Abstract
The epidemic of heart failure continues apace, and development of novel therapies with clinical efficacy has lagged. Now, important insights into the molecular circuitry of cardiovascular autophagy have raised the prospect that this cellular pathway of protein quality control may be a target of clinical relevance. Whereas basal levels of autophagy are required for cell survival, excessive levels – or perhaps distinct forms of autophagic flux – contribute to disease pathogenesis. Our challenge will be to distinguish mechanisms that drive adaptive versus maladaptive autophagy and to manipulate those pathways for therapeutic gain. Recent evidence suggests this may be possible. Here, we review the fundamental biology of autophagy and its role in a variety of forms of cardiovascular disease. We discuss ways in which this evolutionarily conserved catabolic mechanism can be manipulated, discuss studies presently underway in heart disease, and provide our perspective on where this exciting field may lead in the future.
Keywords: heart failure, cardiac hypertrophy, remodeling
Introduction
According to World Health Organization estimates, cardiovascular diseases are the number one cause of death globally [1], a sad ranking which is expected to persist into the future. Costs deriving from cardiovascular disease morbidity and mortality are staggering, estimated to exceed $500 billion in the US alone [1]. In light of these sobering facts, there is urgent need to identify novel mechanisms of disease pathogenesis and therapeutic targets capable of stemming inexorable progression of disease.
For three decades, it has been recognized that lysosomal pathways of protein degradation are prevalent in most forms of heart disease [2]. Until recently, however, it has been difficult to discern the role(s) of these catabolic pathways: whether they promote or antagonize disease pathogenesis. Now, based on molecular discoveries in yeast, a model has emerged of an intricate cascade of events leading to cargo sequestration and delivery to lysosomes. This process, termed autophagy, is an evolutionarily conserved mechanism of protein and organelle catabolism present within all eukaryotic cells [3, 4]. To date, 32 autophagy-related (ATG) genes have been identified which regulate autophagosome processing, including a subset termed the “core autophagy machinery” essential for autophagosome formation [5]. Now, armed with specifics regarding the molecular anatomy of the autophagic machinery, it is becoming possible to determine the role(s) of autophagy in numerous pathological processes [6], including cardiovascular disease [7, 8].
Autophagy, cellular cannibalization
Autophagy is a catabolic process whereby cells respond to energy stress by recycling intracellular components: proteins, ribosomes, lipids, and even entire organelles. [9]. In the presence of ample nutrient supply, anabolic reactions predominate within the cell, and autophagy is maintained at low levels critical for normal cellular homeostasis and survival [10, 11]. For example, basal levels of autophagic flux are required to degrade long-lived proteins, lipid droplets, and dysfunctional organelles, particularly in post-mitotic cells (e.g. cardiomyocytes, neurons), where the capacity for regeneration is limited. In response to starvation, induced either by inadequate nutrient supply or by defects in growth factor signaling pathways, autophagy is rapidly activated. The result is engulfment and degradation of portions of the cytoplasm.
Products derived from autophagic breakdown play a dual role: provision of biosynthetic substrates and replenishment of intracellular energy. With respect to the latter, the major contribution of autophagy to ATP pools derives from degradation of amino acids and fatty acids [12, 13]. This ATP is then used to build new macromolecules and to support cellular processes, including autophagy itself [14]. In addition, autophagic breakdown of RNA yields nucleosides, which are then degraded to ribose-phosphate. Six ribose-phosphate molecules are energetically equivalent to five glucose-phosphates and can yield ATP either aerobically or anaerobically [12, 13].
Apart from conditions of nutrient scarcity, enhanced levels of autophagy are observed in other clinically important circumstances, including neurodegenerative disorders, cancer, misfolded protein accumulation, microbial invasion, and cardiovascular diseases [7–9]. When these cells are exposed to stress, such as starvation and hypoxia, autophagic mechanisms are triggered to liberate energy substrates and eliminate defective organelles. However, taken too far, excessive and uncontrolled autophagic activation leads to depletion of essential molecules and organelles, provoking autophagic cell death [7, 8, 15, 16].
Three distinct types of autophagy have been described: microautophagy, chaperone-mediated autophagy, and macroautophagy [10]. Microautophagy refers to a process of engulfment of cytosolic materials directly into lysosomes. In chaperone-mediated autophagy, misfolded proteins are translocated by heat shock protein 70 (Hsp 70) to the lysosome for clearance. Macroautophagy (hereafter termed autophagy) is the major pathway to degrade and recycle long-lived proteins and the exclusive means of clearing dysfunctional organelles.
Whereas we have known for years that lysosomal pathways participate in the pathogenesis of virtually all forms of heart disease, it is only recently that we are equipped with the knowledge and tools to manipulate and dissect the autophagic apparatus. With this capability in hand, it has become apparent that autophagy in the heart can confer both adaptive and maladaptive actions depending on the context. Now, much work is underway to tease apart “good” autophagy from “bad” autophagy and to define underlying mechanisms. Together, these studies raise the exciting prospect of targeting and titrating the autophagic response in cardiac myocytes to effect therapeutic gain.
Molecular anatomy of autophagy
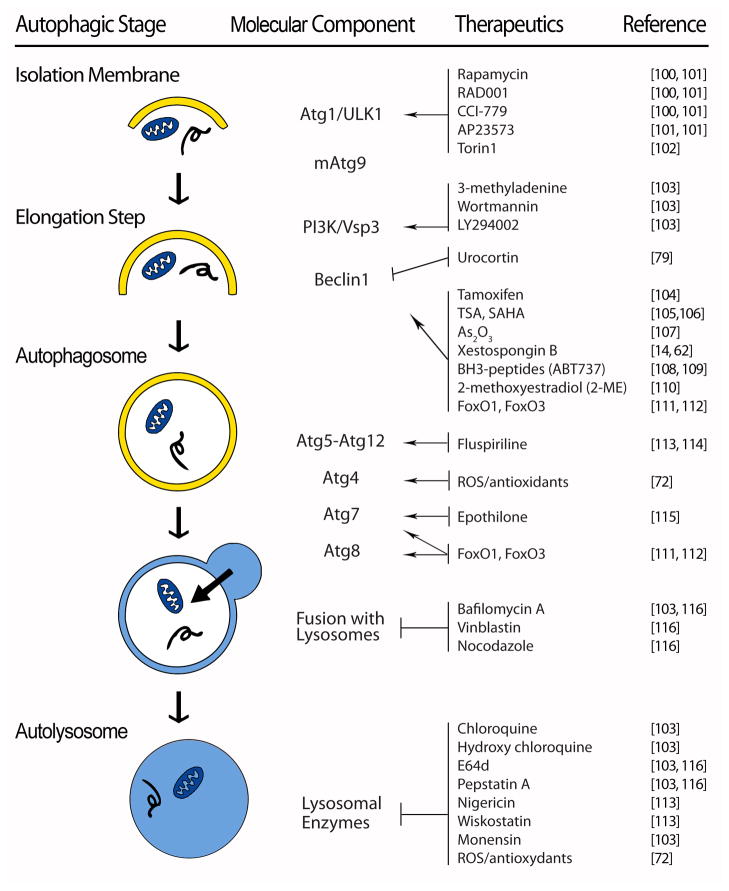
In overview, autophagy is a mechanism whereby cytoplasmic components are sequestered in a double-membrane vesicle (autophagosome) for delivery to the lysosome for breakdown [17]. However, the details of the process itself are quite intricate, involving membrane dynamics, vesicle trafficking, and cargo degradation. The autophagic cascade has been divided into distinct stages, viz. induction, cargo recognition and selection, vesicle formation, autophagosome-vacuole fusion, cargo breakdown and release of degradation products, and termination (Figure 1) [18–20]. The molecular architecture of the autophagic pathway comprises four subgroups:
Figure 1. Therapeutic manipulation of core autophagic machinery.
Therapeutic agents that target specific molecular components of the core autophagic machinery. Abbreviations: TSA – Trichostatin A; SAHA – Suberoylanilide hydroxamic acid; ROS – reactive oxygen species
Atg1 kinase complex
This signaling complex governs early steps in autophagosome formation and is regulated by nutrient availability via mTOR (TORC) [21]. In yeast, Tor integrates information from multiple upstream signaling pathways to negatively regulate Atg1 [22, 23]. In the setting of Tor suppression, either pharmacological (e.g. rapamycin) or as a result of nutrient scarcity, Atg1 kinase activity is triggered. As a consequence, the affinity of Atg1 for both Atg13 and Atg17 increases, promoting the formation of a trimeric, Atg1/Atg13/Atg17 complex. This, in turn, leads to recruitment of other Atg proteins to initiate autophagosome formation. (ULK1/2 is the mammalian orthologue of yeast Atg1 [24, 25].)
In mammalian cells exposed to nutrient-rich conditions, mTOR phosphorylates and inactivates ULKs [26, 27]. Upon starvation or rapamycin treatment, ULK1 and ULK2 are activated and phosphorylate Atg13 and FIP200, which are essential for the induction of autophagic flux [27].
mAtg9 signaling pathway
Double-membrane autophagosomes are assembled at the preautophagosomal structure (PAS) by addition of new membrane material. Then, the phagophore enlarges by Atg9-dependent delivery of new membrane [22]. Atg9 shuttles between the PAS and multiple sites within the cell, acting as a carrier to transfer membrane from the donor sites to the expanding phagophore [22, 28]. Anterograde movement of Atg9 to the PAS is governed by Atg11, Atg23, and Atg27; conversely, Atg1-Atg13, Atg2-Atg18, and Atg14 are involved in retrograde movement of Atg9 back to peripheral sites. Atg9 co-localizes with the Atg2-Atg18 complex at the PAS to promote retrieval of Atg9 from that site [9, 22, 23].
Class III phosphatidylinositol-3-kinase (PI3K)/Vps34 complex
Multiple Atg proteins are recruited to the phagophore to participate in autophagosome formation. Among them, Atg18, Atg20, Atg21, and Atg24 are recruited to the PAS via binding to phosphatidylinositol-3-phosphate (PIP) generated by Vps34 (PI3K in mammalian cells) [9, 22, 23]. Vps34 forms two distinct PI3K complexes: complex I (Vps34, Vps15/p150, Atg6/beclin 1, mAtg14) and complex II (Vps34, Vps15, Atg6, and Vps38). PI3K complex I activity (PI3P generation) is required for the targeting of several Atg proteins, such as Atg18, to the PAS. The autophagy-promoting activity of Beclin 1 is inhibited by Bcl-2 under nutrient-rich conditions; dissociation of Beclin 1 from Bcl-2 is required for the induction of autophagy [9].
Two ubiquitin-like protein conjugation systems
Two cascades with features similar to the ubiquitin-conjugation cascade contribute to phagophore expansion and formation of the autophagosome. Atg12 is activated by a ubiquitin E1-like enzyme, Atg7, and subsequently transferred to a ubiquitin E2-like enzyme, Atg10. Atg12 is next covalently conjugated to Atg5, and the resulting Atg5-Atg12 complex interacts with Atg16. In the other ubiquitin-like cascade, LC3 (mammalian homolog of Atg8) is cleaved by Atg4 to expose a carboxyl terminal glycine. This processed LC3-I is then activated by Atg7, an E1-like enzyme. After being transferred by the E2-like enzyme Atg3, LC3-I is cleaved, covalently linked to a phosphatidylethanolamine molecule, and localized to the phagophore membrane. This cleaved/lipidated isoform, termed LC3-II, migrates faster than LC3-I on SDS-PAGE, and its levels correlate with autophagosome abundance. Thus, the autophagic flux pathway is governed by interlacing circuitry comprising two kinase systems (Atg1-Atg13, class III PI3K), two ubiquitin-like systems (Atg5-Atg12, LC3-II-PE), and a retrieval/maturation system.
Lysosomes
Expansion of the phagophore culminates in self-sealing to complete autophagosome formation; this structure, in turn, fuses with a lysosome to form an autolysosome. This process is mediated by the same machinery that is involved in homotypic vacuole membrane fusion. In mammalian cells, autophagosome-lysosome fusion requires the lysosomal membrane protein LAMP-2 and the small GTPase Rab7 [22]. After fusion, degradation of the inner vesicle, along with its contents, is dependent on a series of lysosomal/vacuolar acid hydrolases, including cathepsin B, D, and L [29]. The small molecule end-products of degradation, including amino acids, sugars, and nucleotides, are released to the cytosol through permeases. However prior to this step, fusion of the autophagosome with early and late endosomes lowers intra-vesicular pH.
Inactivation of the LAMP-2 gene is the causative lesion associated with Danon disease in humans [30]. In the absence of functional LAMP-2, fusion of autophagosomes with lysosomes is blocked, leading to accumulation of long-lived proteins, accretion of unprocessed autophagosomes, and consequent myopathy.
In addition to the aforementioned core Atg proteins, secretory and endocytic pathways, along with the cytoskeleton, are also required during autophagy, providing membrane substrate, facilitating autophagosome transport, and enabling clearance of degraded autophagic cargo. In light of this, both the molecular circuitry of the autophagic pathway itself and downstream lysosomes are points of potential therapeutic manipulation (Figure 1).
Signaling pathways governing autophagy
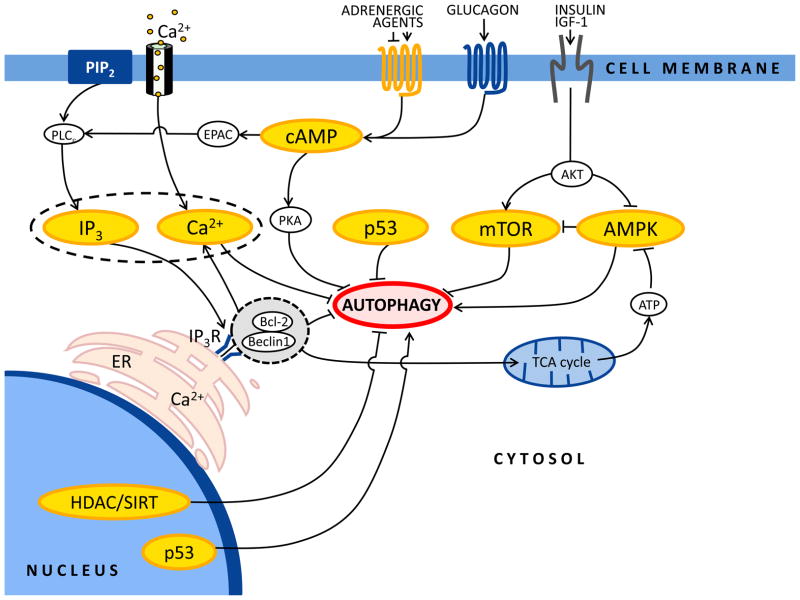
mTORC1
A central nexus of autophagy control is mTOR, a protein kinase that regulates cell growth and metabolism in response to nutrients, growth factors, ATP, and stress [31] (Figure 2). mTOR exists as two distinct multiprotein complexes, TORC1 and TORC2. TORC1 (comprising mTOR, Raptor, and mLST8) is rapamycin sensitive and mediates the temporal control of cell growth by transcription, translation, and autophagy. TORC2 (formed by mTOR, Rictor, mLST8, and Protor) is rapamycin insensitive and governs spatial control of cell growth by regulating the actin cytoskeleton. Abundance of ATP and growth factors activates TORC1 to maintain macroautophagy at low basal levels [31]. In contrast, TORC1 inhibition by nutrient starvation or rapamycin (a macrolide molecule that blocks mTOR through its interaction with FKBP12) triggers robust macroautophagy [31]. Upstream of TORC1, a regulated balance between protein phosphatases and kinases controls its activation. The pro-growth insulin/IGF-1 (insulin-like growth factor-1) pathway inhibits autophagy through activation of class I PI3Ks. This group of enzymes produces phosphatidylinositol-3,4,5-trisphosphate [PIP3], a molecular signal that activates Akt, and ultimately mTOR, thereby inhibiting autophagy [32]. Conversely, the tumor suppressor PTEN (phosphatase and tensin homologue) antagonizes the insulin/IGF-1 pathway via its PIP3 phosphatase activity and stimulates autophagy [33]. TORC1 regulatory factors are Rheb (Ras homolog enriched in brain, a protein which directly associates with TOR to promote its signaling activity) and the tuberous sclerosis complex 1 (TSC1) and TSC2 proteins (which together inhibit Rheb through the GAP activity of TSC2) [31]. As a result, the TSC-Rheb-TOR complex is a point of convergence of multiple signals that ultimately regulate autophagic activity [31].
Figure 2. Main regulatory pathways governing autophagy.
Simplified scheme of the major cellular pathways governing autophagic responses in ventricular cardiomyocytes. See text for details. Arrow denotes stimulation, and T-shaped indicators denote inhibition.
IP3 receptors
Inositol-1,4,5-trisphosphate (IP3) and its receptor (IP3R) have emerged as endogenous negative regulators of autophagy [34, 35]. IP3R plays a major role by fostering microdomains of intracellular Ca2+ accumulation to transmit specialized signals from intracellular Ca2+ stores to specific sites within the cell, such as mitochondria [36, 37]. Inhibition of the IP3R with the specific antagonist xestospongin B, or knockdown of different IP3R isoforms with small interfering RNAs, triggers robust autophagy [35, 38]. For example, xestospongin B and nutrient starvation induce autophagy by disrupting a molecular complex formed by the IP3R and Beclin 1. Recent work has shown that the IP3R at the endoplasmic reticulum provides Ca2+ to mitochondria constitutively, which in turn promotes conversion of pyruvate into acetyl-coA, tricarboxylic acid cycle activity, and production of ATP by the electron transport chain [37]. When the IP3R is not activated, energy levels drop, AMPK is activated, and autophagy is engaged to preserve energy homeostasis and cell survival [37].
AMPK
This protein is a heterotrimeric kinase comprising αβγ subunits which is a critical integrator of multiple signals in the control of energy balance [39]. In some contexts, AMPK serves as a positive regulator of autophagy mainly via inhibition of the mTOR complex [40]. AMPK is activated in response to low ATP levels through the upstream LKB1 kinase. Activated AMPK, in turn, promotes the inhibition of Rheb by the TSC1/TSC2 complex and consequent inhibition of mTORC1 activity. TSC1/TSC2 phosphorylation events mediated by AMPK and AKT have opposite effects on mTORC1 and connect mTORC1 with energy and growth factor signaling, respectively. However, AMPK can also regulate mTORC1 by an alternative mechanism, as this kinase directly phosphorylates Raptor, leading to inhibition of mTORC1 [40]. The upstream kinase CaMKKβ also phosphorylates AMPK in an AMP-independent and Ca2+-dependent manner [39]. Cytokines and increases in intracellular Ca2+ each activate AMPK and autophagy via this mechanism [40].
We have found that AMPK can also serve as an inhibitor of cardiomyocyte autophagy under conditions of ample energy supply (A.N., S.L., J.A.H. unpublished observations). Manipulations of AMPK activity in cultured neonatal rat cardiomyocytes strongly supported a link between AMPK and suppression of autophagy. For example, strong suppression of autophagy was observed when AMPK was activated by the AMPK activator, AICAR. Conversely, compound C, an inhibitor of AMPK, activated autophagy. Further, knockdown of AMPK using two sequence-independent siRNA constructs triggered robust activation of autophagy (A.N., S.L., J.A.H. unpublished observations). Clearly, the role(s) of AMPK in the governance of cardiomyocyte autophagy are complex and warrant careful scrutiny.
PKA
Cyclic adenosine 3′,5′-monophosphate (cAMP) is a universal second messenger that directs numerous physiological events following activation of G protein-coupled receptors [41]. Its primary effector is cAMP-dependent protein kinase A (PKA), a heterotetramer consisting of two regulatory and two catalytic subunits [41]. Following dissociation, the catalytic subunits phosphorylate multiple substrates to regulate a wide range of cellular processes [41]. For example, PKA governs cell growth in response to extracellular nutrients and cellular stress. In the setting of high concentrations of glucose, Ras assumes a GTP-bound active form that up-regulates cAMP concentrations with consequent PKA activation. PKA is a negative regulator of autophagy, acting primarily on Atg1, Atg8 and Atg13 [42] (Figure 2). cAMP also inhibits autophagy in a PKA-independent manner via Epac/Rap2B/phospholipase C-ε [43].
p53
Accumulating evidence indicates that the human tumor suppressor protein p53 modulates autophagy in a dual fashion, depending on its subcellular localization [44]. p53 functions as a nuclear transcription factor and can induce autophagy through transcriptional effects, including transactivation of the human Damage-Regulated Autophagy Modulator (DRAM) family of genes [44, 45]. Indeed, p53-dependent induction of autophagy occurs in response to DNA damage, Arf activation, or re-expression of p53 in p53-negative tumor cells [44]. On the other hand, cytoplasmic p53 can act as a repressor of autophagy via poorly characterized mechanisms [44]. Inhibition of p53 triggers autophagy in enucleated cells, indicating that the cytoplasmic, non-nuclear pool of p53 is capable of governing autophagy [44].
Chromatin remodeling enzymes
During pathological cardiac remodeling, both anabolic and catabolic pathways are activated, and complex cascades of protein modification and protein degradation are triggered. Among the major post-translational modifications that take place is protein acetylation, a powerful regulator of function that may rival protein phosphorylation in terms of ubiquity and importance [46]. In the case of histone proteins, acetylation of ε-lysine groups leads to relaxation of chromatin structure, enhanced accessibility to DNA-binding proteins, and consequent activation of transcription. This epigenetic mechanism is a powerful regulator of tumor responses to chemotherapy and adaptation to environmental triggers (e.g. hypoxia).
Acetylation and deacetylation reactions are controlled by the antagonistic actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs). HDACs are divided into five classes based on phylogenetic and functional criteria [47]: class I (HDACs 1, 2, 3, 8), class IIA (HDACs 4, 5, 7, 9), class IIB (HDACs 6, 10), class III (sirtuins), and class IV (HDAC 11). Class I, II, and IV HDACs are termed “classical” HDACs and are targeted by small molecule inhibitors currently in clinical development for cancer [48–50] and which have been shown to have efficacy in animal models of heart disease [51]. Sirtuin-1 is required for the autophagic response to nutrient deprivation but not for autophagy triggered by downstream signals such as the inhibition of mTOR or p53 [52]. HDAC6 interacts with ubiquitin to modulate aggresome function and autophagy [53].
Recently, HDAC biology has been shown to be critically important in governing myocardial development, metabolism, and responses to stress [46]. Furthermore, a series of animal models, including those from our laboratory, have attributed potent cardioprotective benefits to HDAC inhibitors (HDACi) in the setting of myocardial ischemia. Other investigations by our group and others, using murine models, have demonstrated HDACs to be important mediators of the myocardial hypertrophic response to hemodynamic stress (e.g. from thoracic aortic constriction, TAC) [54]. Furthermore, we found that this pathologic left ventricular remodeling could be blunted by administration of the HDAC inhibitor Trichostatin A (TSA). The downstream effect of TSA in heart involves activation of pro-survival kinase pathways [55, 56]. At the same time, HDACi can provoke both mitochondria-mediated apoptosis and caspase-independent autophagic cell death in tumors [50, 57]. Collectively, this suggests a novel window of benefit with this class of therapies not seen with other drugs.
We have demonstrated previously that cardiomyocyte autophagy elicited by pressure-overload stress is maladaptive and hence is a potential target for therapeutic intervention [58]. Given this, combined with strong evidence of cardioprotective actions of HDACi [51, 54], we hypothesized that HDAC-dependent pathological autophagy may contribute to the disease process. We further posited that suppression of pathological autophagy by HDACi may contribute to their beneficial effects. To test this, we recently employed small molecule HDAC inhibitors in a TAC model of afterload stress. Consistent with prior observations [54], HDACi was, in fact, capable of profoundly suppressing load-induced cardiomyocyte autophagy [59]; further, we found that this autophagic response is required for much of the pathological hypertrophic growth response [59]. Our studies went on to demonstrate that blunting of autophagy with HDACi is capable of reversing pre-existing systolic dysfunction and myocyte hypertrophy, a scenario with potential clinical implications. (McKinsey et al in this issue sheds additional light on HDACi as a cardioprotective therapeutic [60].)
Based on these and other data, we suggest that HDACi has potential in the therapeutic targeting of cardiac autophagy. Further studies, however, are needed to define the actions of HDACs and HDACi in the heart. Historically, HDACs were thought to act via histone acetylation-dependent control of gene expression. Accumulating evidence, however, demonstrates that HDAC-mediated deacetylation targets exist well beyond histone proteins [60, 61]. For example, the report from the Gupta lab in this issue [62] reviews NAD-dependent class III HDACs, collectively called sirtuins, which are suggested to manifest activity at cytoplasmic protein targets. Indeed, the genomic and cytoplasmic actions of HDACs and HDACi are areas of ongoing investigation.
Autophagy in cardiovascular biology
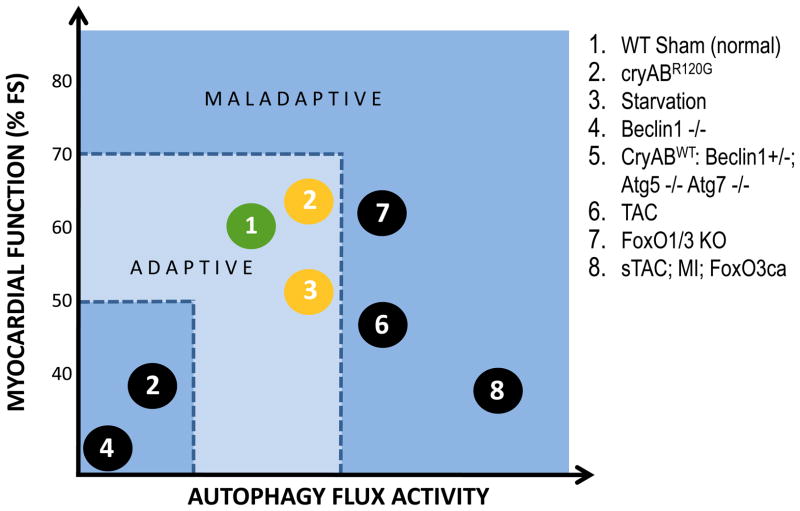
Activation of autophagic flux pathways occurs across a spectrum. At one end, low-level constitutive autophagic flux is fundamental to cell survival. At the other end of the spectrum, over-active autophagy can deplete a cell of elements required for life, thereby triggering cell death. In between these two extremes, the actions of autophagy are complex and potentially pro- or anti-survival (Figure 3).
Figure 3. Relationship between autophagic flux activity and myocardial function.
Working model of instances where changes in cardiac function (expressed as % fractional shortening, FS) were associated with alterations in autophagic flux. 1) WT – wild type; 2) cryABR120G – αB-crystallin mutant; 3) starvation – 48 h food deprivation; 4) Beclin 1 −/− knockout; 5) CryABWT – over-expresser of WT αB-crystallin, Autophagy-related gene (Atg) 5 and 7 knockout; 6) TAC – afterload stress induced in WT mice by thoracic aortic constriction; 7) forkhead box transcription factors, O FoxO1 knockout, FoxO3 knockout; 8) sTAC – severe TAC, MI – myocardial infarction induced in WT mice by ligation of left anterior descending artery. FoxO3 – constitutive over-expression of FoxO3;
Basal autophagy
Cardiomyocyte function and survival rely critically on the presence of basal levels of cardiomyocyte autophagy. Indeed, autophagic recycling of damaged cellular components in nutrient-rich conditions constitutes a major means of protein and organelle quality control, ridding the cell of defective (e.g. misfolded or oxidized) proteins and dysfunctional organelles. This fact is highlighted by the observation that abrogation of autophagic pathways in adult heart by conditional inactivation of either the Atg5 or Atg7 genes triggers rapid-onset cardiac hypertrophy, left ventricular dilation, and diminished cardiac output [63, 64]. Danon disease, a condition marked by severe and progressive myopathy, stems from defective fusion of autophagosomes with lysosomes [65, 66]. In early cardiac development, Atg5 disruption provokes in utero defects and embryonic lethality [67]. At the other end of the age spectrum, age-related declines in the efficiency of autophagic clearance likely contribute to progressive accumulation of defective proteins and organelles which ultimately lead to functional deterioration over time [64, 68]. Normal aging is associated with loss of cardiac function mainly due to impaired relaxation during diastole [69]. Varying formulations of caloric restriction (CR) can prolong life span and improve LV diastolic function; underlying mechanisms may involve induction of autophagy [70–72], possibly by reduced insulin/PI3K signaling [71, 72]. Together, these facts highlight the vital housekeeping role for cardiomyocyte autophagy as a mechanism of protein and organelle surveillance and quality control.
Afterload-induced autophagy
While the critical necessity of basal levels of autophagy is well established, the role(s) of stress-activated autophagy in cardiac disorders is more complex. As noted earlier, conditions of clear-cut pathological stress, such as nutritional paucity or hypoxia, elicit rapid increases in autophagic flux which serve to clear defective organelles and misfolded proteins and replenish scarce nutrients [9, 21, 73]. Consistent with this, suppression of autophagy during fasting reduces intracellular ATP levels and diminishes cardiac performance [67]. Beyond this, cardiac pathology elicited by multiple stressors, including elevated afterload, chronic ischemia, and ischemia/reperfusion (I/R), are associated with robust inductions of autophagy [8]. At present, a consensus is emerging in the field that induction of autophagy can either antagonize disease pathogenesis or contribute to the progression of disease depending on the context and amplitude of induction [7, 8]. For example, activation of autophagy is protective during ischemia, when the cell is “starved” of energy [74]. By contrast, activation of autophagy is maladaptive in the load-stressed heart [58] and during post-ischemic reperfusion [74] (Figure 3).
In a surgical model of pressure overload in vivo, the amplitude of autophagic flux correlates with the degree of pressure stress [58, 59]. When load stress-induced autophagy is augmented by cardiomyocyte-specific over-expression of Beclin 1, a molecular element critical to autophagic flux [75], a rapid transition to cardiac failure was observed. Conversely, diminishing the autophagic response by 50% in Beclin 1 haploinsufficient mice attenuated pathological remodeling induced by afterload stress [58]. Such dualism of autophagy can be explained in part by evidence that excessive autophagy can lead to depletion of key molecules and organelles, triggering autophagic cell death [7, 8, 16].
By contrast, cardiomyocyte autophagy is adaptive in a model of proteotoxic cardiomyopathy, where a mutation in the gene coding for the protein chaperone αB-crystallin confers a dominant-negative effect to inhibit protein folding [76]. In this context, autophagic flux is activated to rid the cell of toxic, misfolded, oxidized proteins. Importantly, these studies of proteotoxicity deriving from afterload stress or chaperone protein dysfunction were performed by manipulating the same gene, BECN1, lending additional credence to the notion that the functional differences in the actions of autophagy derive from differences in the underlying cardiomyopathic stimuli.
Autophagy in myocardial ischemia and ischemia/reperfusion
Induction of autophagic flux in response to ischemic insult has been reported in multiple systems [56, 74, 77]. Despite this, there is disagreement regarding whether the effects of autophagy in ischemia/reperfusion (I/R) injury are protective or maladaptive. For example, during mild ischemic stress, activation of cardioprotective autophagy depends on AMP-activated protein kinase (AMPK)-mediated inhibition of mammalian target of rapamycin (mTOR) [74, 78]. Pharmacological inhibition of autophagy in ischemia-mimicking conditions (e.g. glucose and oxygen withdrawal) enhances cardiomyocyte death, suggesting pro-survival effects [79]. However, when oxygen and nutrients are restored, myocyte autophagy is up-regulated dramatically in vivo (rat [80], rabbit [81], swine [82]), in cultured cell lines (H9c2 [83], HL-1[84]), and in primary cultured neonatal cardiomyocytes [74, 79]. Ischemia, where nutrient and oxygen supply to the myocardium are limited, is a state reminiscent of starvation, a context where autophagy is adaptive. Reperfusion, by contrast, is a very different situation and the associated activation of autophagic flux can be adaptive or detrimental (depending on the model system) and involves Beclin 1 independent of the AMPK/mTOR pathway [74]. Short, repetitive ischemic episodes – which elicit beneficial ischemic preconditioning effects [82, 85] – induce autophagy, as well, and when this autophagic response is suppressed, the protective effects of preconditioning are lost [82, 85].
Cardiac autophagy as a therapeutic target
Despite significant advances in cardiovascular therapeutics, both pharmacologic and device-based, the incidence of heart failure remains distressingly high [1]. A significant reason for this apparent failure is the fact that our understanding of cardiac plasticity and pathological remodeling is incomplete [86]. Recent scientific advances, however, have raised the tantalizing prospect of targeting the myocyte autophagic reaction as a novel means of achieving therapeutic gain.
In the setting of cell growth, both anabolic and catabolic processes are activated. During the initial phase, the former predominates and cell growth ensues. Ultimately, however, a new steady state emerges where levels of autophagic flux are increased. And depending on the strength of the growth stimulus – and the genetic context where autophagy is either suppressed completely, suppressed partially, or amplified – the resulting autophagic activity is either adaptive or maladaptive. Indeed, consensus is coalescing around the notion that cardiomyocyte autophagy triggered by elevations in afterload has both adaptive and maladaptive features. Complete abrogation of autophagic flux is incompatible with cell survival. Activation of autophagy in the setting of pressure stress may be beneficial up to a point, but over-activation of autophagic flux is maladaptive. At one level, this is not surprising, as the dual nature of autophagy is a recurring theme in other organ systems and disease states [87]. Indeed, we have postulated that the (patho)physiological impact of autophagy exists as a continuum, and a window of optimal autophagic activation (“adaptive” zone of autophagy) is critical to the maintenance of cellular homeostasis and function (Figure 3). Another (not mutually exclusive) model holds that different types of autophagic flux exist within the cardiomyocyte – e.g. selective versus nonspecific, mitochondria-targeting, etc. – which contribute to the differential actions of autophagy.
In any event, the prevalence of autophagic activation in the vast majority of cardiac disorders suggests the existence of a common cellular pathway which can be targeted for therapeutic gain. Pause is warranted, however, as the challenge we face will be to remain cognizant of the widespread actions of autophagy in numerous cell types. Further, we must envision tuning the autophagic response within a physiological range without abolishing it altogether. Happily, some evidence suggests this may be possible [59]. Indeed, a growing number of drugs in clinical use already or in development hold promise in this regard (Figure 1, Tables 1, 2).
Table 1.
Therapeutic manipulation of autophagy through regulatory signaling pathways.
| Signaling pathway | Therapeutics | CVS effect | Autophagy |
|---|---|---|---|
|
| |||
| IP3/Ca2+ | • IP3 receptor antagonist: xestospongins[38] | Activation | |
| • Decrease IP3 levels: carbamazepine, lithium, IMPase inhibitors[117, 118] | ND | ||
| • L-type Ca2+ channel antagonists (verapamil, nitrendipine (in vitro)[43] | |||
|
| |||
| • Myoinositol[117, 118] | ND | Inhibition | |
|
| |||
| cAMP | • Glucagon (in vivo)[119] | Yes[119] | Activation |
| • AR agonists & antagonists: adrenaline (in vivo)[119], isoproterenol & salbutamol (in vitro)[120], propranolol & carvedilol (in vivo)[121, 122] | Yes[119–122] | ||
| • Minoxidil & clonidine (in vitro)[43] | ND | ||
|
| |||
| • β-AR antagonists (in vitro)[120] | Yes[120] | Inhibition | |
|
| |||
| p53 | • Etoposide induces nuclear p53[123] | ND | Activation |
|
| |||
| • Nutilin-3, HDM2, RITA[124] | ND | Inhibition | |
|
| |||
| AMPK | • AMPK activator: Metformin (in vitro e in vivo)[125, 126] | Yes[126] | Activation |
|
| |||
| • AMPK inhibitors: Ara A (in vitro)[74], compound C (in vitro)[90], IGF-1 (in vitro & in vivo)[78] | Yes[74, 78] | Inhibition | |
| • AMPK activators: AICAR[127, 128] | ND | ||
|
| |||
| mTOR | mTOR inhibitors: | Activation | |
| • Rapamycin (in vivo)[129], torin 1[130], | Yes[129] | ||
| • Perhexiline, amiodarone, niclosamide (in vitro)[131] | |||
|
| |||
| Akt activators | ND | Inhibition | |
| • IGF-1 (in vitro)[132] | |||
|
| |||
| Chromatin remodeling | • HDAC inhibitors: Trichostatin A (TSA)[59], suberoylanilide hydroxamic acid (SAHA)[133], valproic acid[134], OSU-HDAC42[135], Curcumin[136], ATRA[137], spermidine[138] | ND | Activation |
| • Sirtuin activator: Resveratrol[138, 139] | |||
|
| |||
| • HDAC inhibitors: TSA[59] | Yes[59] | Inhibition | |
AR = adrenergic receptor; HAT = histone acetyltransferases; HDAC = histone deacetylase; ND = not determined
Table 2.
Autophagy-related patents and putative applications in cardiovascular disease.
| Patent category | Therapeutics/Target | Remarks | Patent Number | Ref |
|---|---|---|---|---|
| Autophagy induction | 8-methylchroman-7-ol derivatives | Atherosclerosis, myocardial ischemia. | US2010173983 | [98] |
| Proteinopathy treatment | Farnesyl transferase inhibitor | Myocardial ischemia, MI, vascular hyperplasia, cardiac hypertrophy, CHF, restenosis, atherosclerosis, hypertension, angina pectoris, | US2010160372 | [97] |
| Autophagy regulation | Compounds that regulates ATG14L and Rubicon which binds Class III PI3K/Vps34- Beclin 1 complex, | Inflammatory cardiac diseases | WO2010030936 | [96] |
| Autophagy induction | Glycosylated anti-tumor ether lipids (GAEL) are small molecules that induce and/or enhance autophagy in cells | Ischemic/reperfusion injury | WO2009092170 | [95] |
| Autophagy regulation | Autophagy modulators identified by a high- throughput phenotypic screen | Reperfusion injury, ischemic cardiac disease | WO2008122038 | [94] |
| Cardiac autophagy death regulation | siRNA against ANT isoforms which selectively regulates autophagic cell death | Treating cardiac ischemia | US20060210535 | [93] |
| Autophagy regulation | A phosphorus-rapamycin analog, AP23573 | Hyperproliferative vascular diseases (restenosis; graft vascular atherosclerosis; and cardiovascular disease, cerebral vascular disease, and peripheral vascular disease, | US20040073024 | [90] |
| Anti-cardiac autophagic degeneration/death | Long-term administration of a colony-stimulating factor (G-CSF) | Ischemic cardiac failure, myocardial fibrosis, left ventricular remodeling | US2006051318 | [92] |
| Cardiac atrophy | Agent that increases the expression of the runx1 gene to prevent or decrease cardiac autophagy | Treatment of a heart condition whereby heart muscle is destroyed | US2006003959 | [91] |
Future of cardiovascular autophagy research
As noted, recent findings by our group demonstrate that, at least in the context of cardiac hypertrophy, HDACi blocks pathological cardiomyocyte autophagy and blunts hypertrophic growth [59]. Given the ever-expanding burden of cardiac pathology worldwide, these findings merit further study in patients with heart disease.
Two HDAC inhibitors, Zolinza® (vorinostat) (a hydroxamic acid derivative structurally related to TSA) and romidepsin (Istoda®) have been granted FDA approval for the treatment of cutaneous T-cell lymphoma. Meanwhile, clinical trials are underway evaluating a number of other small molecules with inhibitory actions on HDACs. Based on these facts, we are moving swiftly to a first-in-man study of HDACi in ischemic heart disease. Large animal studies of surgical I/R are presently underway, as is a safety trial in patients with stable coronary disease presenting for coronary angiography. With these data in hand, we anticipate moving quickly toward a proof-of-concept trial in patients presenting with ST-segment elevation myocardial infarction (STEMI).
Meanwhile, HDACi is being evaluated actively in the context of a number of cancers. As noted earlier, HDACi promotes tumor cell death in some instances and antagonizes it in others. Thus, in addition to HDACi trials aimed at promoting cell death, two anti-autophagic drugs, chloroquine and hydroxychloroquine, are being tested in combination with various other therapeutic agents (Figure 1). The rationale here is that tumor cell autophagy promotes chemotherapy resistance in some instances, and blocking this response may re-sensitize cancer cells to toxic effects of concomitant therapy.
Only 11 patents were found relating to autophagy in the cardiovascular field (Table 2). Gottlieb et al described the use of fluorescent cadaverine to label autophagic vesicles in cardiac myocytes [88]. Tanida et al pioneered the use of GFP-GABARAP and GFP-GATE16 transgenes as markers of autophagosomes in the heart [89]. The remaining 9 patents disclosed the use of autophagy activators or inhibitors for the treatment of several cardiovascular diseases, including myocardial ischemia, myocardial infarction, vascular hyperplasia, cardiac hypertrophy, congestive heart failure, cardiomegaly, restenosis, atherosclerosis, hypertension and angina pectoris [90–98].
Concluding remarks
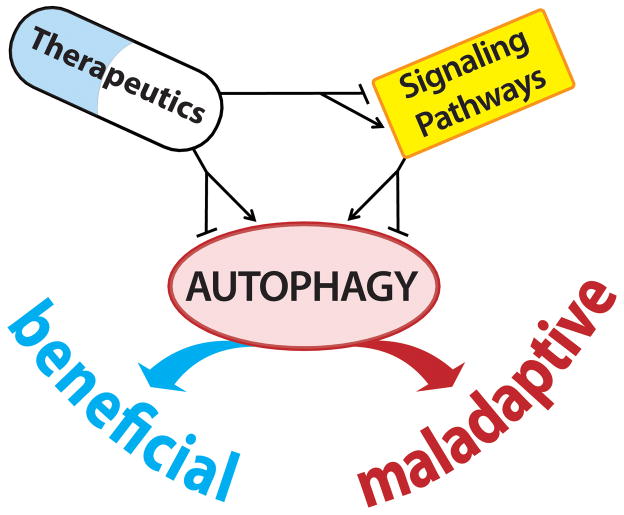
Autophagic “self-eating” is a critical pro-survival response in cardiomyocytes exposed to diverse forms of pathological insult. In this light, the growing number of cardioprotective therapies affecting autophagic activity is encouraging. For example, a recent report showed that chloramphenicol succinate activates autophagy in I/R-stressed swine heart, and this therapy was associated with benefit [99]. Looking to the future, it is possible to envision harnessing therapeutic agents already in clinical use to modulate cardiac autophagy for therapeutic benefit. This modulation of the autophagic processes can be achieved by either direct manipulation of the autophagic “core” machinery or through the exploitation of signaling pathways linked to autophagy (Figure 4). As the benefits afforded by autophagic activation depend on pathological context, vigilance for extra-cardiac effects will be critical. Further, it will be critical to titrate the stress-triggered autophagic response within an “optimal” zone, where proteostasis is promoted and yet the fundamentally important role of basal autophagy-dependent protein quality control is maintained. A comprehensive view of myocardial autophagy will be obligatory, as strategies for suppressing excessive activation of pathological pathways must always be precisely regulated to avoid disrupting homeostatic mechanisms. Major challenges remain, but patients with heart disease are likely to benefit from these efforts.
Figure 4. Therapeutic manipulation of cardiac autophagy.
Targeting cardiac autophagy using FDA-approved therapeutics could be accomplished by direct regulation of the core autophagic machinery (Figure 1) or by manipulation of key upstream regulatory pathways (Figure 2 and Table 1) (see text for details).
Acknowledgments
This work was supported by grants from the NIH (HL-075173, JAH; HL-080144, JAH; HL-090842, JAH), AHA (0640084N, JAH), ADA (7-08-MN-21-ADA, JAH), the AHA-Jon Holden DeHaan Foundation (0970518N, JAH), and the Fondo Nacional de Desarrollo Cientifico y Tecnologico, Chile (FONDECYT 1080436, SL; FONDAP 15010006, SL). SL is on a sabbatical leave at the University of Texas Southwestern Medical Center, Dallas, Texas, USA.
Footnotes
Disclosures
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, et al. Heart Disease and Stroke Statistics--2011 Update: A Report From the American Heart Association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decker RS, Poole AR, Griffin EE, Dingle JT, Wildenthal K. Altered distribution of lysosomal cathepsin D in ischemic myocardium. The Journal of clinical investigation. 1977;59:911–21. doi: 10.1172/JCI108713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klionsky DJ. The molecular machinery of autophagy and its role in physiology and disease. Semin Cell Dev Biol. 2010;21:663. doi: 10.1016/j.semcdb.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothermel BA, Hill JA. Autophagy in load-induced heart disease. Circulation research. 2008;103:1363–9. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang ZV, Rothermel BA, Hill JA. Autophagy in hypertensive heart disease. The Journal of biological chemistry. 2010;285:8509–14. doi: 10.1074/jbc.R109.025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 11.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Developmental cell. 2008;15:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabinowitz JD, White E. Autophagy and metabolism. Science (New York, NY. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loos B, Lochner A, Engelbrecht AM. Autophagy in heart disease: A strong hypothesis for an untouched metabolic reserve. Medical Hypotheses. 2011 doi: 10.1016/j.mehy.2011.03.021. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 16.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Duve C, Pressman B, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604–17. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 19.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Developmental cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–6. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–55. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 22.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 24.Chan EYW, Kir S, Tooze SA. siRNA Screening of the Kinome Identifies ULK1 as a Multidomain Modulator of Autophagy. Journal of Biological Chemistry. 2007;282:25464–74. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 25.Hara T, Takamura A, Kishi C, Iemura S-i, Natsume T, Guan J-L, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. The Journal of Cell Biology. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, et al. Nutrient-dependent mTORC1 Association with the ULK1-Atg13-FIP200 Complex Required for Autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. The Journal of Cell Biology. 2010;190:1005–22. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal Turnover, but Not a Cellular Level, of Endogenous LC3 is a Marker for Autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka Y, Guhde G, Suter A, Eskelinen E-L, Hartmann D, Lullmann-Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–6. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 31.Neufeld TP. TOR-dependent control of autophagy: biting the hand that feeds. Curr Opin Cell Biol. 2010;22:157–68. doi: 10.1016/j.ceb.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. The Journal of biological chemistry. 2000;275:992–8. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 33.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, et al. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. The Journal of biological chemistry. 2001;276:35243–6. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 34.Sarkar S, Rubinsztein DC. Inositol and IP3 levels regulate autophagy: biology and therapeutic speculations. Autophagy. 2006;2:132–4. doi: 10.4161/auto.2387. [DOI] [PubMed] [Google Scholar]
- 35.Criollo A, Vicencio JM, Tasdemir E, Maiuri MC, Lavandero S, Kroemer G. The inositol trisphosphate receptor in the control of autophagy. Autophagy. 2007;3:350–3. doi: 10.4161/auto.4077. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Zhang T, Bossuyt J, Li X, McKinsey TA, Dedman JR, et al. Local InsP3-dependent perinuclear Ca2+ signaling in cardiac myocyte excitation-transcription coupling. The Journal of clinical investigation. 2006;116:675–82. doi: 10.1172/JCI27374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–83. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vicencio JM, Ortiz C, Criollo A, Jones AW, Kepp O, Galluzzi L, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell death and differentiation. 2009;16:1006–17. doi: 10.1038/cdd.2009.34. [DOI] [PubMed] [Google Scholar]
- 39.Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–78. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 40.Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009;196:65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Enns LC, Ladiges W. Protein kinase A signaling as an anti-aging target. Ageing Research Reviews. 2010;9:269–72. doi: 10.1016/j.arr.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Graef M, Nunnari J. Mitochondria regulate autophagy by conserved signalling pathways. The EMBO journal. 2011 doi: 10.1038/emboj.2011.104. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–5. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Ryan KM. p53 and autophagy in cancer: Guardian of the genome meets guardian of the proteome. European Journal of Cancer. 2010 doi: 10.1016/j.ejca.2010.10.020. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 46.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGee-Lawrence ME, Westendorf JJ. Histone deacetylases in skeletal development and bone mass maintenance. Gene. 2011;474:1–11. doi: 10.1016/j.gene.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–68. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 49.Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. Journal of Cellular Biochemistry. 2009;107:600–8. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks PA. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin Investig Drugs. 2010;19:1049–66. doi: 10.1517/13543784.2010.510514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berry JM, Cao DJ, Rothermel BA, Hill JA. Histone deacetylase inhibition in the treatment of heart disease. Expert Opin Drug Saf. 2008;7:53–67. doi: 10.1517/14740338.7.1.53. [DOI] [PubMed] [Google Scholar]
- 52.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–8. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- 53.Simms-Waldrip T, Rodriguez-Gonzalez A, Lin T, Ikeda AK, Fu C, Sakamoto KM. The aggresome pathway as a target for therapy in hematologic malignancies. Mol Genet Metab. 2008;94:283–6. doi: 10.1016/j.ymgme.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, et al. Suppression of class I and II histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–88. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao TC, Cheng G, Zhang LX, Tseng YT, Padbury JF. Inhibition of histone deacetylases triggers pharmacologic preconditioning effects against myocardial ischemic injury. Cardiovascular research. 2007;76:473–81. doi: 10.1016/j.cardiores.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 56.Granger A, Abdullah I, Huebner F, Stout A, Wang T, Huebner T, et al. Histone deacetylase inhibition reduces myocardial ischemia-reperfusion injury in mice. The FASEB Journal. 2008;22:3549–60. doi: 10.1096/fj.08-108548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:18030–5. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–93. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–8. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKinsey TA. Isoform-selective HDAC inhibitors: Closing in on translational medicine for the heart. Journal of Molecular and Cellular Cardiology. 2011 doi: 10.1016/j.yjmcc.2010.11.009. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 61.Finkel T, Deng C-X, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–91. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sundaresan NR, Pillai VB, Gupta MP. Emerging roles of SIRT1 deacetylase in regulating cardiomyocyte survival and hypertrophy. Journal of Molecular and Cellular Cardiology. 2011 doi: 10.1016/j.yjmcc.2011.01.008. In Press, Corrected Proof. [DOI] [PMC free article] [PubMed]
- 63.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 64.Taneike M, Yamaguchi O, Nakai A, Hikoso S, Takeda T, Mizote I, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010:6. doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- 65.Maron BJ, Roberts WC, Arad M, Haas TS, Spirito P, Wright GB, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301:1253–9. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ruivo R, Anne C, Sagne C, Gasnier B. Molecular and cellular basis of lysosomal transmembrane protein dysfunction. Biochim Biophys Acta. 2009;1793:636–49. doi: 10.1016/j.bbamcr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 67.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 68.Demontis F, Perrimon N. FOXO/4E-BP Signaling in Drosophila Muscles Regulates Organism-wide Proteostasis during Aging. Cell. 2010;143:813–25. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boluyt MO, Converso K, Hwang HS, Mikkor A, Russell MW. Echocardiographic assessment of age-associated changes in systolic and diastolic function of the female F344 rat heart. J Appl Physiol. 2004;96:822–8. doi: 10.1152/japplphysiol.01026.2003. [DOI] [PubMed] [Google Scholar]
- 70.Wohlgemuth SE, Julian D, Akin DE, Fried J, Toscano K, Leeuwenburgh C, et al. Autophagy in the heart and liver during normal aging and calorie restriction. Rejuvenation research. 2007;10:281–92. doi: 10.1089/rej.2006.0535. [DOI] [PubMed] [Google Scholar]
- 71.Inuzuka Y, Okuda J, Kawashima T, Kato T, Niizuma S, Tamaki Y, et al. Suppression of phosphoinositide 3-kinase prevents cardiac aging in mice. Circulation. 2009;120:1695–703. doi: 10.1161/CIRCULATIONAHA.109.871137. [DOI] [PubMed] [Google Scholar]
- 72.Shinmura K, Tamaki K, Sano M, Murata M, Yamakawa H, Ishida H, et al. Impact of long-term caloric restriction on cardiac senescence: Caloric restriction ameliorates cardiac diastolic dysfunction associated with aging. Journal of Molecular and Cellular Cardiology. 2011;50:117–27. doi: 10.1016/j.yjmcc.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 73.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–21. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circulation research. 2007;100:914–22. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 75.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008;27 (Suppl 1):S137–48. doi: 10.1038/onc.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proceedings of the National Academy of Sciences. 2008;105:9745–50. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qian J, Ren X, Wang X, Zhang P, Jones WK, Molkentin JD, et al. Blockade of Hsp20 phosphorylation exacerbates cardiac ischemia/reperfusion injury by suppressed autophagy and increased cell death. Circulation research. 2009;105:1223–31. doi: 10.1161/CIRCRESAHA.109.200378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Troncoso R, Vicencio J, Nemchenko A, Kawashima Y, del Campo A, Parra V, et al. unpublished data. 2010 [Google Scholar]
- 79.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. Journal of Molecular and Cellular Cardiology. 2006;40:846–52. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 80.Huang C, Liu W, Perry CN, Yitzhaki S, Lee Y, Yuan H, et al. Autophagy and protein kinase C are required for cardioprotection by sulfaphenazole. American journal of physiology. 2010;298:H570–9. doi: 10.1152/ajpheart.00716.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Decker RS, Wildenthal K. Lysosomal alterations in hypoxic and reoxygenated hearts. I. Ultrastructural and cytochemical changes. The American journal of pathology. 1980;98:425–44. [PMC free article] [PubMed] [Google Scholar]
- 82.Yan L, Vatner DE, Kim S-J, Ge H, Masurekar M, Massover WH, et al. Autophagy in chronically ischemic myocardium. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13807–12. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gurusamy N, Lekli I, Gorbunov NV, Gherghiceanu M, Popescu LM, Das DK. Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. Journal of Cellular and Molecular Medicine. 2009;13:373–87. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing Macroautophagy Protects against Ischemia/Reperfusion Injury in Cardiac Myocytes. Journal of Biological Chemistry. 2006;281:29776–87. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 85.Huang C, Yitzhaki S, Perry CN, Liu W, Giricz Z, Mentzer RM, Jr, et al. Autophagy induced by ischemic preconditioning is essential for cardioprotection. J Cardiovasc Transl Res. 2010;3:365–73. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358:1370–80. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 87.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of Mammalian Autophagy in Physiology and Pathophysiology. Physiological Reviews. 2010;90:1383–435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 88.Gottlieb R, Cole T, Perry-Garza C, Carreira R, Bartlett B, Finley K. Compositions for labeling and identifying autophagosomes and methods for making and using them. 2010045270. International patent WO. 2010 Apr 22;
- 89.Tanida M, Tada N, Kinami H. Transgenic non-human animal. 2008099601. Japan patent JP. 2008 May 1;
- 90.Metcalf C, Rozamus L, Wang Y, Berstein D. Phosphorus-containing compounds and uses thereof. 20040073024. United states patent US. 2004 Apr 15;
- 91.Burden S, Littman D. Methods and agents for maintaining muscle mass and for preventing muscle atrophy and biomarkers for monitoring same. 2006003959. United States patent US. 2006 Jan 5;
- 92.Fujiwara H, Takemura G. Medical composition for treating ischemic cardiac failure. 2006051318. United States patent US. 2006 Mar 9;
- 93.Borgne A, Rebouillat D, Jacotot E. Means for regulating the expression of human isoforms of ANT. 20060210535. United States patent US. 2006 Sept 21;
- 94.Bradner JJPS, Perlstein E, Rubinsztein D, Sarkar S, Schreiber S. Regulating autophagy. 2008122038. International patent WO. 2008 Oct 9;
- 95.Arthur G, Bittman R. The use of glycosylated antitumor ether lipids to induce and/or enhance autophagy for treatment of diseases. 2009092170. International patent WO. 2009 Jul 30;
- 96.Yue Z, Zhong Y. Novel autophagy regulators ATG14L and Rubicon. 2010030936. International patent WO. 2010 Mar 18;
- 97.Lansbury P, Jr, Justman C, Grammatopoulos T, Lynch B, Liu Z. Treatment of proteinopathies using a farnesyl transferase inhibitor. 2010160372. United States patent US. 2010 Jun 24;
- 98.Brown D, Husband A, Mor G. Method for inducing autophagy. 2010173983. United States patent US. 2010 Jul 8;
- 99.Sala-Mercado JA, Wider J, Undyala VV, Jahania S, Yoo W, Mentzer RM, Jr, et al. Profound cardioprotection with chloramphenicol succinate in the swine model of myocardial ischemia-reperfusion injury. Circulation. 2010;122:S179–84. doi: 10.1161/CIRCULATIONAHA.109.928242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cecconi F. Peptides and peptidomimetics: their relevance in autophagy regulation in biomedicine. Gastroenterology and Hepatology from bed to bench. 2009;2 [Google Scholar]
- 102.Thoreen CC, Sabatini DM. Rapamycin inhibits mTORC1, but not completely. Autophagy. 2009;5:725–6. doi: 10.4161/auto.5.5.8504. [DOI] [PubMed] [Google Scholar]
- 103.Chen S, Rehman SK, Zhang W, Wen A, Yao L, Zhang J. Autophagy is a therapeutic target in anticancer drug resistance. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbcan.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 104.de Medina P, Payre B, Boubekeur N, Bertrand-Michel J, Terce F, Silvente-Poirot S, et al. Ligands of the antiestrogen-binding site induce active cell death and autophagy in human breast cancer cells through the modulation of cholesterol metabolism. Cell death and differentiation. 2009;16:1372–84. doi: 10.1038/cdd.2009.62. [DOI] [PubMed] [Google Scholar]
- 105.Park SH, Wang X, Liu R, Lam KS, Weiss RH. High throughput screening of a small molecule one-bead-one-compound combinatorial library to identify attenuators of p21 as chemotherapy sensitizers. Cancer Biol Ther. 2008;7:2015–22. doi: 10.4161/cbt.7.12.7069. [DOI] [PubMed] [Google Scholar]
- 106.Martin AP, Park MA, Mitchell C, Walker T, Rahmani M, Thorburn A, et al. BCL-2 family inhibitors enhance histone deacetylase inhibitor and sorafenib lethality via autophagy and overcome blockade of the extrinsic pathway to facilitate killing. Mol Pharmacol. 2009;76:327–41. doi: 10.1124/mol.109.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kanzawa T, Zhang L, Xiao L, Germano IM, Kondo Y, Kondo S. Arsenic trioxide induces autophagic cell death in malignant glioma cells by upregulation of mitochondrial cell death protein BNIP3. Oncogene. 2005;24:980–91. doi: 10.1038/sj.onc.1208095. [DOI] [PubMed] [Google Scholar]
- 108.Cecconi F. Peptides and peptidomimetics: their relevance in autophagy regulation in biomedicine. 2009 [Google Scholar]
- 109.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy. 2010:6. doi: 10.4161/auto.6.7.12113. [DOI] [PubMed] [Google Scholar]
- 111.Sengupta A, Molkentin JD, Yutzey KE. FoxO transcription factors promote autophagy in cardiomyocytes. The Journal of biological chemistry. 2009;284:28319–31. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–75. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 113.Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19023–8. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leduc F, Thipphavong S, Matzinger F, Dennie C, Sundaresan S. Unusual presentation of a complication after pulmonary wedge resection for coccidioma. Ann Thorac Surg. 2009;88:2011–3. doi: 10.1016/j.athoracsur.2009.04.082. [DOI] [PubMed] [Google Scholar]
- 115.Shen S, Kepp O, Martins I, Vitale I, Souquere S, Castedo M, et al. Defective autophagy associated with LC3 puncta in epothilone-resistant cancer cells. Cell Cycle. 2010;9:377–83. doi: 10.4161/cc.9.2.10468. [DOI] [PubMed] [Google Scholar]
- 116.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell death and differentiation. 2007;14:1029–39. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 118.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kondomerkos DJ, Kalamidas SA, Kotoulas OB, Hann AC. Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histology and histopathology. 2005;20:689–96. doi: 10.14670/HH-20.689. [DOI] [PubMed] [Google Scholar]
- 120.Aranguiz-Urroz P, Canales J, Copaja M, Troncoso R, Vicencio JM, Carrillo C, et al. Beta(2)-adrenergic receptor regulates cardiac fibroblast autophagy and collagen degradation. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbadis.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 121.Bahro M, Pfeifer U. Short-term stimulation by propranolol and verapamil of cardiac cellular autophagy. J Mol Cell Cardiol. 1987;19:1169–78. doi: 10.1016/s0022-2828(87)80527-8. [DOI] [PubMed] [Google Scholar]
- 122.Zhang JL, Lu JK, Chen D, Cai Q, Li TX, Wu LS, et al. Myocardial autophagy variation during acute myocardial infarction in rats: the effects of carvedilol. Chin Med J (Engl) 2009;122:2372–9. [PubMed] [Google Scholar]
- 123.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8204–9. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–87. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Harhaji-Trajkovic L, Vilimanovich U, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V. AMPK-mediated autophagy inhibits apoptosis in cisplatin-treated tumor cells. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2009.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sasaki H, Asanuma H, Fujita M, Takahama H, Wakeno M, Ito S, et al. Metformin prevents progression of heart failure in dogs: role of AMP-activated protein kinase. Circulation. 2009;119:2568–77. doi: 10.1161/CIRCULATIONAHA.108.798561. [DOI] [PubMed] [Google Scholar]
- 127.Viana R, Aguado C, Esteban I, Moreno D, Viollet B, Knecht E, et al. Role of AMP-activated protein kinase in autophagy and proteasome function. Biochem Biophys Res Commun. 2008;369:964–8. doi: 10.1016/j.bbrc.2008.02.126. [DOI] [PubMed] [Google Scholar]
- 128.Paiva M, Gonçalves L, Providência L, Davidson S, Yellon D, Mocanu M. Transitory Activation of AMPK at Reperfusion Protects the Ischaemic-Reperfused Rat Myocardium Against Infarction. Cardiovascular Drugs and Therapy. 2010;24:25–32. doi: 10.1007/s10557-010-6222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Buss SJ, Muenz S, Riffel JH, Malekar P, Hagenmueller M, Weiss CS, et al. Beneficial effects of Mammalian target of rapamycin inhibition on left ventricular remodeling after myocardial infarction. Journal of the American College of Cardiology. 2009;54:2435–46. doi: 10.1016/j.jacc.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 130.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Balgi AD, Fonseca BD, Donohue E, Tsang TC, Lajoie P, Proud CG, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4:e7124. doi: 10.1371/journal.pone.0007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jia G, Cheng G, Gangahar DM, Agrawal DK. Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol Cell Biol. 2006;84:448–54. doi: 10.1111/j.1440-1711.2006.01454.x. [DOI] [PubMed] [Google Scholar]
- 133.Oh M, Choi I-K, Kwon HJ. Inhibition of histone deacetylase1 induces autophagy. Biochemical and Biophysical Research Communications. 2008;369:1179–83. doi: 10.1016/j.bbrc.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 134.Fu J, Shao C-J, Chen F-R, Ng H-K, Chen Z-P. Autophagy induced by valproic acid is associated with oxidative stress in glioma cell lines. Neuro Oncol. 2009;12:328–40. doi: 10.1093/neuonc/nop005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu Y, Yang P, Shun C, Wu M, Weng J, Chen C. Autophagy potentiates the anticancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010:6. doi: 10.4161/auto.6.8.13365. [DOI] [PubMed] [Google Scholar]
- 136.Teiten M, Eifes S, Dicato M, Diederich M. Curcumin—The Paradigm of a Multi-Target Natural Compound with Applications in Cancer Prevention and Treatment. Toxins. 2010;2:128–62. doi: 10.3390/toxins2010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Isakson P, Bjørås M, Bøe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010;116:2324–31. doi: 10.1182/blood-2010-01-261040. [DOI] [PubMed] [Google Scholar]
- 138.Morselli E, Galluzzi L, Kepp O, Criollo A, Maiuri MC, Tavernarakis N, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany NY) 2009;1:961–70. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chung S, Yao H, Caito S, Hwang JW, Arunachalam G, Rahman I. Regulation of SIRT1 in cellular functions: Role of polyphenols. Arch Biochem Biophys. 2010;501:79–90. doi: 10.1016/j.abb.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]