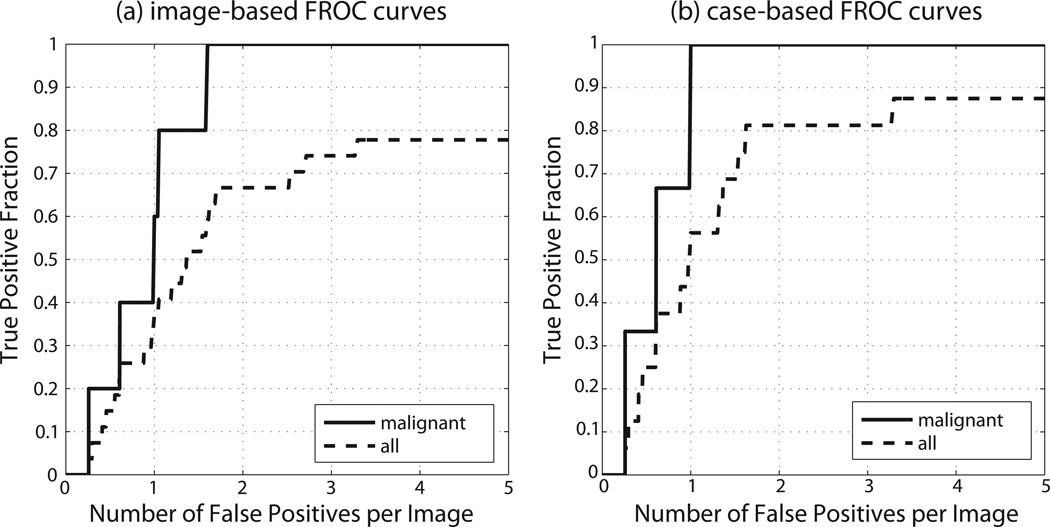
Experiment 2b showed that when this simple modification is applied, the performance of the mammography-trained system tested on DBT reconstructed slices improved dramatically and was in fact comparable to the performance of the same system when tested on mammograms. Performance of the system for an example case is shown in Fig. 6. The complete FROC curves of the system are presented in Fig. 7. For malignant masses, the case-based sensitivity reached 1 (i.e. all malignant masses were detected) for 1 FP/I. The image-based sensitivities for malignant masses were 0.6 and 1 for 1 and 2 FPs/I, respectively. For all lesions, the case-based sensitivities were 0.563 and 0.813 for 1 and 2 FPs/I, respectively. The image-based sensitivities for all lesions were 0.370 and 0.667 for 1 and 2 FPs/I, respectively.
Figure 6.
Illustration of the CAD steps when the system is applied in DBT while operating at 1 FP/slice: (a) original DBT slice at reduced resolution with an annotated malignant mass (b) the likelihood map output of the template matching CAD step (c) CAD mark generated during the final step
Figure 7.
DBT: FROC performance
4. Conclusions
In this study we proposed and evaluated a new system for detection of breast masses that is based on template matching with mutual information. We also assessed if this system, developed using mammograms only, can be successfully applied as-is (without retraining) to DBT reconstructed slices. The conclusions of our study are the following:
The proposed CAD system performs well when tested on mammograms
The sensitivity of our CAD system is better for malignant masses than for benign masses.
When the system is trained using mammograms only and tested “blindly” on digital breast tomosynthesis slices its performance is significantly lower than when it is tested on mammograms
The inferior performance is caused mostly by the radically different breast edge characteristics between digital breast tomosynthesis reconstructed slices and digitized mammograms
When a simple preprocessing step is applied to the system trained on mammograms to account for that difference, the performance of our CAD system for digital breast tomosynthesis reaches levels comparable to the performance for mammograms. Therefore, we conclude that:
Application of mutual information-based template matching schemes developed for mammography to digital breast tomosynthesis shows high promise.
5. Discussion
Regarding the performance of our system for mammograms, we found that it is comparable and often better than the performance reported in the recent literature in the field. For example Wei et al. [25] reports case-based sensitivity of 0.636 and 0.801 at 1 FP/I for their single and dual CAD systems respectively for average masses. The performance for subtle masses was notably lower. The evaluation was performed using digital mammograms. Freixenet et al. [9] report sensitivities of around 0.48 and 0.8 for 1 and 2 FPs/I respectively. In the recent review paper [2] the authors compared 7 previously proposed approaches to mass detection using two databases: a relatively easy database of digitized mammograms (MIAS) and a private full field digital mammography database. Comparison to our results shows that our system outperforms 6 out of 7 methods evaluated by Oliver et al. [2].
However, direct comparison of the algorithms proposed by different groups is very difficult since they often use different validation schemes and, even more importantly, different databases that may drastically differ in difficulty. For our evaluation, we used a large variety of mammograms including challenging cases such as pure architectural distortions. Cases including masses with associated architectural distortion or associated calcifications were also included in the analysis. Finally, to ensure robustness, the CAD testing performance was based on a 10-fold crossvalidation scheme and not on a single split.
Regarding the performance of our system for DBT, please note that a CAD system for DBT will operate on entire volumes rather than on single slices. The information from single slices can be fused in order to take advantage of the information present in the entire volume. In our study, rather than present the volume-based performance, we present the slice-based performance. Therefore comparison to previously published DBT CAD systems (e.g. [28, 29, 30, 18]), is difficult. However, good slice-based performance is a clear indication of a system’s high potential for competitive performance in the entire volume as well, since reliable results in single slices can be effectively combined. Evaluating DBT performance on the entire volumes would introduce another factor to our analysis, namely an arbitrarily chosen method for fusing the CAD results from different slices. Our results could change depending on which method is chosen. In our experiments we eliminate this additional factor by reporting the performance on single slices only. The performance evaluation on the entire volumes will be a subject of further investigation. Furthermore, please note, that the performance of the system as evaluated on DBT slices could further improve if DBT data were used in the training of the system. However, since the focus of this paper was on translation of domain knowledge from one modality to another, we leave such investigation for future research.
Our experiments demonstrated that a system developed using only mammograms can be successfully applied to tomosynthesis reconstructed slices when a small modification in preprocessing is applied. We believe that the simple modification used in this paper is sufficient to show that the system in fact has a high potential for DBT, even though it does not have direct knowledge of the DBT appearance of mass and normal parenchyma. Please note, however, that although excluding a stripe along the breast edge allowed us to demonstrate the potential of translating knowledge from mammography to DBT, it is only a temporary solution since it is bound to miss all masses in these regions (the prevalence of abnormalities in those regions in our DBT database was low). Therefore, the final solution to the problem of inferior performance of the system in those regions will require further algorithm development.
Finally, please note that the two modalities (mammography and DBT) approached in this paper are inherently similar to each other since they are both based on x-ray attenuation principles. However, the resulting images (i.e., mammograms and DBT slices) have many differences with respect to image formation, image characteristics, and visual presentation. Because of those differences, modality-specific CAD systems are typically developed to address the unique challenges for each modality and acquisition protocol. The motivation for our study was to evaluate whether a mutual information-based CAD framework can handle such differences while preserving detection performance. Future studies could consider the possibility of extending the idea to other breast imaging modalities such as ultrasound or breast MRI.
In conclusion, we presented a CAD system that uses a mutual information template matching scheme with an intelligently selected set of templates for automated screening of full mammograms. The CAD system was quite effective in the detection of malignant masses with performance comparable to that of previously published (and often more elaborate) CAD systems. Furthermore, the system was able to perform robustly when applied to breast tomosynthesis data without retraining of the system. A flexible CAD system that can successfully transfer knowledge across modalities offers many advantages in this era of rapid advancements in breast cancer screening.
We propose a computer-aided diagnosis system for breast cancer detection.
The proposed CAD system performs well when tested on mammograms.
“Blind” application to digital breast tomosynthesis (DBT) results in poor performance.
After applying a pre-processing step to DBT slices the performance improves.
Translation of domain knowledge in CAD from mammography to DBT is promising.
Acknowledgments
This study was supported in part by grants NIH/NCI R56-CA101911, NIH/NCI R01-CA112437 and a research grant from Siemens Healthcare. We would like to thank Dr. Jay A. Baker for interpretation of the DBT data. We would also like to thank Sushravya Raghunath for her contribution to the preprocessing steps of our CAD system.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Caution: Investigational Device. Limited by U.S. Federal law to investigational use. The information about this product is preliminary. The product is under development and is not commercially available in the U.S.; and its future availability cannot be ensured.
References
- 1.Cheng H, Shi X, Min R, Hu L, Cai X, Du H. Approaches for automated detection and classification of masses in mammograms. Pattern Recognition. 2006;39:646–668. [Google Scholar]
- 2.Oliver A, Freixenet J, Mart J, Prez E, Pont J, Denton ERE, Zwiggelaar R. A review of automatic mass detection and segmentation in mammographic images. Med Image Anal. 2010;14(2):87–110. doi: 10.1016/j.media.2009.12.005. URL http://dx.doi.org/10.1016/j.media.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Lai SM, Li X, Biscof WF. On techniques for detecting circumscribed masses in mammograms. IEEE Transactions on Medical Imaging. 1989;8(4):377–386. doi: 10.1109/42.41491. URL http://dx.doi.org/10.1109/42.41491. [DOI] [PubMed] [Google Scholar]
- 4.te Brake GM, Karssemeijer N. Single and multiscale detection of masses in digital mammograms. IEEE Transactions on Medical Imaging. 1999;18(7):628–639. doi: 10.1109/42.790462. URL http://dx.doi.org/10.1109/42.790462. [DOI] [PubMed] [Google Scholar]
- 5.Oliver A, Freixenet J, Peracaula M, Marti J. Mass segmentation using a pattern matching approach with a mutual information based metric. Front. Art. Intell. Appl. 2005;131:123–130. [Google Scholar]
- 6.Constantinidis A, Fairhurst M, Rahman A. A new multi-expert decision combination algorithm and its application to the detection of circumscribed masses in digital mammograms. Pattern Recognition. 2001;34:1527–1537. [Google Scholar]
- 7.Tourassi GD, Vargas-Voracek R, Catarious J David M, Floyd J Carey E. Computer-assisted detection of mammographic masses: A template matching scheme based on mutual information. Medical Physics. 2003;30:2123–2130. doi: 10.1118/1.1589494. [DOI] [PubMed] [Google Scholar]
- 8.Tourassi GD, Haarawood B, Singh S, Lo JY, Floyd CE. Evaluation of information-theoretic similarity measures for content-based retrieval and detection of masses in mammograms. Medical Physics. 2007;34:140–150. doi: 10.1118/1.2401667. [DOI] [PubMed] [Google Scholar]
- 9.Freixenet J, Oliver A, Mart R, Llad X, Pont J, Prez E, Denton ERE, Zwigge-laar R. Eigendetection of masses considering false positive reduction and breast density information. Medical Physics. 2008;35(5):1840–1853. doi: 10.1118/1.2897950. [DOI] [PubMed] [Google Scholar]
- 10.Tourassi GD, Harrawood B, Singh S, Lo JY. Information-theoretic cad system in mammography: Entropy-based indexing for computational efficiency and robust performance. Medical Physics. 2007;34:3193–3204. doi: 10.1118/1.2751075. [DOI] [PubMed] [Google Scholar]
- 11.Mazurowski MA, Habas PA, Zurada JM, Tourassi GD. Decision optimization of case-based computer aided decision systems using genetic algorithms with application to mammography. Physics in Medicine and Biology. 2008;53:895–908. doi: 10.1088/0031-9155/53/4/005. [DOI] [PubMed] [Google Scholar]
- 12.Mazurowski MA, Zurada JM, Tourassi GD. Selection of examples in case-based computer-aided decision systems. Physics in Medicine and Biology. 2008;53:6079–6096. doi: 10.1088/0031-9155/53/21/013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazurowski MA, Zurada JM, Tourassi GD. An adaptive incremental approach to constructing ensemble classifiers: application in an information-theoretic computer-aided decision system for detection of masses in mammograms. Medical Physics. 2009;36:2976–2984. doi: 10.1118/1.3132304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tourassi GD, Mazurowski MA, Harrawood BP, Krupinski EA. Exploring the potential of context-sensitive cade in screening mammography. Medical Physics. 2010;37:5728–5736. doi: 10.1118/1.3501882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pluim JPW, Maintz JBA, Viergever MA. Mutual-information-based registration of medical images: a survey. IEEE Transactions on Medical Imaging. 2003;22(8):986–1004. doi: 10.1109/TMI.2003.815867. URL http://dx.doi.org/10.1109/TMI.2003.815867. [DOI] [PubMed] [Google Scholar]
- 16.Tourassi GD, Sharma AC, Singh S, Saunders RS, Lo JY, Samei E, Harra-wood BP. Knowledge transfer across breast cancer screening modalities: A pilot study using an information-theoretic cade system for mass detection; Proceeding of International Workshop on Digital Mammography; 2008. [Google Scholar]
- 17.Heath M, Bowyer K, Kopans D, Kegelmeyer WP, Moore R, Chang K, MunishKumaran S. Ch. Current status of the digital database for screening mammography. Digital Mammography; Proceedings of the Fourth International Workshop on Digital Mammography; Dordrecht: Kluwer Academic Publishers; 1998. pp. 457–460. [Google Scholar]
- 18.Singh S, Tourassi GD, Baker JA, Samei E, Lo JY. Automated breast mass detection in 3d reconstructed tomosynthesis volumes: a featureless approach. Medical Physics. 2008;35(8):3626–3636. doi: 10.1118/1.2953562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ike RC, III, Singh S, Harrawood B, Tourassi GD. Effect of roi size on the performance of an information-theoretic cad system in mammography: multi-size fusion analysis; Proceedings of SPIE; 2008. p. 691527. [Google Scholar]
- 20.Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Transactions on Medical Imaging. 1997;16(2):187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- 21.Sahiner B, Chan H-P, Petrick N, Wei D, Helvie MA, Adler DD, Goodsitt MM. Classification of mass and normal breast tissue: a convolution neural network classifier with spatial domain and texture images. IEEE Transactions on Medical Imaging. 1996;15:598–610. doi: 10.1109/42.538937. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki K, Shiraishi J, Abe H, MacMahon H, Doi K. False-positive reduction in computer-aided diagnostic scheme for detecting nodules in chest radiographs by means of massive training artificial neural network. Academic Radiology. 2005;12(2):191–201. doi: 10.1016/j.acra.2004.11.017. URL http://dx.doi.org/10.1016/j.acra.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Giger ML, Doi K, MacMahon H. Image feature analysis and computer-aided diagnosis in digital radiography. 3. automated detection of nodules in peripheral lung fields. Medical Physics. 1988;15(2):158–166. doi: 10.1118/1.596247. [DOI] [PubMed] [Google Scholar]
- 24.Catarious DM, Baydush AH, Floyd CE. Incorporation of an iterative, linear segmentation routine into a mammographic mass cad system. Medical Physics. 2004;31(6):1512–1520. doi: 10.1118/1.1738960. [DOI] [PubMed] [Google Scholar]
- 25.Wei J, Chan H-P, Sahiner B, Zhou C, Hadjiiski LM, Roubidoux MA, Helvie MA. Computer-aided detection of breast masses on mammograms: dual system approach with two-view analysis. Medical Physics. 2009;36(10):4451–4460. doi: 10.1118/1.3220669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skalak DB. Prototype and feature selection by sampling and random mutation hill climbing algorithms; Proceedings of the Eleventh International Conference on Machine Learning; 1994. pp. 293–301. [Google Scholar]
- 27.Chakraborty D. Handbook of medical imaging. Bellingham, Wash: 2000. Ch. The FROC, AFROC and DROC variants of the ROC analysis; pp. 771–796. [Google Scholar]
- 28.Chan H-P, Wei J, Sahiner B, Rafferty EA, Wu T, Roubidoux MA, Moore RH, Kopans DB, Hadjiiski LM, Helvie MA. Computer-aided detection system for breast masses on digital tomosynthesis mammograms: preliminary experience. Radiology. 2005;237(3):1075–1080. doi: 10.1148/radiol.2373041657. URL http://dx.doi.org/10.1148/radiol.2373041657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiser I, Nishikawa RM, Giger ML, Wu T, Rafferty EA, Moore R, Kopans DB. Computerized mass detection for digital breast tomosynthesis directly from the projection images. Medical Physics. 2006;33(2):482–491. doi: 10.1118/1.2163390. [DOI] [PubMed] [Google Scholar]
- 30.Chan H-P, Wei J, Zhang Y, Helvie MA, Moore RH, Sahiner B, Hadjiiski L, Kopans DB. Computer-aided detection of masses in digital tomosynthesis mammography: comparison of three approaches. Medical Physics. 2008;35(9):4087–4095. doi: 10.1118/1.2968098. [DOI] [PMC free article] [PubMed] [Google Scholar]