SUMMARY
(De)acetylation of histone and non-histone proteins is an important post-translational modification affecting many cellular processes. Here we report that NuA4 acetylation of Sip2, one of three regulatory β subunits of Snf1 complex (yeast AMP-activated protein kinase), decreases as cells age. We used mutations at four acetylation sites, K12, 16, 17 and 256, to study acetyl-Sip2 function. Sip2 acetylation, controlled by antagonizing NuA4 acetyltransferase and Rpd3 deacetylase, enhances interaction with Snf1, the catalytic subunit of Snf1 complex. Sip2-Snf1 interaction inhibits Snf1 activity, thus decreasing phosphorylation of a downstream target, Sch9 (homolog of Akt/S6K), ultimately leads to slower growth but extends replicative lifespan. Sip2 acetylation mimetics are more resistant to oxidative stress. We further demonstrate that the anti-aging effect of Sip2 acetylation is independent of extrinsic nutrient availability and TORC1 activity. We propose a novel protein acetylation- phosphorylation cascade that regulates Sch9 activity, controls intrinsic aging and extends replicative lifespan in yeast.
INTRODUCTION
Reversible acetylation and deacetylation of histones are important in regulating chromatin structure and controlling transcription of genes crucial for maintenance of cell viability (Lin et al., 2008). Among the histone deacetylases, Sir2 (yeast homologue of mammalian SIRT1 (Donmez and Guarente, 2010)) and Rpd3 (yeast homologue of mammalian HDAC1 (Willis-Martinez et al., 2010)) are especially important in lifespan regulation in yeast (Chang and Min, 2002; Jiang et al., 2002). Sir2 extends replicative lifespan partially via deacetylating histone H4 lysine 16 that compromises transcriptional silencing (Dang et al., 2009; Imai et al., 2000; Sinclair and Guarente, 1997). However, the lifespan discrepancies between substrate histone mutants and the acetyltransferase/deacetylase mutants (Dang et al., 2009), suggest possible roles of non-histone acetylation substrates in mediating lifespan regulation by these (de)acetylating enzymes.
In a previous study, we identified many non-histone substrates of the NuA4 complex, of which the catalytic subunit, Esa1, is the only essential histone acetyltransferase in yeast (Lin et al., 2009). We show here that NuA4 catalytic mutants have a replicative lifespan defects caused by impaired acetylation of Sip2, a known replicative lifespan regulator (Ashrafi et al., 2000). Sip2 is one of three β regulatory subunits of the Snf1 complex, and the only β subunit implicated in yeast replicative aging (Ashrafi et al., 2000; Lin et al., 2003). The Snf1 complex contains a catalytic α subunit, Snf1, an AMP-activated serine/threonine protein kinase; a γ subunit, Snf4; and one of the three β regulatory subunits, Sip1, Sip2, or Gal83, each with a distinctive substrate specificity (Schmidt and McCartney, 2000). Besides being required for transcription of glucose-repressed genes and utilization of carbon sources other than glucose (Amodeo et al., 2007), Snf1 is also a key player in the response to cellular stress (Sanz, 2003). Importantly, Snf1 activity increases in aged cells, even when ambient glucose is abundant (Ashrafi et al., 2000; Hedbacker and Carlson, 2008). Null mutations in SIP2, the SNF1 repressor, decrease lifespan; these can be rescued by deletion of SNF4, the SNF1 activator (Guarente and Kenyon, 2000).
Here we report that Sip2 acetylation is controlled by the NuA4 and its counteracting deacetylase, Rpd3 (Chang and Pillus, 2009). We examined replicative lifespan in various SIP2 acetylation mutants and found that Sip2 acetylation extends yeast lifespan; besides, constitutive Sip2 acetylation mimetics nearly totally rescue the lifespan shortening phenotype of the NuA4 catalytic mutant, indicating the critical role of Sip2 acetylation in lifespan modulation. We further investigated glucose limitation, peroxide sensitivity and age-associated changes in Sip2 acetylation, established the role of Sip2 acetylation in controlling Snf1 interactions and activities. Finally we established that Sch9, the yeast homologue of Akt and S6K (Madia et al., 2009), was a common downstream target of two distinct replicative lifespan regulating pathways: the intrinsic aging defense pathway described here controlled by Snf1 kinase and the extrinsic nutrient-sensing pathway regulated by TORC1.
RESULTS
Sip2 Acetylation Is Controlled by NuA4 and Rpd3
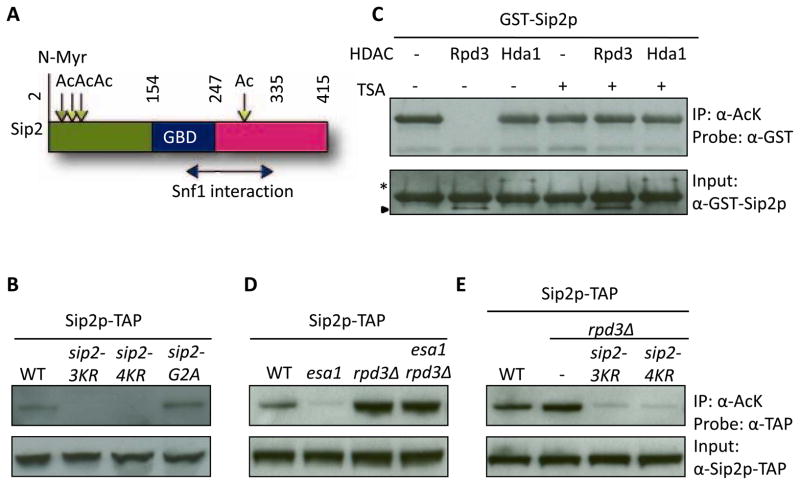
To investigate the function of Sip2 acetylation, we identified four acetylated lysine residues (K12, 16, 17 and 256) (Figure 1A), using tandem mass spectrometry (Figures S1A–C). We then created unacetylable lysine-to-arginine mutant constructs at these four sites in various combinations by site-directed mutagenesis, and introduced these mutant SIP2 constructs into the endogenous chromosomal locus (Toulmay and Schneiter, 2006). Using a previously described reverse IP approach (Lin et al., 2009), we showed that Sip2 is hypoacetylated when the first three (K12, K16, K17; 3KR) or all four (K12, K16, K17 and K256; 4KR) lysines are mutated (Sip2-3KR or -4KR; Figures S1D, Figure 1B). We also created a chromosomally integrated SIP2 glycine 2-to-alanine mutant (sip2-G2A) to mimic the short-lived, non-N-myristoylated species of Sip2 (Ashrafi et al., 2000). We found that sip2-G2A did not have acetylation defects (Figure 1B). An in vitro deacetylation reaction carried out with purified Rpd3-TAP and Hda1-TAP (Figure S2A) revealed that Rpd3 treatment abolished the acetylation signal of Sip2 (Figure 1C). We demonstrated the activity of purified Hda1-TAP by showing that it deacetylated lysine 14 of FLAG-Htz1 purified from hda1Δ strain (Figure S2B) (Lin et al., 2008). As will be shown below, mutagenesis of the Sip2 acetylation sites affects replicative aging. The findings are consistent with a previous report that Rpd3, but not Hda1, is involved in replicative lifespan regulation (Kim et al., 1999). Previously we have shown that the acetylation signals of Sip2 are virtually completely dependent on NuA4/Esa1 both in vivo and in vitro (Lin et al., 2009). Importantly, simultaneous deletion of RPD3 reversed the hypoacetylation caused by NuA4 catalytic subunit temperature sensitive (Ts) allele esa1-531 when cells were shifted to the non-permissive temperature (Figure 1D). Further, the increased acetylation in rpd3Δ was abolished by sip2-3KR and 4KR mutations (Figure 1E). These results support the simple hypothesis that K12, 16, 17 and 256 in Sip2 are acetylated by Esa1, and are deacetylated by Rpd3.
Figure 1. Sip2 Is Acetylated at Four Lysine Sites by Esa1, While Rpd3 Is the Counteracting Deacetylase.
(A) Cartoon of Sip2 structural domains (adapted from Hedbacker, et al. (Hedbacker and Carlson, 2008)). Mass spectrometry identified four acetylated lysine residues of Sip2, K12, K16, K17 and K256. Numbers indicate amino acid residues. Vertical straight line, myristoylation site; Downward arrows, acetylation sites; right-left arrow, regions mapped by deletion analysis as sufficient for Snf1 interaction (Amodeo et al., 2007). Ac, acetylation; N-Myr, N-myristoylation; GBD, glycogen-binding domain.
(B) Chromosomally integrated sip2-3KR and sip2-4KR, but not sip2-G2A mutants, are hypoacetylated in vivo. The sip2-G2A mutant blocks myristoylation. sip2-3KR, sip2-K12/16/17R; sip2-4KR, sip2-K12/16/17/256R.
(C) Rpd3, but not Hda1, removes Sip2 acetylation in vitro. Deacetylation reaction is inhibited by addition of trichostatin A (TSA). Asterisk indicates Hda1-TAP; arrowhead indicates Rpd3-TAP.
(D) Sip2 is hypoacetylated in strains carrying the Ts allele of ESA1, esa1-531, but is hyperacetylated in rpd3Δ; deletion of RPD3 rescues the acetylation defect of esa1-531.
(E) Increased Sip2 acetylation in rpd3Δ is blocked when lysines are mutated to arginines.
Sip2 Acetylation Mimetics Increase Replicative Lifespan
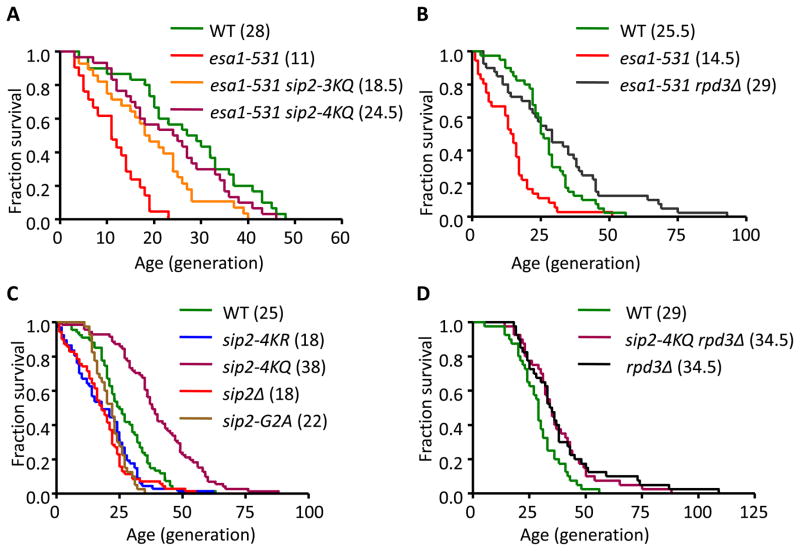
We next examined replicative lifespan of the SIP2 acetylation mutants (refer to Table S1 for detailed statistical results). We found that the esa1-531 strain showed a shortened replicative lifespan when grown at a semi-permissive temperature, 30°C. Importantly, in an esa1-531 strain, the lysine-to-glutamine mutants of SIP2 (sip2-3KQ and sip2-4KQ), mimicking acetylated Sip2, reversed the shortened lifespan of esa1-531 (Figure 2A). Consistent with the above findings, deletion of RPD3 also reversed the shortened lifespan of esa1-531 (Figure 2B). We also found that sip2-4KQ significantly increased the lifespan, whereas sip2-4KR and sip2Δ strains decreased lifespan to a similar extent when compared to wild type (WT) SIP2+ strains (Figure 2C). Because the N-myristoylation on glycine 2 of Sip2 is essential for long lifespan (Ashrafi et al., 2000; Lin et al., 2003), the chromosomally integrated sip2-G2A mutant was used as a positive control for shortened lifespan (Figure 2C). Finally, we confirmed that simultaneous sip2-4KQ mutant did not further extend the lifespan of the null mutant of RPD3 (Figure 2D). These results provide strong evidence supporting Sip2 as the critical downstream target of Esa1 and Rpd3 in regulating replicative lifespan in yeast.
Figure 2. Sip2 Acetylation Increases the Cellular Replicative Lifespan.
(A) Survival curves of the indicated strains with each median lifespan in parentheses. The fractions of live cells are plotted as a function of age in generations. Strains carrying esa1-531 exhibit lifespan shortening that are rescued by SIP2 acetylation mimetics (sip2-3KQ and sip2-4KQ).
(B) Deletion of RPD3 reverses the short lifespan of esa1-531.
(C) sip2-4KR mutants, as well as the sip2-G2A mutants and deletion of SIP2, decrease cellular lifespan, whereas sip2-4KQ mutants increase lifespan.
(D) Simultaneous sip2-4KQ mutants on the background of rpd3Δ fail to further increase lifespan of rpd3Δ. Statistical significance was determined by Mantel-Cox log-rank test and the details are presented in Table S1.
Sip2 Acetylation and Calorie Restriction
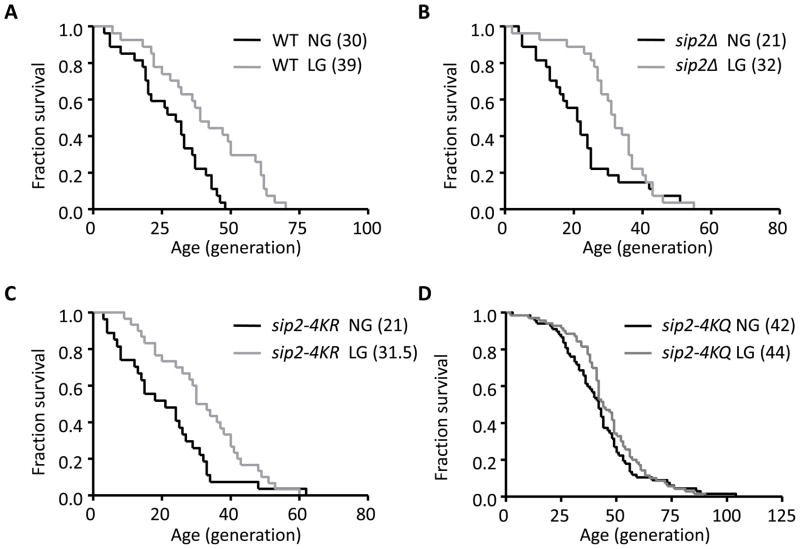
Since carbon substitutions greatly influence longevity in yeast (Barker et al., 1999; Kaeberlein et al., 2005), we next asked whether different carbon sources affect Sip2 acetylation status. We monitored Sip2 acetylation (Ac-Sip2) levels after treating cells with 2% glucose, 2% galactose, 0.05% glucose or 2% glycerol plus 3% ethanol. The only significant difference in acetylation states was observed when cells were treated with 0.05% glucose (low glucose; LG); although Sip2 protein abundance was upregulated, the acetylation signal decreased significantly (Figures S3A, B). This contradicts a simple model that calorie restriction extends replicative lifespan via Sip2 acetylation. Indeed, calorie restriction was shown to increase replicative lifespan through Sir2 (Lin et al., 2002) and other mechanisms (Kaeberlein et al., 2005). We therefore examined whether low glucose (LG) affected lifespan in the SIP2 acetylation mutants, and found that LG, when compared to 2% glucose (normal glucose, NG), increased lifespan by ~50% in the sip2-4KR and sip2Δ strains, and to somewhat a lesser extent in WT (~30%) (Figures 3A–3C, Table S1). However, LG failed to significantly increase the lifespan of the sip2-4KQ mutant (Figure 3D, Table S1). Since glucose limitation significantly improved the replicative lifespan of sip2-4KR and sip2Δ, these findings imply that calorie restriction extends lifespan through a mechanism largely independent of Sip2 acetylation. The fact that calorie restriction failed to significantly extend the lifespan of sip2-4KQ, which was similar to that reported in rpd3Δ (Jiang et al., 2002), suggests the existence of a common effector(s) downstream of the Sip2 acetylation and calorie restriction modulating replicative lifespan in yeast.
Figure 3. Glucose Limitation and Replicative Lifespan in SIP2 Acetylation Mutants.
Survival curves of WT (A), sip2Δ (B), sip2-4KR (C) and sip2-4KQ (D) grown in normal (2%, NG) vs. low glucose (0.05%, LG) media. The fractions of live cells are plotted as a function of age in generations. Median lifespan is shown in parentheses. Glucose limitation significantly increases lifespan in WT, sip2Δ, and sip2-4KR, but not in sip2-4KQ, as determined by Mantel-Cox log-rank test. The statistical results were summarized in Table S1.
Aging Decreases Sip2 Acetylation and Interaction with Snf1
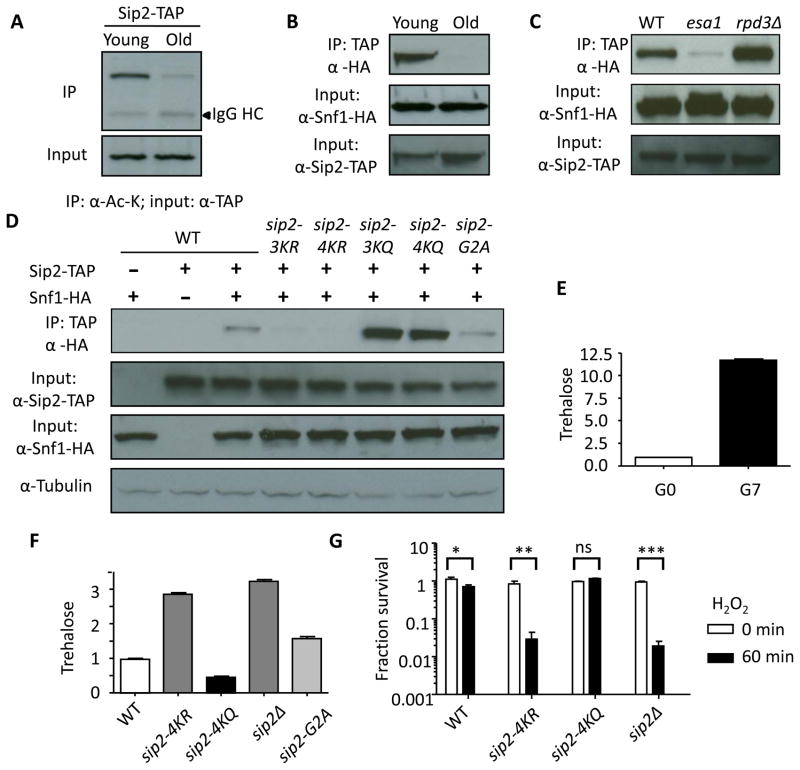
To investigate the effect of replicative aging on Sip2 acetylation, we examined Ac-Sip2 in young and old cells by sorting biotin-labeled mother cells (Smeal et al., 1996). The sorting efficiency was confirmed by showing that old cells contained a mean of 7 bud scars, whereas young cells had none (Figure S3C). We found that Ac-Sip2 levels decreased in old cells (Figure 4A). Co-immunoprecipitation of Sip2-TAP and Snf1-HA demonstrated that although the interaction between Sip2 and Snf1 was readily seen in young cells, it became undetectable in old cells (Figure 4B).
Figure 4. Sip2 Acetylation and Physical Interaction between Sip2 and Snf1 Decrease as Cells Age.
(A) Sip2 acetylation is significantly decreased in old cells.
(B and C) Physical interaction between Sip2 and Snf1 assessed by coimmunoprecipitation is decreased in old cells (B) and esa1-531 strain but increased in rpd3Δ strain (C).
(D) Interaction between Sip2 and Snf1 is decreased in sip2Δ, and deacetylation mimetics (sip2-3KR and sip2-4KR), but is not changed in sip2-G2A mutants. The interaction significantly increases in acetylation mimetics (sip2-3KQ and sip2-4KQ mutants).
(E) Cellular trehalose is significantly increased when cells age. Error bars show standard error of the mean (n=3).
(F) Trehalose levels are significantly higher in sip2-4KR, sip2-G2A and sip2Δ, when compared to WT; but lower in sip2-4KQ. Error bars show standard error of the mean (n=3).
(G) Fraction of survival of WT, sip2-4KR, sip2-4KQ and sip2Δ, before and 1h after treatment with 1.5 mM H2O2, are plotted on a log scale. Error bars show standard error of the mean (n=2). Statistical significance was assessed by two-way ANOVA with post-hoc test. ns, non-significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine whether Sip2 acetylation dictates physical interaction between Sip2 and Snf1, we examined their association in SIP2 acetylation mutants. Physical interaction between Sip2 and Snf1 significantly decreased in esa1-531, but increased in rpd3Δ (Figure 4C). More importantly, whereas the sip2-3KR and sip2-4KR mutations almost abolished the interaction with Snf1, the sip2-3KQ and sip2-4KQ mutations markedly enhanced it (Figure 4D), supporting the idea Sip2 acetylation is required for Snf1 interaction and defining the N-terminus of Sip2 as an enhancer of Snf1 interaction (Figure 1A). In contrast, the myristoylation-null mutation sip2-G2A (Lin et al., 2003) did not affect the physical interaction between Sip2 and Snf1 (Figure 4D), suggesting that myristoylation regulates Snf1 activity via a different mechanism. Since Sip2 is a negative regulator of Snf1 (Ashrafi et al., 2000), these results at least partially explain why Snf1 activity, which is stimulated under carbon stress, increases with age and hence has detrimental effects on lifespan, even in the presence of abundant ambient glucose (Lin et al., 2003).
To establish the connection between aging and Sip2 acetylation status in cells, we measured trehalose levels, a general stress indicator in yeast, in young and old cells. This is based on the previous findings that during the quiescent phase, trehalose is stored in favor over glycogen presumably to fulfill its numerous stress-protectant functions (Shi et al., 2010). Indeed, we found that young cells contained significantly less trehalose than old cells (Figure 4E). Glucose limitation, a form of nutrient stress, also significantly increased trehalose levels as reported before (Figure S3D) (Pluskal et al., 2011). Measurement of trehalose levels revealed that esa1-531 contained elevated trehalose levels that were partially reversed by concomitant RPD3 deletion (Figure S3E). Sip2-4KR rpd3Δ and sip2Δ rpd3Δ increased trehalose levels compared to rpd3Δ (Figure S3F). Also, sip2-4KR, sip2-G2A and sip2Δ mutants had increased trehalose levels compared to WT, whereas sip2-4KQ had a lower level of trehalose (Figure 4F), supporting a role of Ac-Sip2 in counteracting stress. We further showed that sip2-4KR and sip2Δ mutants were much more sensitive to hydrogen peroxide (H2O2), a form of oxidative stress and a mediator of aging (Giorgio et al., 2007), when compared to WT (Figure 4G); on the contrary, sip2-4KQ was resistant to H2O2 (Figure 4G).
Consistent with previous findings that trehalose plays a key role in fueling cell cycle progression during growth and division (Shi et al., 2010), we found that growth rates significantly decreased in the rpd3Δ strain (Figure S4A) and the sip2-4KQ strain on various genetic backgrounds (Figure S4B and S4C). Conversely, sip2-4KR and sip2Δ mutants partially rescued the rpd3Δ growth defect (Figure S4A). Cell volumes were similarly affected by the mutations; all the slower growing variants were smaller than normal cells (Figure S4D).
Sch9 Is the Downstream Target of Sip2-Snf1
Based on the above results and literature information, we proposed that Snf1 might have a downstream target that, when phosphorylated by Snf1, fuels rapid cell growth but results in shortened lifespan. In a previous phosphorylation proteome microarray study, 80 in vitro Snf1 substrates were identified (Ptacek et al., 2005). Among these, Sch9 (an Akt/S6K homolog) kinase was a promising candidate because it had already been shown to play an important role in yeast aging (Kaeberlein et al., 2005). In addition to Snf1, Sch9 is the in vitro substrate of two additional kinases (of 87 kinases tested), Tos3 (the upstream kinase of Snf1 (Kim et al., 2005)) and Pho85 (Ptacek et al., 2005). Sch9 is also a well-known in vivo kinase substrate of TORC1 (target of rapamycin complex 1); and, similar to TORC1, it negatively regulates lifespan in response to nutrient availability (Kaeberlein et al., 2005; Urban et al., 2007). A kinase itself, Sch9 phosphorylates a critically important ribosomal protein, Rps6 (Urban et al., 2007); rps6Δ mutants grow slowly, and are small, but have increased replicative lifespan through general inhibition of translation machinery (Chiocchetti et al., 2007) and protein synthesis (Huber et al., 2009).
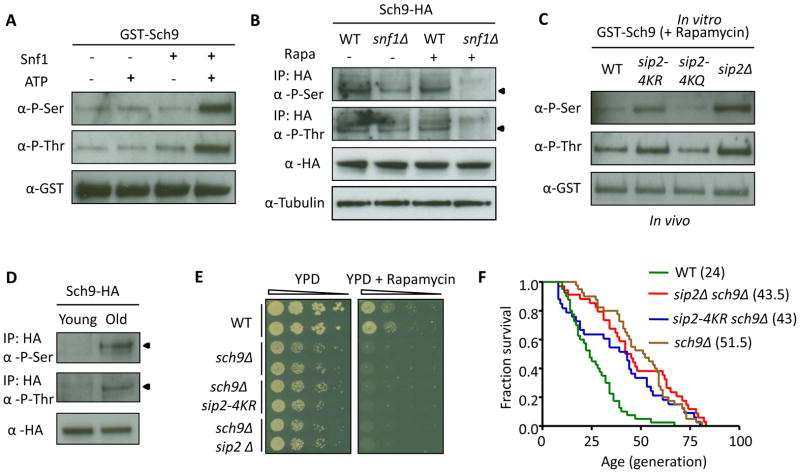
We first showed that Sch9 is an in vitro and in vivo substrate of Snf1. We purified GST-Sch9 and assessed the phosphorylation signal by performing immunoblotting with anti-phosphoserine and anti-phosphothreonine antibodies (Jablonowski et al., 2004). As a proof of concept, the signal of phosphorylated GST-Sch9 nearly totally disappeared after treatment with calf intestine phosphatase, while sodium pervanadate (a phosphatase inhibitor) restored the phosphorylation signal (Figure S5A). We carried out an in vitro kinase assay with purified Snf1-TAP (Figure S2A) from a sip2Δ strain, and showed serine and threonine phosphorylation of GST-Sch9 in vitro by Snf1 kinase (Figure 5A). We further confirmed that endogenous Sch9-HA phosphorylation markedly decreased in snf1Δ especially after treatment with rapamycin to block TORC1 activity (Figure 5B, and whole gel images in Figure S5B). Moreover, a mutation that abolishes the kinase activity of Snf1 (snf1-K84R) fails to revert the decreased phosphorylation of endogenous Sch9-HA in a snf1Δ strain (Figure S5C), which suggests that Sch9 is a bona fide in vivo substrate of Snf1.
Figure 5. Acetylation of Sip2 Affects Snf1 Kinase Activity and Results in Hypo-Phosphorylation of Sch9.
(A) Snf1 phosphorylates GST-Sch9 in vitro at both serine and threonine sites. In vitro kinase assays were performed by incubating purified GST-Sch9 with or without Snf1 or ATP as indicated, and analyzed by phosphoserine antibody (α-P-Ser) and phosphothreonine antibody (α-P-Thr) to detect Sch9p phosphorylation.
(B) Endogenous Sch9-HA phosphorylation decreases in snf1Δ mutants after rapamycin (200 ng/ml) treatment to suppress the TOR pathway. Arrowheads indicate the Sch9-HA band.
(C) GST-Sch9 phosphorylation increases in sip2-4KR and sip2Δ, but decreases in sip2-4KQ, compared to WT after rapamycin (200 ng/ml) treatment.
(D) Endogenous Sch9-HA phosphorylation significantly increases in the old cells. Arrowheads indicate the Sch9-HA band.
(E) SCH9 is epistatic and thus downstream to SIP2 in regulating cellular growth. Ten-fold dilutions of the indicated strains were spotted and grown on YPD plates without (2 days, 30°C) or with rapamycin (25 ng/ml, 4 days, 30°C).
(F) Deletion of SCH9 rescues the lifespan shortening of sip2Δ and sip2-4KR.
To detect Sch9 phosphorylation states in different SIP2 mutants, we purified GST-Sch9 proteins from WT, sip2-4KR, sip2-4KQ and sip2Δ strains, and assessed their phosphorylation signals. Both serine and threonine phosphorylation increased slightly in sip2-4KR and sip2Δ compared to WT and sip2-4KQ (Figure S5D); the differences were enhanced by inhibiting TORC1 with rapamycin (Figure 5C), with the lowest level of phosphorylation observed in the sip2-4KQ mutant. Finally, we confirmed that endogenous Sch9-HA phosphorylation markedly increased as cells aged (Figure 5D), which is consistent with aging-associated increase in Snf1 kinase activity.
To demonstrate that Sch9 is the critical downstream mediator of Sip2 in regulating growth and lifespan, we analyzed epistasis. The growth defects of sch9Δ cells were not rescued by sip2-4KR and sip2Δ (Figure S5E) as expected for a downstream effector. We showed that sch9Δ was hypersensitive to rapamycin as reported (Urban et al., 2007), which remained unaltered in the concomitant SIP2 mutants (Figure 5E). We also observed that the phenotype of shortened lifespan in the sip2Δ and sip2-4KR mutant cells was nearly completely reversed by deleting SCH9 (sip2Δ sch9Δ and sip2-4KR sch9Δ in Figure 5F). Consistent with these observations, sch9Δ cells also showed comparable lifespan extension as both double-mutant strains (Figure 5F). These data support Sch9 as the downstream target of Sip2-Snf1.
Sip2-Snf1 and TORC1 Function in Parallel Upstream of Sch9
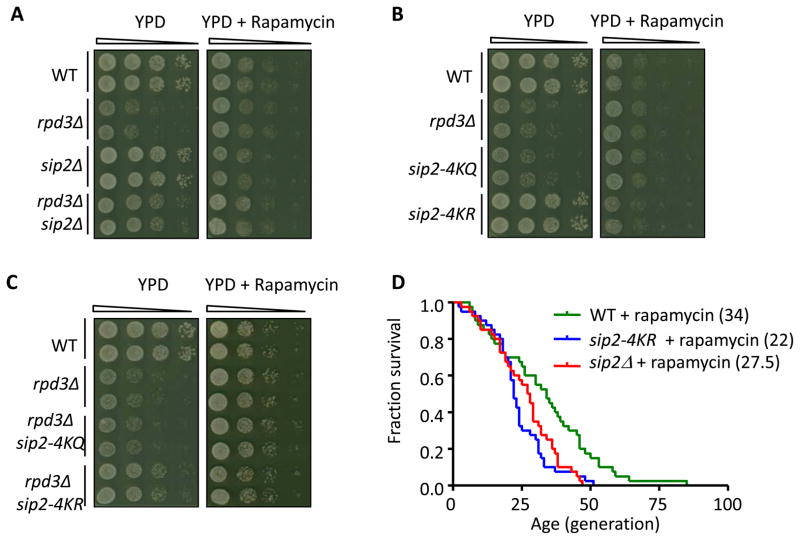
We further examined whether Sip2-Snf1 and TORC1 function in parallel upstream of Sch9 in regulating growth by comparing growth patterns of Sip2 acetylation mutants in YPD and rapamycin. Both rpd3Δ and sip2-4KQ showed growth defects in YPD, consistent with reduced Sch9 activity in these mutants. The growth differences among the mutants and WT were dampened by rapamycin treatment (Figures 6A–C), through suppressing TORC1, an essential activator of Sch9; rapamycin did not further inhibit growth of the already Sch9-suppressed rpd3Δ and sip2-4KQ. Both sip2-4KR and sip2Δ partially rescued the growth defects of rpd3Δ, possibly through Snf1 activation of the downstream Sch9. Again, the growth differences disappeared when treated by rapamycin. In contrast, the growth defects of rpd3Δ were not rescued by sip2-G2A, another short-lived mutant (Figure S6A).
Figure 6. Sch9 Is a Parallel Downstream Target of TORC1 and Sip2-Snf1.
(A–C) Comparison of growth in YPD with and without rapamycin to reveal genetic interactions between RPD3 and SIP2. Ten-fold dilutions of the indicated strains were spotted and grown on YPD plates without (2 days, 30°C) or with rapamycin (25 ng/ml, 4 days, 30°C).
(D) The lifespan of sip2-4KR and sip2Δ in rapamycin is shorter than that of WT in rapamycin (25 ng/ml).
Sip2-Snf1 and TORC1 might similarly function in parallel upstream of Sch9 to regulate replicative lifespan. We first confirmed that rapamycin increased lifespan of WT (Figure S6B) (Kaeberlein et al., 2005). We further showed that lifespan of sip2-4KR and sip2Δ, although mildly increased after rapamycin treatment (Figure S6C, D), remained significantly shorter than WT in rapamycin, suggesting the importance of Sip2 acetylation in controlling cellular lifespan independent of TORC1 activity (Figure 6D). Lifespan of sip2-4KQ and rpd3Δ were not extended by rapamycin treatment (Figure S6E), consistent with the fact that Sch9 serves as a common downstream effector. These results support a protein acetylation/phosphorylation signaling cascade distinct from the one mediated by TORC1 regulating cellular growth and lifespan through Sch9 as the common kinase effector.
Finally, we showed that SCH9 mutants of seven known PDK (phosphoinositide-dependent protein kinase)- and TORC1-dependent phosphorylation sites (Urban et al., 2007) still showed increased phosphorylation signal upon treatment with Snf1 kinase (Figure S6F), supporting the hypothesis that Snf1 catalyzes Sch9 phosphorylation at sites distinct from those used by PDK and TORC1. These results also corroborate our previous findings (Figure 3F) that glucose limitation and Sip2 acetylation seem to operate on the same effector protein in affecting replicative lifespan.
DISCUSSION
Yeast longevity studies have provided instructive models for understanding basic cellular processes in human aging (Bitterman et al., 2003). Among the conserved longevity pathways, acetyltransferase and deacetylases are well established as links between cell metabolism, growth and aging processes (Chang and Min, 2002; Nakamura et al., 2010). Similar to their opposite effects on chromatin silencing (Rundlett et al., 1996; Zhou et al., 2009), mutations in deacetylases RPD3 and SIR2 showed disparities in extending replicative lifespan previously thought to be related to distinct histone tail targets (Chang and Min, 2002). However, although histone acetylation changes do occur in aging yeast cells, specific histone acetylation site mutants, such as those of H4K16 and H3K56, do not phenocopy the effects of the corresponding acetyltransferases and deacetylases on lifespan regulation (Dang et al., 2009; Ehrentraut et al., 2010). This suggests a role for critical non-histone acetylation substrates in lifespan regulation (Close et al., 2010).
We report here an in-depth replicative lifespan analysis of a non-histone substrate, Sip2, a β-regulatory subunit of yeast AMPK (Snf1 complex) previously identified in a large scale proteomic screen of NuA4 acetyltransferase (Lin et al., 2009). Four functionally important acetylation sites were identified; one falls within the C-terminal half within a region of Sip2 previously cocrystallized in a heterotrimer with Snf1 and its γ-subunit (Amodeo et al., 2007), whereas the remaining three sites cluster in the N-terminus in a region absent from the structure (and not phylogenetically conserved). Because mutation of these three lysines is sufficient to confer most of the interaction, lifespan and other phenotypes observed in vivo, we suggest that Sip2 N-terminal acetylation might change Sip2 conformation, allowing binding to Snf1 and/or the γ-subunit to stabilize the Sip2-Snf1 complex.
NuA4 acetyltransferase, along with the corresponding Sip2 deacetylase Rpd3 are directly involved in replicative lifespan regulation. Not only did Sip2 acetylation-mimic mutants live significantly longer, they nearly totally rescued the severely shortened lifespan of NuA4 catalytic mutants. This lifespan extension validates their importance as key substrates regulating aging. Furthermore, unacetylable Sip2 mutants, like a complete deletion, led to a shortened lifespan compared to WT, further supporting the critical role of Sip2 acetylation in Sip2 function. The fact that Sip2 (de)acetylation mimetics phenocopy the corresponding catalytic mutants of NuA4 and Rpd3 strongly supports the importance of Sip2 as the critical downstream non-histone substrate of these enzymes in controlling replicative lifespan.
Sip2 is a regulatory subunit of the Snf1 complex, homologous to the AMPK complex in higher organisms which plays a critical role in metabolism and other cellular processes in response to energy supply (Scott et al., 2009). As a key regulator of energy homeostasis controlled by AMP/ATP ratios and various upstream kinases (Lee et al., 2007; Sanz, 2008), we show here that Snf1 kinase activity can be independently controlled by acetylation of Sip2. These findings help interpret the fact that Snf1, required for transcription of glucose-repressed genes (Hedbacker and Carlson, 2008), and a key player in response to cellular stress (Sanz, 2003), exhibits increased activity with aging and detrimental effects on lifespan (Lin et al., 2003). Our results suggest that Sip2 acetylation enhances physical interaction with Snf1 and thereby antagonizes catalytic activity. An increase in the disaccharide trehalose is observed as a common response to glucose limitation and replicative aging, representing two distinct stressors in yeast (Shi et al., 2010; Wang et al., 2010), Decreased Sip2 acetylation observed in these distinct stress conditions helps explain increased Snf1 activity. The antagonistic effect of Sip2 acetylation on Snf1 activity explains how Sip2 suppresses the detrimental effects of Snf1 on replicative lifespan extension (Lin et al., 2003). Hydrogen peroxide (H2O2), yet another form of stress and aging mediator, accumulates as aging occurs (Giorgio et al., 2005; Migliaccio et al., 1999). We showed here that Sip2 acetylation was critical for H2O2 resistance, supporting the hypothesis that Sip2 acetylation protects yeast cells from oxidative stress.
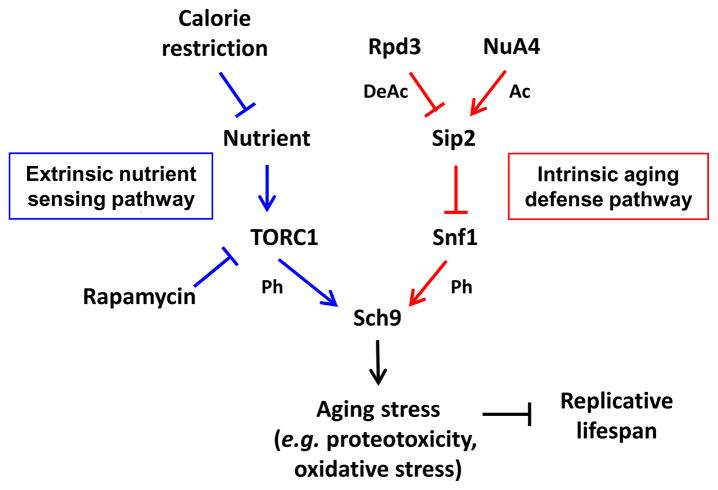
Through in vitro and in vivo kinase assays and epistasis analyses, we confirmed that Sch9, the yeast homolog of mammalian Akt/S6K (Powers, 2007), is a critical downstream effector of both TORC1 (Kaeberlein et al., 2005) and the Snf1 complex. In addition to being boosted by sensing nutrient accessibility via TORC1, we observed that Sch9 can be independently phosphorylated and activated by the Snf1 kinase. This age-dependent regulation of Snf1 kinase activity is governed by progressive deacetylation of Sip2, which releases Snf1 from Sip2 inhibition in old cells. However, it is possible that this progression is only one dimension of this regulatory pathway and that there might be dynamic changes in Sip2 acetylation state in response to certain stresses in younger cells. The Snf1-mediated phosphorylation and activation of Sch9 associated with replicative aging processes might ultimately lead to dysregulated protein homeostasis, representing a type of “intrinsic aging”. “Intrinsic aging” might represent exhaustion of a metabolite or regulatory factor, or the unavoidable accumulation of damage (proteotoxicity, oxidative, metabolic, DNA damage, etc.) during aging (Cohen and Dillin, 2008; Haigis and Yankner, 2010; Silva and Conboy, 2008; Vijg and Campisi, 2008). This “intrinsic aging” is counteracted by a series of post-translational modifications on several critical enzymes that works in parallel to an “extrinsic nutrient-sensing pathway” through common downstream machinery to coordinate vegetative growth and replicative lifespan in the yeast cells (Figure 7).
Figure 7. Model for the Effects of Sip2 Acetylation on Lifespan Regulation.
Red lines indicate the intrinsic aging defense pathway identified in this study. Blue lines indicate the extrinsic nutrient-sensing pathway regulating Sch9 activity. Not shown here for clarity is evidence of a connection between the two pathways in that we also observed a modest decrease in Sip2 acetylation in glucose limitation. Ac, lysine acetylation; DeAc, lysine deacetylation; Ph, phosphorylation of serine and threonine.
AMPK activation promotes longevity under calorie restriction in metazoans (Evans et al., 2011; Mair et al., 2011; Williams et al., 2009). The finding that AMPK in yeast seemingly plays an opposite role to that observed previously in metazoans is intriguing. However, AMPK activity has also been found to increase during senescence in various cellular models, including fibroblasts (Wang et al., 2003), skeletal muscle cells (Thomson et al., 2009), and aortic endothelial cells (Zu et al., 2010). Although transient activation of AMPK protects cells against various internal and external stresses (Narbonne and Roy, 2009), prolonged activation of AMPK is paradoxically correlated with irreversible senescence and has detrimental effects on normal physiological functions in mammalian cells (Jones et al., 2005; Marino et al., 2008; Wang et al., 2011). Also, in a normal mouse brain, AMPK activity increased with age (Liu et al., 2011). These findings on senescent mammalian cells may be related to the “end-stage” phenotype observed in old yeast mother cells.
To the best of our knowledge, this is the first study linking non-histone protein acetylation with regulation of cellular growth and replicative lifespan. This acetylation-phosphorylation signaling cascade is largely independent of calorie restriction, but could potentially integrate information of ambient nutrient availability and intracellular metabolic status through Sch9, a major downstream substrate of TORC1 kinase. Whether calorie restriction is the only means of lifespan extension remains a debated issue (Colman and Anderson, 2011). Previous reports suggest that calorie restriction might lead to unwelcome health concerns, especially in elderly and non-obese subjects (Dirks and Leeuwenburgh, 2006). Thus modulators of the “intrinsic aging” might represent more attractive pharmacologic candidates than calorie restriction mimetics for lifespan extension. Although details might differ between yeast and metazoans, we note that a similar signaling cascade potentially exists in higher organisms because all the major components are evolutionarily conserved.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
Yeast strains used in this study are derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and listed in Table S2. Integrated site-directed mutagenesis was performed as previously described (Toulmay and Schneiter, 2006) and verified by sequencing. The primers used for site-directed mutagenesis and sequencing are listed in Table S3. Refer to Extended Experimental Procedures in Supplemental Information for mass spectrometer, in vivo acetylation, enzyme purification and in vitro deacetylation.
Replicative Lifespan Determination and Statistical Analysis
Replicative lifespan was carried out as described (Kaeberlein et al., 1999). In brief, cells were freshly streaked to YPD (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) from glycerol stock and recovered at room temperature for two days, then were lightly patched on a new YPD plate and grown overnight. An appropriate number of individual cells were randomly picked under a microscope and aligned in isolated areas with a micromanipulator. After about 2h of incubation at 30°C, virgin cells (newly budded cells) were separated and left at the original site, and the mother cells were removed to the grave yard. The lifespan of these cells were determined by counting the total number of daughter cells produced. The old mother cells were defined dead when the budding process stopped and the refractility was completely lost. For lifespan analyses in low glucose (LG), cells were grown on YEP plate (1% yeast extract, 2% peptone, 2% agar) containing 0.05% dextrose. For lifespan analyses in rapamycin, cells were grown on YPD containing 25 ng/ml rapamycin. The nonparametric Mantel-Cox log-rank test was performed using GraphPad Prism (version 5.00 for Windows, GraphPad Software, San Diego California USA) to assess statistical significance of lifespan differences, as listed in Table S1.
Sorting Old Cells
Yeast old mother cells (generation 7, G7) were isolated from young cells (generation 0, G0) as described (Smeal et al., 1996). In brief, fresh cells were grown at 24°C in YPD (1% yeast extract, 2% peptone, 2% dextrose) to an A600 of ~0.6 and then 10 A600 of the cells was labeled with 7 mg freshly prepared biotin (EZ-link Sulfo-NHS-LC-LC; Pierce), cultured to 1 L YPD from A600 0.01 to A600 0.8 at 24°C in YPD, followed by affinity purification using Dynabeads® MyOne™ and DynaMag™-50 (Invitrogen) according to the manufacturer’s suggestions. An aliquot of sorted cells was stained with 0.1 mg/ml Calcofluor White (Sigma-Aldrich, F3543) and observed using a Zeiss Axioskop fluorescent microscope equipped with a Cool Snap FX camera for bud scar counting.
Trehalose Assay
A trehalose assay was performed as previously described (Shi et al., 2010). In brief, 10 A600 units of cells grown in synthetic dropout media to exactly A600 1.2 were pelleted and quickly washed with 1.2 ml ice-cold H2O and then resuspended in 0.3 ml 0.25 M Na2CO3. The samples were boiled for 4 hours and then 0.18 ml of 1 M acetic acid and 0.72 ml of 0.2 M sodium acetate was added to each sample. For controls, half (0.6 ml) of each sample was transferred to a microfuge tube and the remaining half (0.6 ml) of the sample was incubated overnight with 0.025 U/ml trehalase (Sigma-Aldrich, T8778) at 37°C. Samples were then centrifuged at top speed for 3 min and assayed for glucose using a Glucose Assay Kit (Sigma-Aldrich, GAGO20).
H2O2 Stress Tolerance Test
The test was performed as previously described (Jamieson, 1992) with slight modification. Exponential phase cultures of WT, sip2-4KR, sip2-4KQ and sip2Δ strains growing at 30°C in YPD (A600 0.25, equals to 5.4 × 106 per ml) were divided into two aliquots (duplicated) and treated for 1h at 30°C with 1.5 mM H2O2. About 200 cells were placed on YPD plates before (0 min) and after (60 min) H2O2 treatment. Number of colonies formed was counted after overnight culture at 30°C. Fraction of survival was calculated by dividing the number of colonies formed after H2O2 treatment by the number before treatment, with the fraction of survival of each strain before treatment being set as 1. Statistical significance was assessed by two-way ANOVA plus post-hoc tests. Refer to Extended Experimental Procedures in Supplemental Information for cell size determination.
In Vitro Kinase Assay
Snf1-TAP in a sip2Δ strain was purified as described (Puig et al., 2001). Kinase reactions were done in kinase buffer (25 mM HEPES pH 7.5, 10 mM MgCl2, 2 mM DTT, 0.1 mM EDTA, 0.1 mM EGTA, 50 mM KCl, with or without 1 mM ATP) for 30 min at 30°C using ~1 μg substrate and ~100 ng Snf1-TAP. Reactions were stopped by boiling in 2X SDS sample buffer and then separated by SDS-PAGE. Phosphorylation signals were assessed by Western blotting using phosphoserine antibody (Qiagen, 37430) and phosphothreonine antibody (Qiagen, 37420) antibodies following essentially the manufacturer’s manual as described (Jablonowski et al., 2004). To verify that the antibodies are phosphorylation-specific, GST-Sch9 purified from WT was treated with calf intestinal alkaline phosphatase (New England Biolabs, M0290S) following the manufacturer’s protocol, with or without adding Na3VO4 (Sigma-Aldrich, S6508), a phosphatase inhibitor. Reactions were separated by SDS–PAGE and probed with phosphoserine and phosphothreonine antibodies as described above. In vitro kinase assay using GST-Sch9 mutated at four serine or three threonine phosphorylation sites previously identified as PDK and TORC1 targets (Urban et al., 2007) was performed to test if these sites were Snf1 targets. Refer to Extended Experimental Procedures in Supplemental Information for In vivo Sch9 phosphorylation.
Supplementary Material
Acknowledgments
We thank Sheng-Ce Tao and Chien-Sheng Chen for their early contribution to this work. We are grateful to Dr. Brian K. Kennedy for providing sch9Δ strain, Dr. Robbie Loewith for providing SCH9 phosphorylation mutants and Dr. Marian B. Carlson for providing snf1-K84R construct. We thank Dr. Fang-Jen Lee for kindly sharing the laboratory space and reagents. We thank Department of Medical Science, National Taiwan University for technical help with sequencing. We thank Dr. Yi-Juang Chern for critical suggestions on this work. Work was supported by National Science Council (NSC 98-2314-B-002 -031 -MY3, to J.-Y.L.), National Taiwan University Hospital (099-001376, to J.-Y.L.), National Taiwan University (99C101-603, to J.-Y.L., Y.-Y.L. and L.-M.C.), Liver Disease Prevention & Treatment Research Foundation (to J.-Y.L. and Y.-Y.L.), Taiwan, and NIH Common Fund grant (U54-RR020839, to H.Z. and J.D.B.), U.S.A.
Footnotes
Supplemental information including Extended Experimental Procedures, three Supplemental Tables and six Figures with associated references are available with this article on line.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature. 2007;449:492–495. doi: 10.1038/nature06127. [DOI] [PubMed] [Google Scholar]
- Ashrafi K, Lin SS, Manchester JK, Gordon JI. Sip2p and its partner snf1p kinase affect aging in S. cerevisiae. Genes Dev. 2000;14:1872–1885. [PMC free article] [PubMed] [Google Scholar]
- Barker MG, Brimage LJ, Smart KA. Effect of Cu, Zn superoxide dismutase disruption mutation on replicative senescence in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;177:199–204. doi: 10.1111/j.1574-6968.1999.tb13732.x. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol Mol Biol Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CS, Pillus L. Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae. Genetics. 2009;183:149–160. doi: 10.1534/genetics.109.103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KT, Min KT. Regulation of lifespan by histone deacetylase. Ageing Res Rev. 2002;1:313–326. doi: 10.1016/s1568-1637(02)00003-x. [DOI] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Close P, Creppe C, Gillard M, Ladang A, Chapelle JP, Nguyen L, Chariot A. The emerging role of lysine acetylation of non-nuclear proteins. Cell Mol Life Sci. 2010;67:1255–1264. doi: 10.1007/s00018-009-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Dillin A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci. 2008;9:759–767. doi: 10.1038/nrn2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM. Nonhuman primate calorie restriction. Antioxid Redox Signal. 2011;14:229–239. doi: 10.1089/ars.2010.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks AJ, Leeuwenburgh C. Caloric restriction in humans: potential pitfalls and health concerns. Mech Ageing Dev. 2006;127:1–7. doi: 10.1016/j.mad.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- Ehrentraut S, Weber JM, Dybowski JN, Hoffmann D, Ehrenhofer-Murray AE. Rpd3-dependent boundary formation at telomeres by removal of Sir2 substrate. Proc Natl Acad Sci U S A. 2010;107:5522–5527. doi: 10.1073/pnas.0909169107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DS, Kapahi P, Hsueh WC, Kockel L. TOR signaling never gets old: Aging, longevity and TORC1 activity. Ageing Res Rev. 2011;10:225–237. doi: 10.1016/j.arr.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, Pelliccia G, Luzi L, Minucci S, Marcaccio M, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nature reviews Molecular cell biology. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Yankner BA. The aging stress response. Molecular cell. 2010;40:333–344. doi: 10.1016/j.molcel.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Jablonowski D, Fichtner L, Stark MJ, Schaffrath R. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol Biol Cell. 2004;15:1459–1469. doi: 10.1091/mbc.E03-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson DJ. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JC, Wawryn J, Shantha Kumara HM, Jazwinski SM. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp Gerontol. 2002;37:1023–1030. doi: 10.1016/s0531-5565(02)00064-5. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Molecular cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kim MD, Hong SP, Carlson M. Role of Tos3, a Snf1 protein kinase kinase, during growth of Saccharomyces cerevisiae on nonfermentable carbon sources. Eukaryot Cell. 2005;4:861–866. doi: 10.1128/EC.4.5.861-866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee SH, Shong M, Kim JM, Kim J, et al. Energy-dependent regulation of cell structure by AMP-activated protein kinase. Nature. 2007;447:1017–1020. doi: 10.1038/nature05828. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J Biol Chem. 2003;278:13390–13397. doi: 10.1074/jbc.M212818200. [DOI] [PubMed] [Google Scholar]
- Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, Qian J, Zhao Y, Boeke JD, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062–2074. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Benashski SE, Persky R, Xu Y, Li J, McCullough LD. Age-related changes in AMP-activated protein kinase after stroke. Age. 2011 doi: 10.1007/s11357-011-9214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia F, Wei M, Yuan V, Hu J, Gattazzo C, Pham P, Goodman MF, Longo VD. Oncogene homologue Sch9 promotes age-dependent mutations by a superoxide and Rev1/Polzeta-dependent mechanism. J Cell Biol. 2009;186:509–523. doi: 10.1083/jcb.200906011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W, Morantte I, Rodrigues AP, Manning G, Montminy M, Shaw RJ, Dillin A. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470:404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Ugalde AP, Salvador-Montoliu N, Varela I, Quiros PM, Cadinanos J, van der Pluijm I, Freije JM, Lopez-Otin C. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Human molecular genetics. 2008;17:2196–2211. doi: 10.1093/hmg/ddn120. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Kawakami K, Kametani F, Nakamoto H, Goto S. Biological significance of protein modifications in aging and calorie restriction. Ann N Y Acad Sci. 2010;1197:33–39. doi: 10.1111/j.1749-6632.2009.05374.x. [DOI] [PubMed] [Google Scholar]
- Narbonne P, Roy R. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature. 2009;457:210–214. doi: 10.1038/nature07536. [DOI] [PubMed] [Google Scholar]
- Pluskal T, Hayashi T, Saitoh S, Fujisawa A, Yanagida M. Specific biomarkers for stochastic division patterns and starvation-induced quiescence under limited glucose levels in fission yeast. Febs J. 2011;278:1299–1315. doi: 10.1111/j.1742-4658.2011.08050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. TOR signaling and S6 kinase 1: Yeast catches up. Cell Metab. 2007;6:1–2. doi: 10.1016/j.cmet.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci U S A. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem Soc Trans. 2003;31:178–181. doi: 10.1042/bst0310178. [DOI] [PubMed] [Google Scholar]
- Sanz P. AMP-activated protein kinase: structure and regulation. Curr Protein Pept Sci. 2008;9:478–492. doi: 10.2174/138920308785915254. [DOI] [PubMed] [Google Scholar]
- Schmidt MC, McCartney RR. beta-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Oakhill JS, van Denderen BJ. AMPK/SNF1 structure: a menage a trois of energy-sensing. Front Biosci. 2009;14:596–610. doi: 10.2741/3266. [DOI] [PubMed] [Google Scholar]
- Shi L, Sutter BM, Ye X, Tu BP. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol Biol Cell. 2010;21:1982–1990. doi: 10.1091/mbc.E10-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva H, Conboy IM. Aging and stem cell renewal. StemBook; Cambridge (MA): 2008. [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell. 1996;84:633–642. doi: 10.1016/s0092-8674(00)81038-7. [DOI] [PubMed] [Google Scholar]
- Thomson DM, Brown JD, Fillmore N, Ellsworth SK, Jacobs DL, Winder WW, Fick CA, Gordon SE. AMP-activated protein kinase response to contractions and treatment with the AMPK activator AICAR in young adult and old skeletal muscle. The Journal of physiology. 2009;587:2077–2086. doi: 10.1113/jphysiol.2008.166512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulmay A, Schneiter R. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast. 2006;23:825–831. doi: 10.1002/yea.1397. [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663–674. doi: 10.1016/j.molcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Vijg J, Campisi J. Puzzles, promises and a cure for ageing. Nature. 2008;454:1065–1071. doi: 10.1038/nature07216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jiang JC, Jazwinski SM. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp Gerontol. 2010;45:621–631. doi: 10.1016/j.exger.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Lopez de Silanes I, Carling D, Gorospe M. Increased AMP:ATP ratio and AMP-activated protein kinase activity during cellular senescence linked to reduced HuR function. The Journal of biological chemistry. 2003;278:27016–27023. doi: 10.1074/jbc.M300318200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liang Y, Vanhoutte PM. SIRT1 and AMPK in regulating mammalian senescence: A critical review and a working model. FEBS letters. 2011;585:986–994. doi: 10.1016/j.febslet.2010.11.047. [DOI] [PubMed] [Google Scholar]
- Williams DS, Cash A, Hamadani L, Diemer T. Oxaloacetate supplementation increases lifespan in Caenorhabditis elegans through an AMPK/FOXO-dependent pathway. Aging Cell. 2009;8:765–768. doi: 10.1111/j.1474-9726.2009.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45:279–285. doi: 10.1016/j.exger.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Zhou BO, Lenzmeier BA, Zhou JQ. Histone deacetylase Rpd3 antagonizes Sir2-dependent silent chromatin propagation. Nucleic acids research. 2009;37:3699–3713. doi: 10.1093/nar/gkp233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y, Liu L, Lee MY, Xu C, Liang Y, Man RY, Vanhoutte PM, Wang Y. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circulation research. 2010;106:1384–1393. doi: 10.1161/CIRCRESAHA.109.215483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.