Abstract
The best characterized examples of crosstalk between two or more different post-translational modifications (PTMs) occur with respect to histones. These examples demonstrate the critical roles that crosstalk plays in regulating cell signaling pathways. Recently, however, non-histone crosstalk has been observed between serine/threonine phosphorylation and the modification of arginine and lysine residues within kinase consensus sequences. Interestingly, many kinase consensus sequences contain critical arginine/lysine residues surrounding the substrate serine/threonine residue. Therefore, we hypothesize that non-histone crosstalk between serine/threonine phosphorylation and arginine/lysine modifications is a global mechanism for the modulation of cellular signaling. In this review, we discuss several recent examples of non-histone kinase consensus sequence crosstalk, as well as provide the biophysical basis for these observations. In addition, we predict likely examples of crosstalk between protein arginine methyltransferases 1 (PRMT1) and Akt, and discuss the future implications of these findings.
Introduction
Over the last decade there have been several examples of crosstalk between two or more different post-translational modifications (PTMs), with many of these being observed within the context of histones. Generally, this crosstalk is thought to fine-tune cell signaling cascades such that a desired outcome is achieved, e.g., transcription of a particular gene or, alternatively, activation of one gene under the control of a transcription factor and repression of another. In recent years, several papers have been published that describe crosstalk in non-histone proteins, with a particular set of crosstalk examples involving serine/threonine phosphorylation and the modification of neighboring arginine and lysine residues. Given that these neighboring arginine and lysines are key substrate recognition elements for many protein kinases (Figure 1), we hypothesize that crosstalk between serine/threonine phosphorylation and arginine/lysine modifications (Figure 2) is a general mechanism to regulate eukaryotic cell signaling. Given the implications of this hypothesis on both human cell signaling and disease, this review will mainly focus on crosstalk between these specific modifications. In particular, we briefly discuss the concept of crosstalk and its roots in chromatin biology, provide several known examples of crosstalk between these modifications, and describe the biophysical reasoning for this communication. In addition, we highlight key features of protein kinase substrate recognition, identify potential substrates for crosstalk between PRMT1 and Akt, and finally discuss future perspectives and the medical relevance of this type of crosstalk.
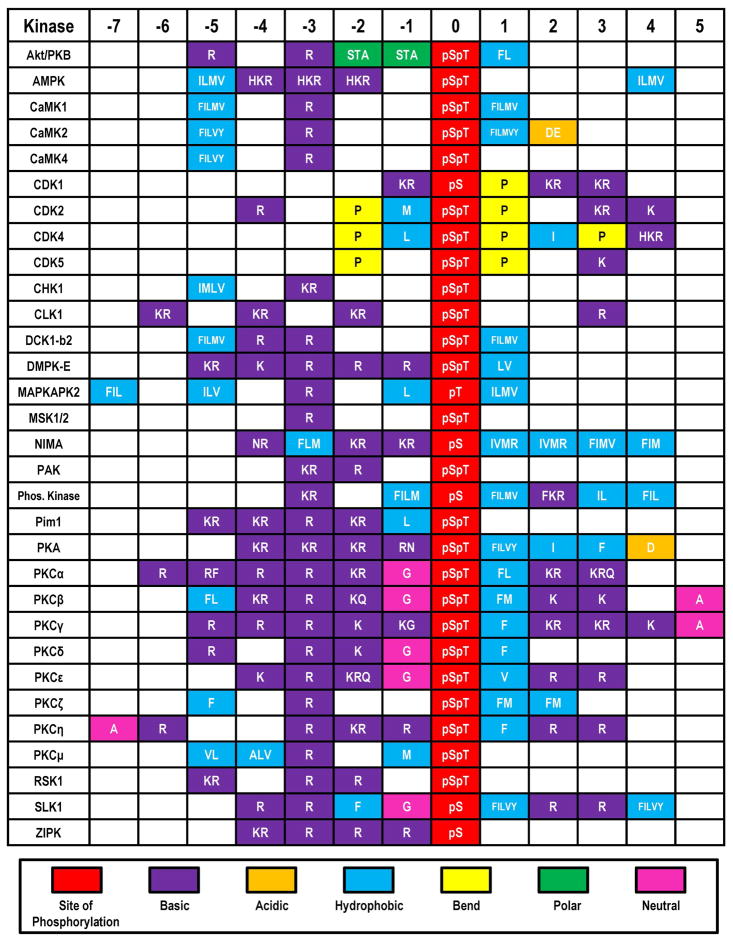
Figure 1.
Serine/Threonine Protein Kinase consensus sequences. A number of serine/threonine protein kinases recognize protein sequences that contain positively charged arginine and lysine residues adjacent to the site of phosphorylation. For example, Akt, prefers substrates that have two arginines (or lysines) at positions −3 and −5 with respect to the modification site that are separated by a variant residue. Each box represents one residue’s position and multiple single letter amino acid codes demonstrate variability within that position. Adapted from (82).
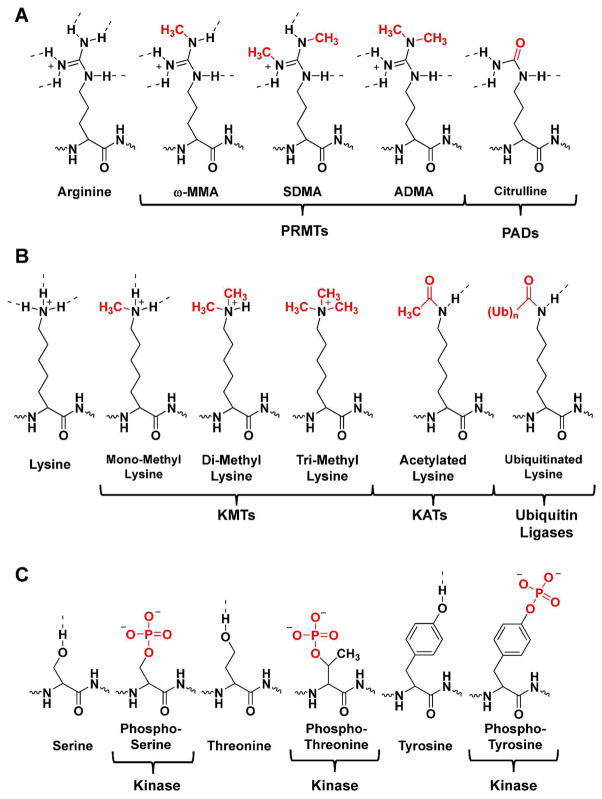
Figure 2.
Selected posttranslational modifications of arginine, lysine, serine, threonine, and tyrosine. (a) Arginine residues can be mono- and dimethylated by the PRMTs to form ω-MMA, ADMA, or SDMA. They can also be converted to citrulline by the PADs. (b) Lysine residues can be mono-, di-, and trimethylated by KMTs, acetylated by KATs, or ubiquitinated by ubiquitin ligases. (c) Serine, threonine, and tyrosine residues can be phosphorylated by kinases.
Crosstalk Models
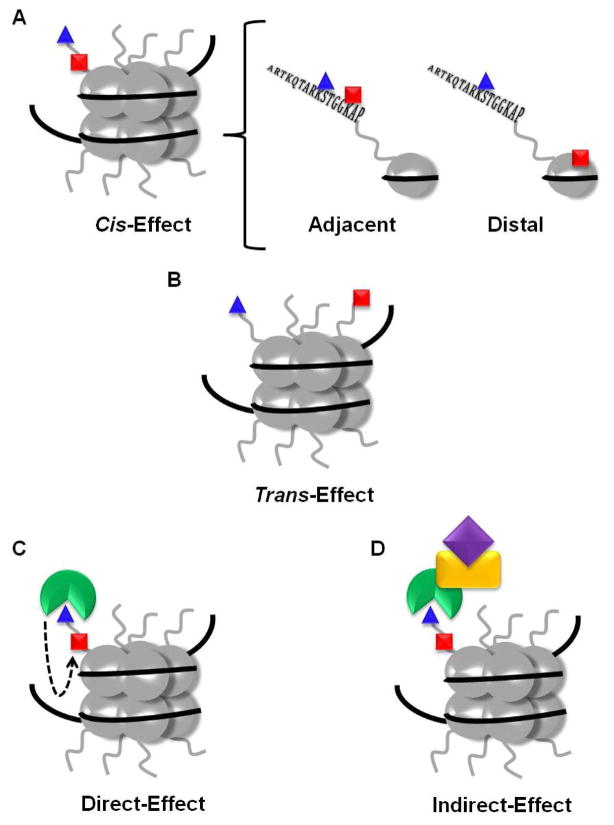
Although crosstalk between two or more PTMs has predominantly been studied within the context of chromatin biology (1, 2), as one would expect, this type of regulatory mechanism extends to non-histone proteins as well. Several models have been proposed for histone crosstalk (3–5), and they are readily applied to non-histone proteins as well (Figure 3). For example, cis crosstalk refers to communication between modifications on the same protein (Figure 3A). Within cis crosstalk lies the possibility for adjacent crosstalk (i.e., between residues that are close to one another in both the primary and tertiary structures) or distal crosstalk (i.e., between residues that are far apart in both the primary and tertiary structures) (Figure 3A). Trans crosstalk is also possible and occurs between modifications on two different proteins (Figure 3B). Functionally, direct crosstalk refers to one PTM directly affecting the modification of a second residue (e.g., modification of one residue prevents the modification of another residue) (Figure 3C). Indirect crosstalk involves modulating a protein-protein interaction via the presence, or lack, of a PTM (e.g., a PTM enhances the binding of a transcription factor leading to the recruitment of other coactivators) (Figure 3D). An early example of direct cis crosstalk (Figure 3A) from the histone field involves the phosphorylation of H3S10 and the acetylation of H3K14. Stimulation of the Ras-MAPK (mitogen activated protein kinase) pathway (6) results in the Rsk-2 (ribosomal S6 kinase) dependent phosphorylation of H3S10 (7), which enhances the acetylation of H3K14 by generating a better substrate for the histone acetyltransferase Gcn5 (general control non-repressed 5) (8–10). Although this is only one example of crosstalk from the histone field, a plethora of others have been published (reviewed in (1, 2, 11)).
Figure 3.
Crosstalk scenarios. (a) The cis-effect refers to crosstalk between two or more modifications located on the same protein. Within the same protein there can be adjacent (i.e., between residues that are close in both primary and tertiary structures) or distal (i.e. between residues that are separated in primary and tertiary structures) crosstalk. (b) The trans-effect refers to crosstalk between two modifications located on two different proteins. (c) Functionally, crosstalk can be direct (i.e., one modification inhibits or enhances the subsequent modification of the same or a different residue) or (d) indirect (i.e., a specific modification inhibits or enhances protein-protein interactions leading to altered downstream effects). Note that while this figure depicts crosstalk involving histones, the same explanations can be applied to non-histone proteins. Adapted from (3).
Crosstalk – Moving Beyond Histones
Arginine Methylation Blocks Phosphorylation
Four examples of direct, adjacent cis crosstalk have emerged involving arginine methylation and serine phosphorylation. First, Yamagata et al. demonstrated that FOXO1 (forkhead box O1) is methylated in vitro and in vivo by PRMT1 (Protein Arginine Methyltransferase 1) (12). FOXO1 is a member of the FOXO family of transcription factors, which play essential roles in cell cycle regulation, apoptosis, the oxidative stress response, and overall cell survival (13). The sites of FOXO1 modification, R248 and R250, are conserved across family members and species. Interestingly, the FOXO family members are phosphorylated by Akt at three conserved residues, one of which, i.e., S253 in FOXO1 (14), lies adjacent to R248 and R250 (mouse numbering). The authors then demonstrated that methylation of these two arginine residues inhibited phosphorylation of S253, a clear example of cis crosstalk. However, the converse was not observed – phosphorylation does not prevent methylation (12).
Functionally, methylation of R248 and R250 blocks the phosphorylation of S253, thereby preventing the phosphorylation dependent nuclear export of FOXO1 (15). Additional experiments indicated that it is the lack of phosphorylation, and not the presence of methylated arginine residues, that is responsible for inhibiting FOXO1 export (12). Because the phosphorylation and nuclear export of FOXO1 is associated with its polyubiquitination and degradation by the proteosome (16, 17), the authors tested the effects of PRMT1 knockdown on FOXO ubiquitination and stability. The results indicated that PRMT1 knockdown enhanced the polyubiquitination of FOXO1, which promoted its degradation by the proteosome (12).
Since it is well established that FOXO family members control the response to oxidative stress (13), the authors further hypothesized that the methylation of FOXO1 would affect this pathway. As expected, hydrogen peroxide led to an increase in the PRMT1-dependent methylation of FOXO1, which in turn blocked the phosphorylation of S253, and nuclear export. As a result, the transcription of a number of FOXO1-dependent genes was increased including, BIM (BCL-2-interacting mediator), an apoptosis inducing protein (12). Consistent with this model, when either PRMT1 or FOXO1 were knocked down by siRNA, no increase in BIM transcription was observed. Although PRMT1 knockdown inhibited apoptosis in response to oxidative stress, inhibition of PI3K-Akt signaling has the reverse effect (12). Taken together, these results demonstrate a functional crosstalk between the methylation of R248 and R250 and phosphorylation of S253 of FOXO1. This mechanism of crosstalk appears to be evolutionarily conserved because methylation of the FOXO1 orthologue in C. elegans, DAF-16, also inhibits its phosphorylation by Akt. Interestingly, this activity in C. elegans appears to play a role in life span extension, as PRMT1 knockouts died significantly earlier than wild type worms, thereby suggesting that PRMT1 inhibition may exert pleiotropic off target effects (18).
Recently, Sakamaki et al. published a second example of cis crosstalk between PRMT1- and Akt. Specifically, the authors investigated whether the phosphorylation of other proteins that contain an Akt consensus sequence (i.e., RXRXXS/T) are modulated by the methylation of adjacent arginine residues (19). Several known Akt substrates (i.e., BAD, PGC-1α, eNOS, p27, GSK3β, and MDM2) were tested as PRMT1 substrates. Only PGC-1α (peroxisome proliferator-activated receptor- coactivator) (12) and BAD (BCL-2 antagonist of cell death) (19), were shown to be methylated by PRMT1. The fact that eNOS, p27, GSK3β, and MDM2 were not methylated (19) suggests that additional PRMT1 recognition elements are required for substrate methylation (vide infra). In BAD, the sites of modification were identified as R94 and R96, and, as was the case with FOXO1 (12), arginine methylation prevented phosphorylation of an adjacent serine residue (i.e., S99), but prior phosphorylation did not affect BAD methylation by PRMT1 (19).
Due to the functional role of a methylation/phosphorylation switch in regulating FOXO1 activity (12), it was probable that crosstalk would also affect the physiological activity of BAD. BAD is a pro-apoptotic member of the BCL-2 protein family and plays a major role in regulating cellular apoptosis (20). Previous studies have demonstrated that several kinases and phosphatases are responsible for altering the phosphorylation state of BAD and thus dictating its location and activity (20–30). For example, when BAD is dephosphorylated, it binds to the pro-survival proteins BCL-XL/BCL-2 and displaces the pro-apoptotic proteins BAK and/or BAX (from BCL-XL/BCL-2) to create activated homodimers that form a pore in the mitochondria, which ultimately leads to apoptosis (20, 21, 31). However, in response to cellular stress, BAD is phosphorylated at S75, S99, and S118. Once phosphorylated, BAD binds to a 14-3-3 protein and is subsequently removed from the mitochondria and sequestered in the cytoplasm. As a consequence, the pro-apoptotic function of BAD is muted (22–27, 32, 33). Based on this model, one would expect that decreased methylation of BAD by PRMT1 would increase phospho-BAD levels, which would lead to enhanced 14-3-3 binding, sequesteration in the cytoplasm, decreased caspase activity, and consequently an increase in cell viability, all of which were observed when PRMT1 was knocked down by siRNA (19). In contrast to the situation with FOXO1, methylation of BAD was not triggered by oxidative stress or known BAD activators. Thus it is unclear whether BAD methylation is constitutive or occurs in response to an unknown stimulant (19). In any case, these observations demonstrate that the methyltransferase activity of PRMT1 is critical for the pro-apoptotic function of BAD through its prevention of Akt mediated phosphorylation of S99 (19).
The previous two examples involved PRMT1, a Type I PRMT that catalyzes the asymmetric dimethylation of arginine residues in proteins. In the third example of direct adjacent cis crosstalk, PRMT5, a Type II PRMT that symmetrically dimethylates arginines, is the responsible enzyme. Specifically, Guo et al. show that FEN1 (flap endonuclease 1) is methylated at R192 by PRMT5 and that methylation of this particular arginine inhibits the phosphorylation of S187 by Cdk2-cyclin E (cyclin dependent kinase-2). As with the previous examples, the reverse scenario was not observed, i.e., phosphorylation does not prevent methylation (34). Since FEN1 phosphorylation prevents its binding to PCNA (proliferating cell nuclear antigen) (35), it is unsurprising that by preventing phosphorylation, methylation promotes the PCNA and FEN1 interaction. However, methylation occurring after phosphorylation did not re-establish this protein complex, confirming that the inhibition of phosphorylation, and not methylation alone, is responsible for the observed effect (34). The interaction between PCNA and FEN1 is responsible for localizing FEN1 to the site of replication. This interaction is important because FEN1 is an exo- and endonuclease involved in essential DNA processes, such as replication and repair (36). A R192K mutant, which cannot be methylated, abrogated both the PCNA/FEN1 interaction and the localization of FEN1 to the site of replication. As a consequence, a buildup of DNA double-stranded breaks was detected, followed by slower progression through the cell cycle, and ultimately mitotic arrest (34). Because other studies have shown that PRMT1 plays a role in the oxidative stress response (12), the authors also investigated the affect of hydrogen peroxide on the methylation of FEN1. The results showed that oxidative stress results in localization of methylated FEN1 to the nucleus. FEN1 that lacked methylation resulted in a decrease in cell survival and an increase in mutations, thus demonstrating a correlation between arginine methylation and DNA repair (34). These observations demonstrate that PRMT5 dependent methylation of the R192 residue of FEN1 plays a critical role in preventing Cdk2-cyclinE dependent phosphorylation of S187 and subsequently allows for proper DNA replication and repair. With respect to human disease, these results suggest that PRMT5 inhibition would synergize with DNA damaging agents as a way to treat cancer.
Phosphorylation Blocks Arginine Methylation
The above crosstalk examples demonstrate a functional role for the inhibition of serine phosphorylation by arginine methylation, but not the inverse. More recently, Sims et al. uncovered such an example of direct adjacent cis crosstalk, where phosphorylation of RNA polymerase II (RNAPII) prevents its methylation at R1810 (37). The CTD (carboxy terminal domain) of RNAPII contains a series of heptad repeats whose consensus sequence is YSPTSPS (38), however, several of these repeats contain arginine or lysine substitutions in the last position (i.e., YSPTSP[R/K]) (37). It is known that S2 and S5 of these sequences can be phosphorylated by P-TEFb (positive transcription elongation factor b) and CAK, (CDK activating kinase), respectively (37). Phosphorylation of these residues activates RNAPII and aids in the recruitment of essential proteins (39, 40) that are important for gene transcription (38). Due to the unique nature of the arginine and lysine substitutions, the authors investigated whether they were specifically modified. The only PTM to be identified was methylation of R1810 by CARM1 (co-activator-associated protein arginine methyltransferases 1) or PRMT4. Interestingly, phosphorylation of S2 and S5 prevented methylation of R1810 but methylation did not prevent phosphorylation. Also, the presence of both methylation and phosphorylation was observed in vivo suggesting that methylation occurs before phosphorylation (37). The functional consequence of a lack of methylation of R1810 is downregulated transcription of small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA) (37).
Distal Crosstalk Between Arginine Methylation and Phosphorylation
Although the main focus of this paper is direct adjacent cis crosstalk within kinase consensus sequences, it is worth noting two additional examples involving distal non-histone crosstalk between arginine methylation and phosphorylation. The first example involves an interesting interplay between the CARM1 dependent methylation of R3 in C/EBPβ (CCAAT/enhancer-binding protein β) (41), a transcription factor, and the phosphorylation of T253 by MAPK (42). Here, phosphorylation of T253 abrogates the interaction between C/EBPβ and CARM1, which abolishes the methylation of R3 (41). Unmethylated C/EBPβ is then free to bind to the SWI/SNF nucleosome remodeling and the Mediator transcriptional co-activator complexes to facilitate the increased transcription of C/EBPβ-dependent genes. Thus, R3 methylation inhibits interactions between C/EBPβ, SWI/SNF, and Mediator, and as a consequence, down regulates the transcription of genes under the control of C/EBPβ.
A second example of distal crosstalk involves the members of the STAT (signal transducer and activator of transcription) family of proteins, which play important roles in cell differentiation, survival, and apoptosis (43). Given that STAT1 is methylated by PRMT1 at R31, a conserved arginine residue (44), Chen et al. investigated whether the corresponding arginine in STAT6, R27, was also modified, and found that this was the case. However, the responsible methyltransferase was not identified (45). Nevertheless, the authors did show that the lack of methylation prevented phosphorylation of a distal tyrosine residue (i.e., Y641) and consequently inhibited nuclear translocation, abrogated DNA binding, and decreased protein stability. It was shown that this crosstalk was not due to the activation of tyrosine phosphatases. Overall, the results demonstrated that arginine methylation of STAT6 is required for phosphorylation of an essential tyrosine residue, which ultimately effects the location of the protein, its DNA binding capabilities, and protein stability (45).
Lysine Methylation Blocks Phosphorylation
In addition to arginine methylation, direct adjacent cis crosstalk has been observed between lysine methylation and serine phosphorylation. Recently, Carr et al. determined that K810 of pRb (retinoblastoma protein), a tumor suppressor protein that plays a critical role in controlling progression through the cell cycle (46–49), is methylated by SET7/9. This residues lies within a Cdk (cyclin dependent kinase) consensus sequence (i.e., [S/T]PX[K/R]), and the authors speculated that an interplay might exist between serine phosphorylation and modification of this lysine. Indeed, lysine methylation directly prevented phosphorylation of not only S807, but S811 as well. Additionally, the inhibition of S807 and S811 phosphorylation by methylation of K810 decreased phosphorylation levels throughout pRb suggesting that methylation has an indirect effect on the phosphorylation of distal sites. The reverse case, however, was not true in that prior phosphorylation of pRb did not affect methylation (46).
From a physiological standpoint, these findings are significant because pRb phosphorylation occurs in a cell cycle dependent manner and is a major mechanism to control cell cycle progression. For example, when pRb is phosphorylated it is incapable of binding E2F, a transcription factor, resulting in the activated transcription of E2F target genes (e.g., the cyclins), which ultimate leads to cell cycle progression (46–53). In contrast, when pRb is hypophosphorylated, it binds to E2F and represses the expression of E2F target genes, leading to cell cycle arrest (46–53). In light of the fact that phosphorylation modulates the functional role of pRb, the authors hypothesized that methylation of pRb would affect the transcriptional activity of E2F in response to DNA damage. Knockout of SET7/9 yielded an increase in the levels of phosphorylated pRb, decreased binding to E2F, and a consequent increase in the expression of several E2F target genes (e.g., DHFR, Cdc2, and Cdc6), with and without the addition of etoposide, a known DNA damaging agent. Overall, these results suggest that methylation plays a critical role in regulating the transcriptional activity of E2F, and that the methylation of K810 in pRb by SET7/9 is important for cell cycle arrest (46).
A Phosphorylation & Lysine Methylation Switch
A unique example of consensus crosstalk was found to exist between the methylation and phosphorylation of DNMT1 (DNA methyltransferase 1); DNMT1 catalyzes the methylation of the C5 position of cytosines in DNA, which is important for establishing and maintaining tissue specific gene transcription. Building on previous work that had shown that K142 of DNMT1 is methylated by the lysine methyltransferase SET7/9 (54), Estève et al. found that an adjacent serine residue (i.e., S143) is phosphorylated by Akt. Further investigation yielded the discovery that, in vivo, phosphorylation of S143 prevents methylation of K142 and vice versa. Interestingly, in vitro, a small percentage of a methylated DNMT1 peptide became phosphorylated. Therefore, it is possible that both modifications may be present at the same time on a small quantity of the total protein concentration. Regardless, a knockout of Akt resulted in increased proteolysis of DNMT1 overtime, although, mRNA levels remained constant. Also, the half-life of phosphorylated DNMT1 was found to be greater than that of the methylated protein (55). These results suggest that methylated DNMT1 is more susceptible to degradation and that phosphorylation can prevent it. Functionally, the communication between these methylation and phosphorylation events appears to be cell cycle dependent, as increases in phosphorylation are observed as the cell cycle progresses from S phase to G2 followed by a decrease in phosphorylation and a corresponding increase in methylation (55). This observation makes sense because DNMT1 is involved in establishing and regulating tissue specific DNA methylation patterns. Overall, the results of these studies suggest that Akt and SET7/9 mediated phosphorylation and methylation of DNMT1 regulate its activity in a cell cycle dependent manner. Although not investigated, it is likely that the activity of DNMT1 is further regulated by phosphatases and lysine demethylases.
Indirect Crosstalk Between Lysine Methylation and Phosphorylation
As discussed in regards to histones, indirect crosstalk involves the enhancement or inhibition of protein-protein interactions due to specific modifications. Levy et al. provide an interesting example of such indirect crosstalk between the methylation and phosphorylation of RelA (56). RelA is a subunit of the transcription factor NF-κB (57, 58). First, they first discovered that RelA is monomethylated at K310 by SETD6 and phosphorylated at S311 by PKC. Interestingly, the authors also demonstrated that the modification of either K310 or S311 does not affect the modification of the other residue, suggesting that both modifications can be present on RelA at the same time (56). Functionally, these two modifications modulate the expression of NF-κB target genes. For example, in the absence of phosphorylation, methylated RelA interacts with GLP (G9a-like protein), a second lysine methyltransferase, and localizes at the promoters of NF-κB target genes (e.g. CCND1, IL8, MYC, IL1A). This interaction facilitates H3K9 methylation by GLP, which results in decreased gene transcription. In contrast, when cells are stimulated with TNF (tumor necrosis factor), RelA is phosphorylated by PKC and, consequently, the interaction between GLP and RelA is inhibited. Thus, without the repressive histone methyltransferase activity of GLP, RelA activates the expression of NF-κB target genes (56). In this case it appears that phosphorylation can override the inhibitory effects of lysine methylation.
A Phosphorylation & Lysine Acetylation Switch
As discussed above, lysine acetylation transforms a positively charged residue into a neutral residue, and thus also likely affects adjacent cis crosstalk. This speculation is confirmed in an example involving phosphorylation and acetylation of ERα (estrogen receptor α), an estrogen responsive transcription factor (59). ERa is phosphorylated at S305 by PKA (60, 61). Adjacent to S305, and present within the same PKA consensus sequence (i.e., XRRXSX or XKKXSX), are K302 and K303, which are acetylated by p300 (62). In light of the fact that K303 is mutated to an arginine in one third of patients with premalignant breast hyperplasias and that this mutation renders the cell hypersensitive to low doses of estrogen (63), Cui et al. decided to focus on crosstalk between the acetylation of K303 and phosphorylation of S305. In addition, they hoped to determine the impact of the K303R mutation on possible ERα crosstalk. The results showed that acetylation of K303 prevents the phosphorylation of S305 and vice versa, thus demonstrating negative crosstalk between these two residues (59). This is in line with substrate specificity studies on p300 that showed that positively charged residues adjacent to the site of acetylation are important for substrate recognition (64). Interestingly, the K303R mutant yielded a higher concentration of phosphorylated protein compared to wild type ERα, suggesting that when an arginine is present at this position the protein is a better substrate for PKA (59).
With regards to the functional consequences, the authors investigated the impact of this crosstalk on the involvement of ERα in transcriptional regulation and cell proliferation. The results demonstrated that the K303R mutant yielded an increase in both transcription and cell proliferation compared to wild type due to its hypersensitivity towards low doses of estrogen. The inhibition of PKA, however, diminished this effect, thus demonstrating that the hypersensitivity and increased activity of the K303R mutant is a result of increased phosphorylation of S305 (59). In addition, a more recent article provided evidence for crosstalk between acetylation of K303 and methylation of K302. Subramanian et al. showed that methylation of K302 of ERα by SET7/9 stabilizes the protein and is essential for the activation of ERα. Interestingly, acetylation of K303 by p300 decreases methylation of K302, however, the K303R mutation increases methylation. The authors suggest that the repression of transcription that is observed with acetylation of K303 may not only be due to prevention of S305 phosphorylation, but due to the degradation of ERα caused by a lack of K302 methylation. The K303R mutation, however, allows for both K302 methylation and S305 phosphorylation, thus upregulating gene transcription (65).
Structural Basis for Crosstalk
Protein kinases typically bind and phosphorylate serine, threonine, or tyrosine residues within a distinct consensus sequence. Of these kinases, Akt is a perfect example of how PTMs within a consensus sequence can alter substrate binding. For Akt, the consensus sequence (i.e., RXR[S/T/A][S/T/A][S/T][F/L]) contains two positively charged arginine residues at the −3 and −5 positions relative to the phosphorylation site (Figure 1). Based on the structure of Akt bound to a peptide whose sequence is derived from GSK3β (66) (Figure 4A), it is apparent that R-5 forms several direct and indirect hydrogen bonds with surrounding Akt residues, including E279, Y316, and E342, as well as T-2 on the peptide itself. In addition, the positive charge of R-5 and the negative charge of E279 are likely capable of forming a salt bridge. R-3 also forms a hydrogen bond and salt bridge with E236, and, although the distance between R-3 and D440 is too great for a hydrogen bond interaction, it is also possible that there are electrostatic attractions between these two residues. In the examples described above, both R-3 and R-5 were methylated and this prevented the phosphorylation of the targeted serine residue (12, 19). The structural basis for this crosstalk is easily discerned. For example, while the formation of an asymmetrically dimethylated arginine would not alter the charge of the residue, it would undoubtedly create steric bulk. This added steric bulk would prevent the formation of key hydrogen bonds, with for example E279 and Y316 in the substrate binding cleft, which would result in an inability to properly bind the substrate and thereby inhibit phosphorylation. Similar effects would be expected for the methylation of R-3. One could imagine that deimination of these two arginine residues by the PADs would likely yield the same result. However, instead of adding steric bulk, abrogation of protein binding would be due to the neutralization of the positively charged arginine residue and the disruption of proper hydrogen bonding. Although the conversion to citrulline would still allow for a lower degree of hydrogen bond formation, the carbonyl oxygen would only be a hydrogen bond acceptor, thus terminating the bidentate interactions between R-3 and E236 and R-5 and E279.
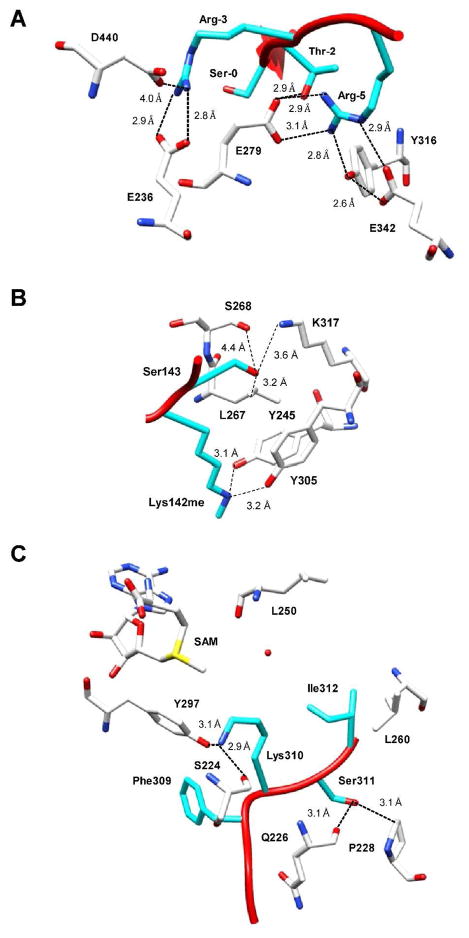
Figure 4.
Structural basis for crosstalk. (a) A structure of Akt (white) bound to a GSK3β (cyan) derived peptide demonstrates that arginine residues in the −5 and −3 positions are critical for Akt substrate recognition. R-5 forms direct and indirect hydrogen bonds with several key residues (i.e. E279, Y316, E342), as well as, with T-2 on the peptide. This residue is also capable of forming a salt bridge with E279. R-3 forms both a hydrogen bond and salt bride with E236. The methylation of both R-5 and R-3 would disrupt these key interactions and thus result in the observed inhibition of serine phosphorylation, which is demonstrated in several examples presented in this review (i.e., FOXO1 (12) and BAD (19)). This figure was prepared with UCSF Chimera using the coordinates for the Akt·GSK3β peptide complex (PDBID 1O6L). (b) A structure of SET7/9 (white) bound to DNMT1 (cyan) provides the structural basis for the inhibition of lysine methylation by phosphorylation of an adjacent serine residue (55). In this case, the potentially phosphorylated residue, i.e. S143 forms a hydrogen bond with K317 and also has van der Waals interactions with L267. The addition of a phosphate group would cause van der Waals respulsions between between the S143 and L267 and thus would prevent methylation. This figure was prepared with UCSF Chimera using the coordinates for the SET7/9·DNMT1 peptide complex (PDBID 3OS5). (c) A structure of SETD6 (white) bound to a RelA peptide (cyan) shows that the potentially phosphorylated S311 forms a hydrogen bond with the backbone carbonyl of Q226 and also has van der Waals interactions with P228. As the authors note, the addition of a phosphate group would likely prevent RelA binding due to sterics and thus abrogate methylation of K310 (69). Note that in all structures a dashed line simply represents the distance between two residues and not necessarily a hydrogen bond. Also note that single letter abbreviations are used to denote residues on the enzyme and three letter abbreviations are used to denote residues on the peptide.
One of the more interesting examples of crosstalk that was presented above involved the inhibition of PKA dependent phosphorylation of ERα by p300 dependent acetylation. Acetylation of K303 prevents the phosphorylation of S305, however, a K303R mutation enhances phosphorylation of S305 (59). As described by Yang et al., PKA also has several conserved glutamate/aspartate residues within the peptide binding cleft that form critical interactions with positively charged residues adjacent to the site of phosphorylation (66). As mentioned previously, acetylation neutralizes the positively charged lysine, and, like methylation, introduces a significant degree of steric bulk. Thus, it is unsurprising that acetylation of K303 would prevent the phosphorylation of S305.
Interestingly, phosphorylation does not block methylation by PRMT1 and PRMT5. Based on the crystal structure of PRMT1 (67), as well as work from our own lab (68), this observation is easily rationalized. For example, while the surface of PRMT1 is highly negatively charged (67), which would suggest that the introduction of a phosphate group would lead to electrostatic repulsions, we have shown that the residues between the site of methylation and distal positively charged residues are relatively unimportant for substrate recognition (68). Although a similar explanation is likely for PRMT5, detailed substrate specificity studies have not been performed on this isozyme. In contrast to the situation with PRMTs 1 and 5, phosphorylation of S2 and S5 in RNAPII blocked the methylation of R1810 by CARM1 (37). Given that detailed substrate specificity studies have also not been performed for this enzyme and no structures of CARM1 bound to cognate peptide substrates are available, it is difficult to speculate on why phosphorylation blocks methylation. Nevertheless, the addition of two phosphate groups adjacent to the site of methylation would likely not only cause a perturbation in the peptide structure itself, due to repulsion between the two phosphate groups, but would also likely disrupt key interactions within the substrate binding cleft.
With regard to phosphorylation blocking lysine methylation or acetylation, as in the cases of DNMT1 (55) and ERα (59), it is likely that transitioning from a neutrally charged hydroxyl group to a bulky and negatively charged phosphate group would affect the binding capability of a methyltransferase or acetyltransferase. This is observed in the structure of the SET7/9-DNMT1 product complex between the SET7/9 lysine methyltransferase and a monomethylated lysine peptide substrate derived from DNMT1 (Figure 4B) (55). In this structure, Ser143 in DNMT1, which can be phosphorylated by Akt, forms a hydrogen bond with a nearby lysine residue, K317, and also has van der Waals interactions with a leucine residue, L267. The authors speculate that the observed inhibition of methylation is due to van der Waal repulsions between the phosphate and leucine residue (55).
Chang et al. recently provided a structural explanation for indirect crosstalk between the methylation by SETD6 at K310 and phosphorylation by PKC at S311 of RelA (Figure 4C) (56, 69). Within the SETD6 active site, the target lysine (i.e. K310) can adopt a bent or linear conformation, however, in both cases key hydrogen bonds are formed with the backbone carboxyl of L250 in the linear conformation or with the hydroxyl of Y297 and the backbone carbonyl of S224 in the bent conformation. S311 also hydroxyl forms a hydrogen bond with the backbone carbonyl oxygen of Q226 and has van der Waal interactions with nearby P228. As the authors note, the addition of a phosphate group would likely prevent RelA binding due to sterics (69).
Predicting Crosstalk
Given that protein kinase substrates can be readily predicted based on the presence/absence of a particular consensus sequence, the most obvious question is whether it is possible to predict crosstalk. The answer appears to be yes. Below we predict potential crosstalk between Akt substrates and PRMT1-mediated methylation. Note that we focused on these two enzymes because of prior precedents with Akt (see above) and our own expertise in predicting PRMT1 substrates (68). Additionally, PRMT1 is responsible for 85% of all PRMT activity in vivo (70, 71), thus it is likely that, if kinase consensus crosstalk is a global mechanism for cellular regulation, PRMT1 would be the principal isozyme involved. Nevertheless, it should be recognized that the same approach could be taken to predict crosstalk between any given kinase with a known consensus sequence and a lysine or arginine modifying enzyme whose substrate specificity determinants are known.
In contrast to kinases, the distinct substrate recognition sequences for PRMT1, and the PRMTs in general, are relatively unknown. In an effort to determine a minimal peptide substrate, based on the N-terminus of histone H4, our lab discovered that positively charged residues distal to the site of methylation are important for substrate recognition and catalysis (68). This makes sense because the surface of PRMT1 is negatively charged and therefore electrostatic interactions between the protein and the substrate are likely present (67, 68). With this knowledge, we investigated whether similarities are present between histone H4 and the recently discovered PRMT1 substrates described above. As shown in Table 1, for histone H4, BAD, and the FOXO family members, two arginine residues, in close proximity to each other, are present distal to the site of methylation in addition to a number of other positively charged residues. This observation coincides with our previous findings (68). Closer examination of Akt substrates that failed to be methylated by the PRMTs, (e.g., eNOS, p27, and GSK3β) (19) shows that they lack distal positively charged residues throughout the intervening sequences (Table 1). The one exception is MDM2. However in this case, the presence of several negatively charged glutamates likely masks the presence of distal positively charged residues. In light of this analysis, we hypothesized that novel Akt/PRMT1 substrates can be predicted based on the identity of residues downstream from the kinase consensus sequences (Table 2). Note that we have stratified the Akt substrates into three groups, i.e., highly probable, likely, and unlikely PRMT1 substrates. Highly probable substrates were selected based on the presence of at least two distal arginine residues separated by one or two variable residue(s), as these RXR and RXXR motifs are a common theme among the known PRMT substrates. These predictions also include at least one other distal positively charged residue in addition to these motifs. Likely substrates include those Akt substrates that possess a number of positively charged residues distal from the predicted sites of methylation, but do not possess the RXR or RXXR motifs, or do not contain an additional positively charged residue. Note that further studies will need to be conducted to determine whether lysine can substitute for arginine in these positions or if the presence of several nonspecific positively charged residues alone is enough for efficient substrate recognition and catalysis. The effect of the presence and position of negatively charged residues on substrate recognition also will need further investigation because, as mentioned above, they likely mask the positively charged residues. Validation of the predicted PRMT1 and Akt substrates would indicate that consensus crosstalk is a general mechanism to control eukaryotic cell signaling.
Table 1.
Tested Substrates for PRMT1 and Akt

|
Known PRMT1 substrate.
Not known to be an Akt Substrate.
Numbers correspond to human FOXO1. Mouse FOXO1 begins at 248.
Sites of methylation have not been identified.
Not a PRMT1 Substrate. The light blue represents sites of methylationde. The red represents known sites of Akt phosphorylation. The purple represents positively charged residues. The orange represents negatively charged residues.
Table 2.
PRMT1 and Akt Crosstalk Predictions
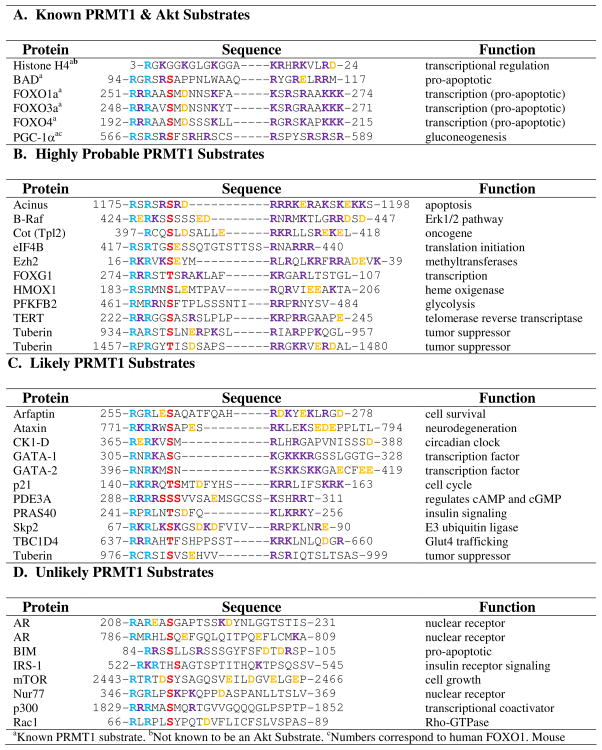
|
Known PRMT1 substrate.
Not known to be an Akt Substrate.
Numbers correspond to human FOXO1. Mouse FOXO1 begins at 248.
Sites of methylation have not been identified.
Not a PRMT1 Substrate. The light blue represents sites of methylationde. The red represents known sites of Akt phosphorylation. The purple represents positively charged residues. The orange represents negatively charged residues.
Future Perspectives
Role of Additional Modifications
Although the discussion so far has focused mainly on arginine and lysine methylation, as well as lysine acetylation, both of these residues are subject to a plethora of additional modifications (Figure 2). As such, it is likely that these modifications will exhibit crosstalk with protein phosphorylation. In support of this notion, the Coonrod group recently showed that Elk-1 (ETS like gene 1), a member of the ETS family of transcription factors, is deiminated by PAD4 and that this modification increases Elk-1 phosphorylation by ERK2. Although it has yet to be established that these two modifications occur in the same consensus sequence, it is known that phosphorylation facilitates a tight interaction between Elk-1 and p300 leading to increased histone acetylation and ultimately the activation of c-Fos (72).
An additional layer of complexity to the crosstalk puzzle is the roles of the de-modifying enzymes (e.g., HDACs, lysine demethylases, and phosphatases) in modulating crosstalk. It is likely that a precisely regulated cycle of modification and de-modification of lysine in accord with the phosphorylation and de-phosphorylation of serine, threonine, or tyrosine residues must exist. The situation with arginine methylation is more complex because an arginine demethylase has yet to be identified. However, it should be clear that the dysregulation of any one of those enzymes could facilitate the onset of a plethora of diseases.
One final layer of complexity is the role of antagonistic PTMs. For example, we and others have shown that deimination/citrullination of an arginine residue can antagonize/prevent the methylation of that same arginine residue (73–78). In addition, serine O-GlcNAcylation has been shown to antagonize phosphorylation of the same serine residue (79, 80). Overall, these individual observations suggest that crosstalk within kinase consensus sequences is potentially quite complex, has multiple levels, and that eukaryotic cell signaling is not well represented by the linear pathways often depicted in textbooks.
Crosstalk and Disease
Crosstalk between protein phosphorylation and modification of arginine and lysine residues within kinase consensus sequences potentially has significant relevance to human disease. This is the case because the modification of these basic residues can potentially have either growth promoting or growth suppressing effects. For example, in the two examples of Akt crosstalk, PRMT1 opposes the effects of Akt-mediated phosphorylation (12, 19). Given that PI3K-Akt signaling is overactive in multiple cancers, these results suggest that the inhibition of PRMT1 would further stimulate the growth promoting and cell survival effects of Akt signaling. Nevertheless, PRMT1 has been shown to be required for the growth promoting effects of estrogen signaling and siRNA knockdown of PRMT1 has been shown to suppress the growth of MCF7 cells (81). As such, it is unclear whether PRMT1 represents a valid target for the development of an anticancer therapeutic, thereby highlighting the critical need for developing bioavailable PRMT1 inhibitors that can be used to specifically address this question. Additionally, the putative roles of arginine and lysine modifying enzymes in regulating kinase signaling highlight the possibility that the effects of inhibitors targeting these enzymes, including those enzymes that remove these modifications, may be due not only to effects on gene transcription but also to effects on kinase signaling pathways. Again, this highlights the need for additional research to examine the links between consensus crosstalk and human disease. Finally, the fact that the mutation of K303 to an arginine residue in ERa is present in one-third of patients with premalignant hyperplasias (63) is highly interesting because it suggest that cancer associated mutations can impinge on crosstalk. Given the numerous PRMTs, KMTs, and HATs/KATs, research in this area is undoubtedly an untapped treasure waiting to be discovered.
Conclusions
In this review we have summarized and presented recent examples of non-histone crosstalk involving arginine and lysine modifications and the phosphorylation of serine residues. These examples have led us to hypothesize that crosstalk within kinase consensus sequences is a general mechanism for controlling cellular signaling. Although several of the examples described above indicated that the serine phosphorylation and lysine/arginine modifications are mutually exclusive, it is interesting to note that phosphorylation does not prevent methylation in all cases (e.g., FOXO1 (12), BAD (19), FEN1 (34), pRb (46)). While the physiological basis for this effect is not known, it is tempting to speculate that, in these specific examples, arginine methylation may prevent repeated phosphorylation/dephosphorylation cycles in order to desensitize cell signaling. Although the substrate specificities of many kinases are known, the sequence requirements for arginine and lysine modifying enzymes remain less clear and their discovery would enable the prediction of additional potential crosstalk candidates. Overall, the dysregulation of this newly uncovered mechanism for cellular signaling likely plays an important role in numerous human diseases.
Acknowledgments
The authors would like to thank W. Ja and T. Kodadek for their critical reading of this manuscript. We also thank X. Cheng for providing the PDB file used to generate Figure 4C.
Abbreviations
- PTM
post translational modification
- PRMT
protein arginine methyltransferase
- MAPK
mitogen activated protein kinase
- Rsk-2
ribosomal S6 kinase
- Gcn5
growth control non-repressed 5
- IE
immediate-early
- COMPASS
complex of proteins associated with Set1
- MLL
mixed lineage leukemia
- Paf1
RNA polymerase II associated factor
- SAM
S-adenosyl methionine
- ω-MMA
omega-monomethylarginine
- ADMA
asymmetric dimethyl arginine
- SDMA
symmetric dimethyl arginine
- PAD
protein arginine deiminase
- KMT
lysine methyltransferase
- HP1
heterochromatin protein 1
- KDM
lysine demethylase
- KAT
lysine acetyltransferase
- CBP
CREB-binding protein
- PCAF
p300/CBP-associated factor
- HDACS
histone deacetylases
- FOXO1
forkhead box O1
- BIM
BCL-2-interacting mediator
- BCL-2
B-cell lymphoma-2
- PI3K
phopshoinositide 3-kinase
- PGC-1α
peroxisome proliferator-activated receptor- coactivator
- BAD
BCL-2 antagonist of cell death
- eNOS
endothelial nitric oxide synthase
- GSK3β
glycogen synthase kinase-3 β
- MDM2
murine double minute-2
- FEN1
flap endonuclease 1
- Cdk-2
cyclin dependent kinase-2
- PCNA
proliferating cell nuclear antigen
- RNAPII
RNA polymerase II
- CTD
carboxy terminal domain
- P-TEFb
positive transcription elongation factor b
- CAK
CDK activating kinase
- CARM1
co-activator associated protein arginine methyltransferase 1
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
- C/EBPβ
CCAT/enhancer-binding protein β
- SWI/SNF
switch/sucrose nonfermentable
- STAT
signal transducer and activator of transcription
- pRb
retinoblastoma protein
- DNMT1
DNA methyltransferase 1
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- GLP
G9a-like protein
- TNF
tumor necrosis factor
- ERα
estrogen receptor α
- ELK1
ETS like gene 1
- ERK2
mitogen activated protein kinase 1
Footnotes
This work was supported by funds provided by the The Scripps Research Institute, Scripps Florida, and National Institute of Health grant GM079357 to P.R.T.
References
- 1.Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 2.Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischle W. Talk is cheap--cross-talk in establishment, maintenance, and readout of chromatin modifications. Genes Dev. 2008;22:3375–3382. doi: 10.1101/gad.1759708. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 5.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 6.Chadee DN, Hendzel MJ, Tylipski CP, Allis CD, Bazett-Jones DP, Wright JA, Davie JR. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- 7.Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 8.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 9.Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 11.Baek Sung H. When Signaling Kinases Meet Histones and Histone Modifiers in the Nucleus. Molecular cell. 2011;42:274–284. doi: 10.1016/j.molcel.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 12.Yamagata K, Daitoku H, Takahashi Y, Namiki K, Hisatake K, Kako K, Mukai H, Kasuya Y, Fukamizu A. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 14.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Gan L, Pan H, Guo S, He X, Olson ST, Mesecar A, Adam S, Unterman TG. Phosphorylation of serine 256 suppresses transactivation by FKHR (FOXO1) by multiple mechanisms. Direct and indirect effects on nuclear/cytoplasmic shuttling and DNA binding. J Biol Chem. 2002;277:45276–45284. doi: 10.1074/jbc.M208063200. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi Y, Daitoku H, Hirota K, Tamiya H, Yokoyama A, Kako K, Nagashima Y, Nakamura A, Shimada T, Watanabe S, Yamagata K, Yasuda K, Ishii N, Fukamizu A. Asymmetric Arginine Dimethylation Determines Life Span in C. elegans by Regulating Forkhead Transcription Factor DAF-16. Cell Metab. 2011;13:505–516. doi: 10.1016/j.cmet.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Sakamaki JI, Daitoku H, Ueno K, Hagiwara A, Yamagata K, Fukamizu A. Arginine methylation of BCL-2 antagonist of cell death (BAD) counteracts its phosphorylation and inactivation by Akt. Proc Natl Acad Sci U S A. 2011;108:6085–6090. doi: 10.1073/pnas.1015328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danial NN. BAD: undertaker by night, candyman by day. Oncogene. 2008;27(Suppl 1):S53–70. doi: 10.1038/onc.2009.44. [DOI] [PubMed] [Google Scholar]
- 21.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 22.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 23.Harada H, Becknell B, Wilm M, Mann M, Huang LJ, Taylor SS, Scott JD, Korsmeyer SJ. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 24.Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 25.Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 26.Tan Y, Demeter MR, Ruan H, Comb MJ. BAD Ser-155 phosphorylation regulates BAD/Bcl-XL interaction and cell survival. J Biol Chem. 2000;275:25865–25869. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 27.Virdee K, Parone PA, Tolkovsky AM. Phosphorylation of the pro-apoptotic protein BAD on serine 155, a novel site, contributes to cell survival. Curr Biol. 2000;10:1151–1154. doi: 10.1016/s0960-9822(00)00702-8. [DOI] [PubMed] [Google Scholar]
- 28.Ayllon V, Martinez AC, Garcia A, Cayla X, Rebollo A. Protein phosphatase 1alpha is a Ras-activated Bad phosphatase that regulates interleukin-2 deprivation-induced apoptosis. EMBO J. 2000;19:2237–2246. doi: 10.1093/emboj/19.10.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiang CW, Harris G, Ellig C, Masters SC, Subramanian R, Shenolikar S, Wadzinski BE, Yang E. Protein phosphatase 2A activates the proapoptotic function of BAD in interleukin- 3-dependent lymphoid cells by a mechanism requiring 14-3-3 dissociation. Blood. 2001;97:1289–1297. doi: 10.1182/blood.v97.5.1289. [DOI] [PubMed] [Google Scholar]
- 30.Wang HG, Pathan N, Ethell IM, Krajewski S, Yamaguchi Y, Shibasaki F, McKeon F, Bobo T, Franke TF, Reed JC. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 31.Dewson G, Kluck RM. Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J Cell Sci. 2009;122:2801–2808. doi: 10.1242/jcs.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L) Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 33.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 34.Guo Z, Zheng L, Xu H, Dai H, Zhou M, Pascua MR, Chen QM, Shen B. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat Chem Biol. 2010;6:766–773. doi: 10.1038/nchembio.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henneke G, Koundrioukoff S, Hubscher U. Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene. 2003;22:4301–4313. doi: 10.1038/sj.onc.1206606. [DOI] [PubMed] [Google Scholar]
- 36.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 37.Sims RJ, 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, 3rd, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Fong N, Bentley DL. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misteli T, Spector DL. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 41.Kowenz-Leutz E, Pless O, Dittmar G, Knoblich M, Leutz A. Crosstalk between C/EBPbeta phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 2010;29:1105–1115. doi: 10.1038/emboj.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakajima T, Kinoshita S, Sasagawa T, Sasaki K, Naruto M, Kishimoto T, Akira S. Phosphorylation at threonine-235 by a ras-dependent mitogen-activated protein kinase cascade is essential for transcription factor NF-IL6. Proc Natl Acad Sci U S A. 1993;90:2207–2211. doi: 10.1073/pnas.90.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calo V, Migliavacca M, Bazan V, Macaluso M, Buscemi M, Gebbia N, Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J Cell Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 44.Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 45.Chen W, Daines MO, Hershey GK. Methylation of STAT6 modulates STAT6 phosphorylation, nuclear translocation, and DNA-binding activity. J Immunol. 2004;172:6744–6750. doi: 10.4049/jimmunol.172.11.6744. [DOI] [PubMed] [Google Scholar]
- 46.Carr SM, Munro S, Kessler B, Oppermann U, La Thangue NB. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 2011;30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 48.Stevens C, La Thangue NB. E2F and cell cycle control: a double-edged sword. Arch Biochem Biophys. 2003;412:157–169. doi: 10.1016/s0003-9861(03)00054-7. [DOI] [PubMed] [Google Scholar]
- 49.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 50.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 51.Goodrich DW, Wang NP, Qian YW, Lee EY, Lee WH. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell. 1991;67:293–302. doi: 10.1016/0092-8674(91)90181-w. [DOI] [PubMed] [Google Scholar]
- 52.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 53.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 54.Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, Pradhan S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, Espejo A, Zee BM, Liu CL, Tangsombatvisit S, Tennen RI, Kuo AY, Tanjing S, Cheung R, Chua KF, Utz PJ, Shi X, Prinjha RK, Lee K, Garcia BA, Bedford MT, Tarakhovsky A, Cheng X, Gozani O. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat Immunol. 2011;12:29–36. doi: 10.1038/ni.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 58.Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cui Y, Zhang M, Pestell R, Curran EM, Welshons WV, Fuqua SA. Phosphorylation of estrogen receptor alpha blocks its acetylation and regulates estrogen sensitivity. Cancer Res. 2004;64:9199–9208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- 60.Cho H, Katzenellenbogen BS. Synergistic activation of estrogen receptor-mediated transcription by estradiol and protein kinase activators. Mol Endocrinol. 1993;7:441–452. doi: 10.1210/mend.7.3.7683375. [DOI] [PubMed] [Google Scholar]
- 61.Chen D, Pace PE, Coombes RC, Ali S. Phosphorylation of human estrogen receptor alpha by protein kinase A regulates dimerization. Mol Cell Biol. 1999;19:1002–1015. doi: 10.1128/mcb.19.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, Lisanti MP, Katzenellenbogen BS, Kato S, Hopp T, Fuqua SA, Lopez GN, Kushner PJ, Pestell RG. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–18383. doi: 10.1074/jbc.M100800200. [DOI] [PubMed] [Google Scholar]
- 63.Fuqua SA, Wiltschke C, Zhang QX, Borg A, Castles CG, Friedrichs WE, Hopp T, Hilsenbeck S, Mohsin S, O’Connell P, Allred DC. A hypersensitive estrogen receptor-alpha mutation in premalignant breast lesions. Cancer Res. 2000;60:4026–4029. [PubMed] [Google Scholar]
- 64.Thompson PR, Kurooka H, Nakatani Y, Cole PA. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J Biol Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- 65.Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol. 2002;9:940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- 67.Zhang X, Cheng X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure. 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osborne TC, Obianyo O, Zhang X, Cheng X, Thompson PR. Protein arginine methyltransferase 1: positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry. 2007;46:13370–13381. doi: 10.1021/bi701558t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang Y, Levy D, Horton JR, Peng J, Zhang X, Gozani O, Cheng X. Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang J, Frankel A, Cook RJ, Kim S, Paik WK, Williams KR, Clarke S, Herschman HR. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J Biol Chem. 2000;275:7723–7730. doi: 10.1074/jbc.275.11.7723. [DOI] [PubMed] [Google Scholar]
- 71.Nicholson TB, Chen T, Richard S. The physiological and pathophysiological role of PRMT1-mediated protein arginine methylation. Pharmacol Res. 2009;60:466–474. doi: 10.1016/j.phrs.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 72.Zhang X, Gamble MJ, Stadler S, Cherrington BD, Causey CP, Thompson PR, Roberson MS, Kraus WL, Coonrod SA. Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate c-Fos Expression in Breast Cancer Cells. PLOS Genetics. 2011;7:e1002112. doi: 10.1371/journal.pgen.1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hidaka Y, Hagiwara T, Yamada M. Methylation of the guanidino group of arginine residues prevents citrullination by peptidylarginine deiminase IV. FEBS Lett. 2005;579:4088–4092. doi: 10.1016/j.febslet.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 74.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 75.Thompson PR, Fast W. Histone citrullination by protein arginine deiminase: is arginine methylation a green light or a roadblock? ACS Chem Biol. 2006;1:433–441. doi: 10.1021/cb6002306. [DOI] [PubMed] [Google Scholar]
- 76.Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol. 2007;367:1118–1129. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- 77.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 78.Guo Q, Bedford MT, Fast W. Discovery of peptidylarginine deiminase-4 substrates by protein array: antagonistic citrullination and methylation of human ribosomal protein S2. Mol Biosyst. 2011;7:2286–2295. doi: 10.1039/c1mb05089c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu Rev Biochem. 2010;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Le Romancer M, Treilleux I, Leconte N, Robin-Lespinasse Y, Sentis S, Bouchekioua-Bouzaghou K, Goddard S, Gobert-Gosse S, Corbo L. Regulation of estrogen rapid signaling through arginine methylation by PRMT1. Mol Cell. 2008;31:212–221. doi: 10.1016/j.molcel.2008.05.025. [DOI] [PubMed] [Google Scholar]
- 82. [Accessed On May 1, 2011]; http://www.kinexus.ca/pdf/graphs_charts/ProteinSerKinaseSpecificity.pdf.