Abstract
Our research has focused on systemic delivery of small interference RNA (siRNA) by branched peptides composed of histidine and lysine, called HK peptides. After studying several HK peptides, one four-branched peptide, H3K(+H)4b, with a predominant repeating pattern of -HHHK-, was found to be an effective carrier of siRNA. Although the unmodified H3K(+H)4b carrier of siRNA targeting an oncogene was previously shown to have promise in a tumor-bearing mouse model, we sought to develop a more effective HK carrier of siRNA in the current study. Our primary goal was to determine whether different ligand (cyclic RGD)-pegylation patterns on the H3K(+H)4b peptide affect siRNA delivery in vitro and in vivo. We compared the unmodified H3K(+H)4b with two modified H3K(+H)4b peptides for their ability to deliver siRNA in a tumor-bearing mouse model; one modified HK peptide, (RGD-PEG)4-H3K(+H)4b, had four cRGD-PEG conjugated to each molecule, while the other peptide, (RGD-PEG)-H3K(+H)4b, had one cRGD-PEG per molecule. Although the modified HK peptides by themselves did not form stable nanoplexes with siRNA, combination of a highly charged unmodified HK peptide, H2K4b, with either of the modified HK peptides did form stable siRNA nanoparticles. For in vitro experiments with MDA-MB-435 cells that expressed luciferase, the H3K(+H)4b siRNA nanoplexes targeting luciferase decreased its activity by 90% compared with negligible down-regulation by the modified H3K(+H)4b nanoplexes (P<0.01). In contrast, the two modified H3K(+H)4b siRNA nanoplexes administered intravenously were more effective than the H3K(+H)4b nanoplexes in silencing luciferase in a tumor xenograft model. The luciferase activity in tumor lysates of mice administered H3K(+H)4b, (RGD-PEG)-H3K(+H)4b, and (RGD-PEG)4-H3K(+H)4b nanoplexes decreased by 18%, 35%, and 75%, respectively. Thus, the siRNA nanoplex incorporating the highly modified peptide, (RGD-PEG)4-H3K(+H)4b, was the most effective at silencing its target in vivo (P<0.01). These studies demonstrate that selectively modified HK polymers are promising candidates for targeting oncogenes with siRNA.
Keywords: non-viral, siRNA, MDA-MB-435 cells, xenograft, systemic, mouse model
Introduction
RNAi silencing is a promising research and therapeutic strategy, utilized to down-regulate aberrant disease-causing genes and to study genes important for signal transduction pathways. The RNAi process is activated by incorporation of a 19 to 23-mer double stranded RNA fragment (siRNA) into the RISC complex 1. Together with the Argonaut 2 endonuclease that is part of the RISC complex, the siRNA recognizes and cleaves the target mRNA in a catalytic manner 2–5. Despite the potential of siRNA as a therapeutic agent, an effective delivery system for siRNA is essential to facilitate specific targeting and cellular uptake in the target tissue, particularly for systemic diseases such as cancer. Indeed, the development of an efficient delivery system for nucleic acids including siRNA has proved elusive and has been the rate-limiting step in developing siRNA-based therapeutics.
Nevertheless, there are many promising carriers for siRNA in various stages of pre-clinical and clinical trials, including synthetic polymers, aptamers, neutral and cationic liposomes, and peptides 6–14. Currently, no systemic carrier of siRNA has fully been proven effective for clinical use against cancer, but evidence points to a need for ligand-mediated tissue targeting and a preference for nanoplex forms. Moreover, it is likely that non-viral carriers will continue to evolve for the foreseeable future, with progressive improvements. To this end, our lab has focused on developing an effective in vitro and in vivo vehicle for small interference RNA. To accomplish this, we have synthesized and screened a number of histidine-lysine rich peptides (HK)15–18 testing their ability to carry and deliver siRNAto give effective gene inhibition. While lysines are essential for binding siRNA, histidines are important for buffering and may aid in the release of siRNA from endosomes. Altering the sequence of histidines and lysines within the branches of the HK polymer can affect its ability to transport siRNA within the cell. After studying several HK peptides, one 4-branched form, H3K(+H)4b, with predominant repeating patterns of -HHHK-, was determined to be an effective carrier of siRNA: H3K(+H)4b siRNA nanoplexes targeting the Raf-1 oncogene inhibited tumor growth by 60% 19.
In the current study, we sought to develop a more effective ligand targeted form of the H3K(+H)4b siRNAnanoplex, particularly for use in vivo. In order to prevent aggregation and to selectively target the tumor with the nanoparticle, the H3K(+H)4b was modified by varying the number and location of cRGD-PEG conjugates. Two modifications of H3K(+H)4b were made: one highly modified HK polymer in which an cRGD-PEG was attached to each branch (four cRGD-PEG per molecule) and the other, a more limited HK modification in which the cRGD-PEG was attached to the (Lys)3 core that generates the branched polypeptide (one cRGD-PEG per molecule). Compared to the unmodified H3K(+H)4b, both modified HK peptides when combined with an unmodified HK peptide were markedly more effective carriers of siRNA to tumors in a murine model. Moreover, the siRNA nanoplex containing the highly modified HK was the most effective in silencing the target gene in tumor xenografts.
Materials and Methods
Animals
Female athymic mice (4–8 wk old) were purchased from NCI Frederick. The experiments were done in accordance with regulations by the Institutional Animal Care and Use Committee of the University of Maryland Baltimore.
Cell line
A human malignant cell line MDA-MB-435, stably expressing firefly luciferase, was cultured in Dulbecco’s minimal essential medium (DMEM) containing 10% fetal calf serum (FCS) and 20mM glutamine.
Polymers
The branched HK polymer was synthesized on a Ranin Voyager synthesizer (PTI, Tucson, AZ) by the biopolymer core facility at the University of Maryland, as previously described. The unmodified four branched H3K(+H)4b and H2K4b polymers with a dominant repeating pattern of –HHK- and -HHHK-, respectively were synthesized as described previously 18. The four terminal branches of the HK polymers emanate from the 3-lysine core: for H3K(+H)4b, the branch sequence is KHHHKHHHKHHHHKHHHK and for H2K4b, the branch sequence is KHKHHKHHKHHKHHKHHKHK. For modified H3K(+H)4b, a cysteine was added to each of the four N-terminal branches or to the C-terminal end of the lysine core. Cyclic (c)RGD-PEG was conjugated to the HK polymer using a synthesis procedure similar to one previously described 12. Briefly, cyclic RGD peptide with the sequence c(RGDfK) (purchased from Peptides International, Louisville, KY) was coupled to 3.4Kd PEG using a heterobifunctional PEG, SCM-PEG-VS, from Nektar Therapeutics (Huntsville, AL), in dry DMSO in equimolar ratio. The cRGD-PEG conjugate was precipitated from DMSO by adding 10 fold excess of anhydrous cold ether. The resulting solid material was dried, characterized and used for coupling to H3K(+H)4b. The cRGD-PEG-VS conjugate obtained was then reacted with H3K(+H)4b consisting of cysteine at the N-terminus of each of the four branches (for RGD-PEG)4-H3K(+H)4b) or C-terminus (for RGD-PEG-H3K(+H)4b ) at pH 7.3 (HEPES buffer), to obtain the cRGD-PEG modified polypeptides. The modified H3K(+H)4b peptides were purified by dialysis and characterized by amino acid analysis. The resulting conjugates on average had one (for RGD-PEG-H3K(+H)4b) or four (for (RGD-PEG)4-H3K(+H)4b)) cRDG-PEG moieties, based on amino acid analysis of the purified conjugates.
siRNA
The siRNA duplex targeting luciferase (Luc) was as follow: sense, 5′-CUG-CAC-AAG-GCC-AUG-AAG- A-dTdT-3′; antisense, 5′-UCU-UCA-UGG-CCU-UGU-GCA-G-dTdT-3′, targeting 5′-CUG-CAC-AAG-GCC-AUG-AAG-A-3′. The 2-O-methyl (2′OMe) siRNA for the luciferase was similar to the unmodified siRNA except that the uridines in the sense strand had 2-OMe modifications. The control siRNA was siGENOME Non-Targeting siRNA #3, sense 5′-AUG-UAU-UGG-CCU-GUA-UUA-G-dTdT-3′; antisense, 5′-CUA-AUA-CAG-GCC-AAU-ACA-C-dTdT-3′ (Dharmacon, Lafayette, CO).
Gel Retardation Assay
The amount of HK polymer (modified and/or unmodified) to neutralize siRNA was determined by the gel retardation assay. Varying amounts of HK peptides were mixed with 1 μg of siRNA and incubated for 30 minutes at room temperature. Specifically, the following HK:siRNA ratios (w/w or w/w/w) were prepared in water: 1) H3K(+H)4b:siRNA (1:1; 1.5:1; 2:1; 2.2:1; 2.5:1); 2) RGD-PEG-H3K(+H)4b/H2K4b:siRNA and 3) (RGD-PEG-H3K(+H)4b)4/H2K4b:siRNA (3:0.6:1; 4:0.6:1; 6:0.6:1; 3:0.8:1; 4:0.8:1; 6:0.8:1). After the HK siRNA nanoplex was loaded onto the gel (3 % agarose), electrophoresis was carried out at a constant voltage of 75 V for 45 minutes in TBE buffer containing ethidium bromide. The siRNA band densities were then visualized under a UV transilluminator at a wavelength of 365 nm. On the basis of fluorescence (UN-SCAN-IT; Silk Scientific, Orem, UT) of siRNA that had migrated into each lane, the stability of HK siRNA nanoplex was determined. A gel retardation assay was also done on each in vivo HK siRNA preparation injected into the mice. Gel retardation assays similar to those described above were also carried out with HK nanoplexes after exposing them to various concentrations of serum to assess their stability.
Particle size and surface charge
Particle size was measured on all HK nanoplexes by dynamic light scattering (DLS) with the N4 plus particle size analyzer (Beckman Coulter Corp., Miami, FL). The zeta potential was measured by Delsa 440 SX instrument (Coulter) on HK siRNA nanoplexes that were used for in vivo studies.
HK:siRNA nanoparticle preparation
For in vitro experiments, HK siRNA nanoplexes were prepared as previously described 18, 19 by briefly mixing siRNA (1 μg) in 50 μl of OptiMEM with various amounts of HK peptides. The siRNA nanoplexes were maintained at room temperature for 30 min prior to size measurements and/or incubation with cells or serum. The ratios for HK to siRNA were based on gel retardation assays with additional ratios tested for in vitro bioluminescent assays.
For tumor xenografts experiments in vivo, mice were treated by injection in the tail vein with HK nanoplexes containing 40 μg of luciferase or control siRNA. After the polymers were mixed with the siRNA, the resulting nanoplex formed at room temperature for 40 minutes. Each mouse was then treated by i.v. injection with 300 μl of the nanoplexes. The ratios of polymers to siRNA for in vivo bioluminescent studies were based on the gel retardation and particle size measurements.
Stability of HK nanoplexes in Serum
With one microgram of siRNA in complex with the optimal amount of unmodified H3K(+H)4b or the two modified cRGD-PEG H3K(+H)4b peptides, the nanoplexes (2 μl) were incubated with increasing concentrations (0%, 10%, 50% and 75%) of mouse serum (total volume 10 μl) for 1 h at room temperature. The gel retardation assay was used to test the stability of the nanoplexes in serum.
Cellular Uptake
Cells (1 × 105) were incubated with different carriers (H3K(+H)4b, (RGD-PEG)-H3K(+H)4b/H2K4b, or (RGD-PEG)4H3K(+H)4b/H2K4b) in complex with Cy3 labeled-siRNA (IDT, Coralville, IA). Four hours later, cells were fixed in formaldehyde for 5 min; the nuclei were stained with chromatin dye Hoechst 33342 (Invitrogen, Carlsbad, CA) in PBS for 5 min. Confocal images were obtained with Zeiss LSM510 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY).
In vitro bioluminescence assays
MDA-MB-435 cells expressing luciferase were plated in a 24-well plate (0.5 ml of DMEM, 10% serum) at a density of 3 × 104 cells/well. Various peptides and peptide combinations (H3K(+H)4b, (RGD-PEG)-H3K(+H)4b, (RGD-PEG)4-H3K(+H)4b, (RGD-PEG)-H3K(+H)4b/H2K4b, and (RGD-PEG)4-H3K(+H)4b, in complex with Luc-siRNA (1 μg), were prepared as described previously, and nanoplexes were incubated with MDA-MB-435 cells for 48 h. The HK to siRNA ratios that were tested for in vitro bioluminescent experiments were as follows: H3K(+H)4b:siRNA, 2.2:1, 4:1, 6:1; (RGD-PEG)-H3K(+H)4b, 2:1, 4:1, 6:1; (RGD-PEG)4-H3K(+H)4b, 2:1, 4:1, 6:1; (RGD-PEG)-H3K(+H)4b/H2K4b, 2:0.8:1, 3:0.8:1; 6:0.8:1; (RGD-PEG)4-H3K(+H)4b/H2K4b, 2:0.8:1, 4:0.8:1, 6:0.8:1. The cells were then exposed to lysis buffer (200 μl) (Promega, Madison, WI) followed by centrifugation at 12,500 rpm for 5 min. The luciferase activities in the supernatant fraction were measured by a Turner TD 20/20 luminometer (Promega).
In vivo bioluminescence experiments
Tumor lysates: MDA-MB-435 xenografts that stably expressed luciferase were established by injecting 2 × 106 cells into midclavicular line of female nude mice (NCI Frederick). After tumor reached about 50 mm3, mice were separated into five treatment groups: untreated, H3K(+H)4b/Luc siRNA (w/w, 2.2:1), (RGD-PEG)H3K(+H)4b/H2K4b/Luc siRNA (w/w/w, 3:0.8:1), and (RGD-PEG)4H3K(+H)4b/H2K4b/Luc siRNA or control siRNA (w/w/w, 4:0.8:1). These HK nanoplexes containing 40 μg of siRNA were administered via the tail veins. Forty-eight hours after inoculation of the nanoplexes, the mice were euthanized and luciferase activity was then measured in tumor homogenates. After determining the most effective carrier, time course and 2′OMeLuc siRNA experiments were performed in a similar manner. Tumor volume was determined using the formula 1/2 × length × width2.
Bioluminescence Imaging: Protocol for preparation and inoculation of HK siRNA nanoplexes in tumor- bearing mice were similar to tumor lysate experiments except that the IVIS-200 optical imaging system (Xenogen Corp., Almeda, CA) measured tumor luciferase activity in real-time in living mice. At the time of imaging, mice were anesthetized with a 2.5% isofluorane/oxygen mixture and injected i.p. with 150 mg/kg of D-Luciferin (Caliper LifeSciences, Hopkinton, MA); photon emission was measured 15 min after the administration of luciferin. To normalize the photon emission (i.e., RLU), the region of interest at times 0 and 48 h were of equal size.
Cytokine Measurements
A multiplex bead-based assay using Luminex technology was used to measure serum levels of mouse INF-γ, TNF- α, and IL-6 as described by manufacturer (Upstate Waltham, MA). Levels of mouse INF-α were measured with an ELISA kit as described by the manufacturer (PBL Biochemical Laboratories, Piscataway, NJ). These assays were done by the cytokine core facility at the University of Maryland School of Medicine.
Statistics
Results comparing multiple groups were analyzed by one-way analysis of variance followed by Bonferoni test (Sigma Plot, 11.0). Student t-test was used, where appropriate, for pair-wise comparisons. Values shown represent means, and error bars represent standard deviation. In all cases, differences were considered to be statistically significant when the P values were <0.05.
Results
Optimization of HK-siRNA Nanoplexes
To determine appropriate HK and siRNA ratios for in vivo studies, gel retardation assays 20–22 and particle size measurements using dynamic light scattering (DLS) were used to facilitate development of siRNA nanoplexes. Moreover, we have previously found that plasmid nanoplexes formed with HK peptides that were approximately 100 to 150 nm in size were effective in transfection of tumor xenografts 23. Therefore, we examined various ratios of HK to siRNA to determine the minimal amount of HK that would form nanoplexes retaining approximately 90% or more of the siRNA in the loading well and that would result in a nanoplex with a diameter of 150 nm or less (Figure 1). For H3K(+H)4b, the lowest ratio to retard siRNA and still be associated with the smallest particle size was 2.2:1 (w/w) (N/P ratio, 2.7:1). At this ratio, about 92% of the siRNA was retarded and the size of the siRNA nanoplex was 149 ± 41 nm. In contrast to the unmodified H3K(+H)4b, both modified HK were unable to retard siRNA, even at very high ratios (i.e., 7:1, N/P). Moreover, at any ratio of the modified HK polymers to siRNA, the particle size of the siRNA nanoplex could not be determined by DLS (data not shown). As a result, we combined an unmodified H2K4b peptide that has a high lysine content and greater condensing properties with the modified H3K(+H)4b polymers to enable greater siRNAbinding. For the modified polymer with only one cRGD-PEG conjugated to its core, the optimal (RGD-PEG)-H3K(+H)4b/H2K4b/siRNA ratio was 3:0.8:1 (w/w/w; N/N/P ratio, 3:1:1); this ratio resulted in 99% retardation of the siRNA and a nanoparticle size of 147±51 nm. For the highly modified HK, the selected (RGD-PEG)4-H3K(+H)4b/H2K4b/siRNA ratio was 4:0.8:1 (w/w/w; N/N/P ratio 2.7:1:1), the minimum amount of HK polymer to produce a stable nanoplex. These selected ratios for unmodified and modified polymers to siRNA were used in subsequent experiments although additional ratios were also evaluated for in vitro gene silencing experiments. At these selected ratios, nanoplexes containing the H3K(+H)4b, (RGD-PEG)-H3K(+H)4b, and (RGD-PEG)4-H3K(+H)4b had zeta potentials of 37, 17.3, and 16.7 mV, respectively. Notably, the amount of H2K4b used in siRNA nanoplexes that also incorporated modified H3K(+H)4b was not sufficient to neutralize the negatively charged siRNA; the H2K4b:siRNA ratio of 0.8 to 1 (w:w) retained only 35% of the siRNA in the gel retardation assay (data not shown).
Fig. 1. Gel Retardation Assay.
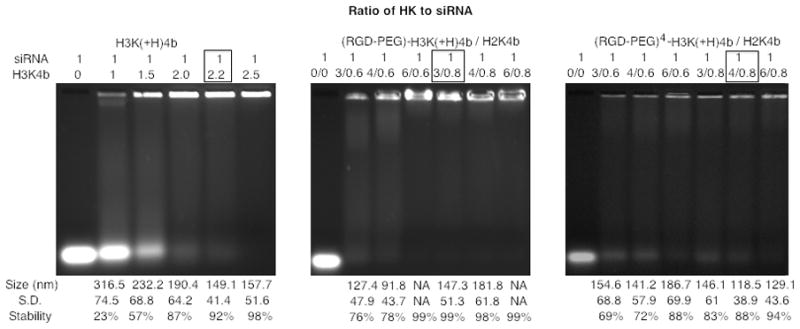
To determine the amount of HK that retards siRNA migration, various ratios of unmodified H3K(+H)4b (a) or modified HK combinations (b, (RGD-PEG)-H3K(+H)4b/H2K4b; c, (RGD-PEG)4-H3K(+H)4b/H2K4b) in complex with siRNA (1 μg) were prepared and then subjected to gel electrophoresis for 30 minutes (3% gel). Different ratios of HK polymers to siRNA are represented above the gel and the particle size is shown below the gel. For the modified HK groups (b,c), modified and unmodified polymers (H2K4b) were mixed for more complete retardation of siRNA. The ratio enclosed in the box represents the selected in vitro and in vivo silencing conditions based on the size of the particle (less than 150 nm) and siRNA retardation. NA represents nanoparticles in which the size could not be determined by dynamic light scattering.
In vitro comparison of siRNA nanoplexes with cRGD-HK conjugates
We also tested the ability of unmodified and modified HK siRNA nanoplexes to down- regulate luciferase stably expressed in MDA-MB-435 cells. Of note, MDA-MB-435 cells have elevated levels of the αvβ3 integrin, the receptor for RGD 24. The unmodified H3K(+H)4b or the two modified H3K(+H)4b with and without H2K4b were used as carriers of a luciferase-targeting siRNA in vitro. Unmodified H3K(+H)4b nanoplexes inhibited luciferase activity by approximately 90%, while the nanoplexes containing only modified HK peptides had minimal activity (Fig. 2a). In addition, the nanoplexes with the modified HK combined with H2K4b had no activity across several HK:siRNA ratios. These functional siRNA assays correlated with fluorescent uptake studies. We observed discrete intracellular particles in cells treated with unmodified H3K(+H)4b siRNA nanoplexes while we found a weak diffuse fluorescence pattern in cells treated with the siRNA control or modified HK siRNA nanoplexes (Fig. 2b). Over a range of time periods (1 to 8 hr) in which the siRNA was incubated with the cells, there was no discernable difference in the intracellular fluorescent patterns between siRNA control and the modified HK siRNA nanoplexes—that is, weak to no intracellular accumulation occurred in these groups (data not shown).
Fig. 2. In vitro comparison of HK polymers as carriers of siRNA.
(a) Several HK or combination of HK carriers (H3K(+H)4b, (RGD-PEG)H3K(+H)4b, (RGD-PEG)4H3K(+H)4b, (RGD-PEG)H3K(+H)4b/H2K4b, (RGD-PEG)4H3K(+H)4b/H2K4b) of Luc siRNA were tested for their abilities to inhibit luciferase expression in MDA-MB-435 cells compared with the untreated group. H3K(+H)4b was the most effective carrier down- regulating expression by nearly 90%, whereas inhibition by other carriers was negligible. The data represent the mean ± SD of luciferase of six determinations for each carrier. *, P < 0.01; H3K(+H)4b vs. modified HK carriers. (b) Comparison of uptake of different HK siRNA nanoplexes. A number of discrete intracellular fluorescent particles of siRNA were observed with unmodified H3K(+H)4b carriers, whereas the combination of modified and unmodified HK siRNA nanoplexes showed diffuse intracellular fluorescence- similar to the fluorescence pattern observed with the siRNA alone. Images were obtained by a Zeiss LSM510 laser scanning confocal microscope. The HK to siRNA ratios were as follows: H3K(+H)4b:siRNA, 4:1; (RGD-PEG)-H3K(+H)4b, 4:1; (RGD-PEG)4-H3K(+H)4b, 4:1; (RGD-PEG)-H3K(+H)4b/H2K4b, 3:0.8:1; (RGD-PEG)4-H3K(+H)4b/H2K4b, 4:0.8:1.
In vivo evaluation of HK polymers as carriers of Luc siRNA
Similar to the in vitro study, we screened numerous HK polymers in complex with Luc siRNA for their ability to inhibit luciferase in tumor xenografts. After MDA-MB-435 tumors expressing luciferase grew to approximately 50 mm3, the mice were divided into various treatment groups and treated by injection with the different HK Luc siRNA nanoplexes. Forty-eight hours later, the mice were euthanized and the tumor lysates were evaluated for luciferase activity. The group treated with (RGD-PEG)4H3K(+H)4b/H2K4b in complex with Luc siRNA was the most effective in silencing the tumor luciferase activity (Fig. 3), with activity reduced by about 75% (P< 0.01; (RGD-PEG)4H3K(+H)4b/H2K4b vs. other carriers and control groups). The unmodified H3K(+H)4b and (RGD-PEG)-H3K(+H)4b/H2K4b nanoplexes reduced tumor luciferase activity by 18 and 35%, respectively (P< 0.05; (RGD-PEG)-H3K(+H)4b/H2K4b vs. unmodified H3K(+H)4b or untreated). Thus, the highly modified HK nanoplex formulation was the most effective. To determine whether reduction of tumor luciferase was based on the specificity of the siRNA mechanism, a non-targeting siRNA in complex to the most effective carrier, (RGD-PEG)4-H3K(+H)4b/H2K4b, was used as the negative control in vivo. There was approximately 20% inhibition for the non-targeting siRNA nanoplex, suggesting that the Luc siRNA specifically inhibited luciferase expression.
Fig. 3. In vivo evaluation of HK polymers to determine most effective carrier for Luc-siRNA.

Four HK siRNA preparations (three HK-Luc siRNA; one HK-control siRNA) were compared for their ability to silence luciferase expression in MDA-MB-435 xenografts. The modified HK peptide, (RGD-PEG)4H3K(+H)4b/H2K4b, of Luc siRNA was the most effective, reducing luciferase expression in tumor xenografts by 75%. HK:siRNA ratios used in this experiment were based on the gel retardation assays. The data represent the mean ± SD of luciferase of six determinations for each carrier. *, P< 0.01; (RGD-PEG)4H3K(+H)4b/H2K4b carrier vs. other carriers and control groups.
We also confirmed the tumor lysate studies with the Xenogen optical imaging system and demonstrated that highly modified H3K(+H)4b nanoplexes significantly suppressed luciferase activity in tumor xenografts (Fig. 4). Mice treated with the highly modified H3K(+H)4b-H2K4b combination in complex with Luc-siRNA had decreased luciferase activity in tumors of 63.14% ± 15.18 compared to luciferase activity of tumors in untreated mice (P = 0.01, n = 3). For untreated mice, the RLU was 7.47 ±1.71×105 at time 0, and 9.43 ± 2.33 ×105 48 h later, whereas for treated mice, the RLU was 7.78 ±1.90 ×105 at time 0, and 3.08 ±0.59 ×105 at 48 h. The regions of interest at time 0 and 48 h for RLU determinations were equal in size.
Fig. 4. Bioluminescence Imaging.
Bioluminescence imaging was performed with the Xenogen IVIS 200 system. Mice bearing tumor xenografts were separated into two groups after the tumor size reached 50 mm3 and this experiment was repeated three times. (a) and (b) show tumor images of representative mice taken before treatment and 48 h after treatment, respectively. The mouse on the right in (a) and (b) was untreated; the mouse on the left of (a) and (b) was treated with (RGD-PEG)4H3K(+H)4b/H2K4b/Luc siRNA nanoplex (designated HK/Luc siRNA) by tail vein injection. The mouse treated with siRNA showed a marked down-regulation of luciferase. (c) shows the mean of the experiments comparing untreated mice with treated mice (n=3). In mice treated with luciferase siRNA, luciferase activity was reduced by 63.14 ± 15.18%. *, P = 0.01; siRNA treated vs. untreated.
Time Course of siRNA-mediated luciferase inhibition in vivo
To determine the duration of Luc silencing in the MDA-MB-435 xenografts, a time course was done with the most effective nanoplex, (RGD-PEG)4-H3K(+H)4b/H2K4b/Luc siRNA. After HK-siRNA nanoplex injection, inhibition of luciferase activity was 71% at 24 h, 75% at 48 h, and 46% at 72 h compared to untreated controls (Fig. 5). These findings suggest that the optimal time interval for dosing may be about 3 days for rapidly dividing tumor cells to obtain maximum tumor growth inhibition 6, 25, 26.
Fig. 5. Time course of siRNA induced luciferase activity reduction.

Tumor-bearing mice were injected with the optimal HK-Luc siRNA nanoplex and luciferase activity was determined from tumor lysates at 24, 48, or 72 h. Compared to activity before treatment, luciferase activity was down-regulated by 71%, 75% and 46% at 24 h, 48 h, and 72 h respectively. *, P< 0.05; 72 h vs. 24 and 48 h time point. The data for each time point represent the mean ± SD of luciferase.
HK siRNA Polyplexes do not induce cytokine response
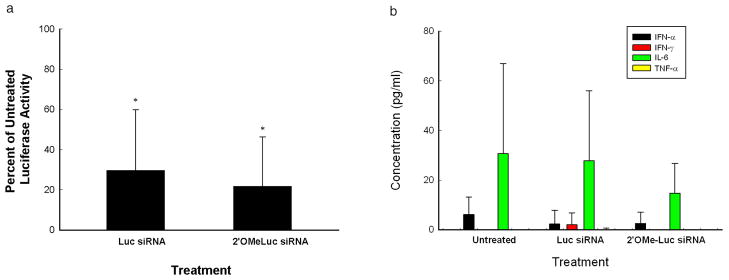
Cationic carriers in complex with siRNA can induce cytokines which in turn can result in cellular toxicity and/or inhibit gene expression in tumors by an indirect mechanism 27–30. Thus, luciferase activity may be reduced due to intra-tumoral cytokine induction and tumor cell apoptosis, non-specific mechanisms other than siRNA-mediated gene silencing. To minimize cytokine induction by the siRNA nanoplex in vivo, the sense strand of the siRNAcan be modified by cautious addition of a methyl group to the 2′-hydroxyl group of the ribose without significantly affecting the biological activity of the siRNA 31. In (RGD-PEG)4-H3K(+H)4b/H2K4b nanoplexes, a 2′O-methylated Luc siRNA was compared to unmodified Luc siRNA. If cytokine induction resulted in a non-specific cytotoxic effect from the nanoplexes, then the unmodified siRNA nanoparticle would likely be more effective at reducing luciferase activity compared to the 2′-OMe luciferase nanoparticle. As shown in Fig. 6a, however, there were no significant differences between the unmodified and 2′-OMe siRNAtumor-bearing treatment groups; the decrease in luciferase activity was 80% in the 2-OMe siRNA group vs. 75% in the unmodified siRNA group. Furthermore, the HK in complex with siRNA (unmodified or 2′-OMe modified) did not induce higher cytokine levels (IL-6, INF- α, INF-γ, and TNF-α) compared to the untreated groups (Fig. 6b). The cytokine levels in three groups were very low in comparison to DOTAP liposomal siRNA complexes, particularly for INF-α (INF-α, 754 pg/ml). Together, these data indicate that modified HK siRNA nanoplexes administered i.v. did not increase the level of cytokines in mice and cytokine induction by the HK siRNA nanoparticles did not have a significant role in decreased expression of luciferase in the tumors.
Fig. 6. Effect of induced cytokines on luciferase activity.
(a) Unmodified and 2′OMeLuc siRNA in complex with the optimal HK combination were compared for their ability to reduce luciferase activity in tumor xenografts. The reduction in luciferase activity was similar in the two groups (75% with unmodified vs. 80% with 2′OMeLuc siRNA), indicating that silencing was not due to induction of cytokines. The data represent the mean ± SD of luciferase activity of five determinations for each carrier. *, P < 0.01; siRNA treated vs. untreated. (b) Serum cytokine levels in MDA-MB-435 bearing mice were measured in untreated, optimal HK-Luc siRNA and optimal HK-2′O Me-Luc siRNA treatment groups. No differences were detected in the cytokine levels among the different treatment groups.
Nuclease Assay
The integrity of HK:siRNA polyplex may be sensitive to the serum components in vivo 32. Pegylated polymers may augment the stability of the polyplex, by reduced binding of serum components. To investigate their stability in mouse serum, HK nanoplexes were incubated in the presence of varying amounts of serum for one hour at room temperature and then analyzed by gel electrophoresis (3% agarose). As shown in Table 1, increased dissociation of siRNA was observed from all carriers as the serum concentration was increased to 50% and above. Among the nanoplexes, the unmodified H3K(+H)4b siRNA nanoplexes were less stable than the modified HK nanoplexes. There was little difference in the stability in serum between the two modified HK nanoplexes suggesting that there may be other factors (e.g., number of cRGD) that affect their ability to silence their targets in vivo.
Table 1.
Stability of HK siRNA nanoplexes in Serum
| [Serum] | 0% | 10% | 50% | 75% |
|---|---|---|---|---|
| Peptides | ||||
| H3K(+H)4b | 89.99% ± 0.77% | 87.18% ± 1.39% | 64.62% ± 3.38%* | 51.46% ± 3.20%* |
| ((RGD-PEG)H3K(+H)4b/H2K4b | 94.93% ± 0.26% | 92.89% ± 2.21% | 83.58% ± 2.46% | 73.32% ± 2.18% |
| (RGD-PEG)4H3K(+H)4b/H2K4b | 88.09% ± 3.33% | 95.12% ± 0.42% | 84.09% ± 0.43% | 75.41% ± 3.37% |
Unmodified H3K(+H)4b and modified H3K(+H)4b/H2K4b carriers in complex with siRNA were incubated at various concentrations of mouse serum (0%, 10%, 50% and 75%) for 1 hr. Gel retardation was then used to measure the stability of the nanoplexes as described in the Methods section. Compared to unmodified H3K(+H)4b siRNA nanoplexes, the two pegylated HK nanoplexes had greater stability at high serum concentration (50%, 75%);
, P < 0.01. The data represent the mean ± SD of measurement of 3 determinations for each carrier.
Discussion
To develop more effective carriers of siRNA, we compared two ligand modified HK peptides with an unmodified HK peptide. The two cRGD-PEG HK peptides differed in the number and location of cRGD-PEG conjugates on the polymer. One highly modified HK polymer with a cRGD-PEG conjugated to each branch was made that we thought would have greater tumor uptake and selectivity. Nevertheless, we were concerned that such a modification on each branch might interfere with binding to siRNA. To investigate this, an HK peptide with only one cRGD-PEG attached to its core was prepared and was expected to bind siRNA more effectively because the branching arms were not modified. Surprisingly, neither modified HK polymers by themselves effectively retarded siRNA, even at elevated N/P ratios (7:1). This data, together with the inability to determine their particle size by DLS, suggest that a stable nanoparticle was not formed.
Nevertheless, combining modified H3K(+H)4b with unmodified H2K4b when mixed with siRNA did form stable nanoparticles that could effectively silence their target in vivo. Because H2K4b alone previously was not an effective carrier of siRNAin silencing its targets18 and reduced amounts of H2K4b were used to form the nanoplex, it is likely that H2K4b has a supportive role in forming the nanoplex, but not a primary role in the siRNA nanoplex silencing role of the target. H2K4b was selected as a helper peptide because of its high content of lysines and its greater ability to condense nucleic acids compared to other HK peptides. It will be interesting to test nanoplexes using other unmodified HK peptides (i.e., H3K(+H)4b) that are significantly more effective as carriers of siRNA, even though they are less efficient in condensing nucleic acids compared with H2K4b. The data in this study are consistent with the notion that H2K4b binds tightly with siRNA and provides a central nidus to which the modified H3K(+H)4b then binds. Such a model is similar to what the results shown by Kim et al. with their carrier siRNA complexes. In that model, although the pegylated siRNA did not effectively silence its cellular target, a combination of PEI with the targeted PEGylated siRNA was effective; PEI was posited to form the central core and stabilize the siRNA nanoplex 8, 20. Although the modified and unmodified HK peptides were mixed together before addition of the siRNA, we plan in future studies to determine whether sequential addition of H2K4b first to the siRNA, followed by the modified H3K(+H)4b might increase the efficacy of the carrier.
Moreover, since both cRGD-PEG modifications of H3K(+H)4b did not form an effective or a defined siRNA nanoparticle without the addition of an unmodified HK polymer (i.e., H2K4b), it is unclear whether any PEG modifications to the H3K(+H)4b polymer will result in a stable siRNA nanoplex. If binding alone were the sole determinant, we would expect that the singly modified H3K(+H)4b would form a more stable and defined particle, but there was no evidence to support this. Consequently, there may be other factors in addition to modification of H3K(+H)4b that interferes with its binding with siRNA. For example, PEGylation of HK may interfere not only with binding, but also with the self-assembly of the nanoparticle; this may explain why unmodified HK peptides were needed to form stable nanoplexes. Of course, neither interference with binding nor nanoplex formation is mutually exclusive, but it is certainly plausible that the role of these mechanisms may vary with the different modified HK peptides. Utilizing fluorescent quenching assays, we are currently investigating whether limited and highly cRGD-PEGylated HK peptides differ in their binding to siRNA.
In contrast to most carriers in vivo that were identified from the most promising carriers in vitro, we found in this study that the most effective HK carrier in vitro was not the optimal carrier in vivo. Of the HK investigated, unmodified H3K(+H)4b was the most effective carrier in vitro, whereas the modified H3K(+H)4b/H2K4b carrier of siRNA had no observed silencing activity in cell culture experiments. In contrast, the silencing efficiency by siRNA in tumor xenografts was significantly greater with the modified HK nanoplexes in vivo and correlated with the number of cRGD-PEG per HK. Increased multivalency based on the ligand conjugated to the drug or complex is known to augment internalization 33, 34 and this provides the likely rationale for greatest activity by the highly modified H3K(+H)4b nanoplex in vivo. It is not completely clear why efficient silencing of luciferase was observed in vivo but not in vitro with the modified HK nanoplexes. Although the literature is consistent with our results that PEGylation of the nanoplex may decrease uptake in vitro and enhance accumulation of the nanoplex within the tumor tissue, addition of cell-specific ligands such as cRGD to PEG usually augments uptake of nanoplexes not only in vivo but also in vitro 12, 35–39. Indeed, previous groups have found that cRGD conjugated nanoparticles (with or without PEG) selectively targeted αvβ3 integrins on the cell surfaces of MDA-MB-435 cells in vitro and in vivo 35, 36, 40–42. Although modification of H3K(+H)4b with cRGD-PEG did not enhance uptake or silencing with siRNA in vitro, the in vivo results demonstrating down-regulation of luciferase in tumors formed by MDA-MB-435 cells suggest that modification of HK greatly augmented silencing activity of the siRNA nanoplex. This is further bolstered by our finding that similarly modified HK were effective and specific carriers of luciferase-expressing plasmids to MDA-MB-435 tumor xenografts compared to other tissues (data not shown). Although it is common to observe carriers that are ineffective in vivo that were effective in vitro, there are few other studies 43–45 demonstrating that more efficient carriers in vivo were ineffective in vitro. The importance of this observation is that it suggests that until alternative screening methods to evaluate carriers are developed for in vivo models, it is likely many other carriers that are particularly effective in vivo will never be identified.
Delivery remains the major obstacle to achieving meaningful RNAi silencing. Consequently, our primary goal in this study was to develop a more effective carrier of siRNA to tumor xenografts. Previously, we determined that unmodified H3K(+H)4b in complex with a Raf-1 siRNA inhibited tumor size by approximately 60% after five injections 19. On the basis of the current results, we anticipate that the (RGD-PEG)4H3K(+H)4b/H2K4b combination will be significantly more effective carrier than the unmodified carrier for siRNA in targeting oncogenes of tumor xenografts. Furthermore, the silencing of luciferase by the cRGD-targeted HK siRNA nanoplex appears to be specific, as evidenced by the minimal silencing by the control siRNA and low induction of cytokines by the HK siRNA nanoparticle.
Our silencing tumor xenograft model examined whether targeted HK siRNA polyplexes could effectively traversed the endothelial cell barrier to silence luciferase expressed by tumor cells. Nevertheless, it is likely that the cRGD HK siRNA polyplexes will also target the more readily accessible αvβ3-expressing endothelial cells of tumors. In addition to cyclic RGD peptides, other tumor-selective ligands could be conjugated to the HK peptides to target tumor cells and silence their oncogenes 46. Although the efficacy of cRGD-PEG targeted HK as a carrier of siRNA targeting tumors and angiogenesis requires further validation, the studies thus far have indicated that modified HK is a promising candidate for systemic delivery of siRNA.
Acknowledgments
The authors thank Dr. Pamela Talalay for her helpful suggestions and careful reading of this manuscript. This work was supported by the National Institutes of Health (R01-CA136938).
Abbreviations
- HK
generic term for histidine-lysine peptides
- H3K(+H)4b or H2K4b
two unmodified 4-branched HK polymers which differ in their lysine to histidine ratios
- cRGD
cyclic RGDfK peptide
- cRGD-PEG
cyclic peptide conjugated with polyethylene glycol
- (RGD-PEG)-H3K(+H)4b
a modified H3K(+H)4b in which one cRGD-PEG is attached to the C-terminal core
- (RGD-PEG)4-H3K(+H)4b
a modified H3K(+H)4b in which cRGD-PEG is attached to each of the four N-terminal branches
- (RGD-PEG)-H3K(+H)4b/H2K4b or (RGD-PEG)4-H3K(+H)4b/H2K4b
combination of modified and unmodified polymers
Footnotes
Conflict of Interest Statement
A. James Mixson has license agreements and/or equity with Aparna Biosciences, Sirnaomics Inc., and Silence Therapeutics
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 4.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293(5532):1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 5.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett DW, Davis ME. Impact of tumor-specific targeting and dosing schedule on tumor growth inhibition after intravenous administration of siRNA-containing nanoparticles. Biotechnol Bioeng. 2008;99(4):975–985. doi: 10.1002/bit.21668. [DOI] [PubMed] [Google Scholar]
- 7.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, et al. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol. 2009;27(9):839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. Local and systemic delivery of VEGF siRNA using polyelectrolyte complex micelles for effective treatment of cancer. J Control Release. 2008;129(2):107–116. doi: 10.1016/j.jconrel.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 9.Merritt WM, Lin YG, Spannuth WA, Fletcher MS, Kamat AA, Han LY, et al. Effect of interleukin–8 gene silencing with liposome-encapsulated small interfering RNA on ovarian cancer cell growth. J Natl Cancer Inst. 2008;100(5):359–372. doi: 10.1093/jnci/djn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat Biotechnol. 2005;23(8):1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 11.Mu P, Nagahara S, Makita N, Tarumi Y, Kadomatsu K, Takei Y. Systemic delivery of siRNA specific to tumor mediated by atelocollagen: combined therapy using siRNA targeting Bcl-xL and cisplatin against prostate cancer. Int J Cancer. 2009;125(12):2978–2990. doi: 10.1002/ijc.24382. [DOI] [PubMed] [Google Scholar]
- 12.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, et al. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32(19):e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Systemic leukocyte-directed siRNA delivery revealing cyclin D1 as an anti-inflammatory target. Science. 2008;319(5863):627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 15.Chen QR, Zhang L, Stass SA, Mixson AJ. Branched co-polymers of histidine and lysine are efficient carriers of plasmids. Nucleic Acids Res. 2001;29(6):1334–1340. doi: 10.1093/nar/29.6.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen QR, Zhang L, Luther PW, Mixson AJ. Optimal transfection with the HK polymer depends on its degree of branching and the pH of endocytic vesicles. Nucleic Acids Res. 2002;30(6):1338–1345. doi: 10.1093/nar/30.6.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leng Q, Mixson AJ. Modified branched peptides with a histidine-rich tail enhance in vitro gene transfection. Nucleic Acids Res. 2005;33(4):e40. doi: 10.1093/nar/gni040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leng Q, Scaria P, Zhu J, Ambulos N, Campbell P, Mixson AJ. Highly branched HK peptides are effective carriers of siRNA. J Gene Med. 2005;7(7):977–986. doi: 10.1002/jgm.748. [DOI] [PubMed] [Google Scholar]
- 19.Leng Q, Scaria P, Lu P, Woodle MC, Mixson AJ. Systemic delivery of HK Raf-1 siRNA polyplexes inhibits MDA-MB-435 xenografts. Cancer Gene Ther. 2008;15(8):485–495. doi: 10.1038/cgt.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SH, Jeong JH, Lee SH, Kim SW, Park TG. PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J Control Release. 2006;116(2):123–129. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 21.Huh MS, Lee SY, Park S, Lee S, Chung H, Choi Y, et al. Tumor-homing glycol chitosan/polyethylenimine nanoparticles for the systemic delivery of siRNA in tumor-bearing mice. J Control Release. 2010;144(2):134–143. doi: 10.1016/j.jconrel.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Ofek P, Fischer W, Calderon M, Haag R, Satchi-Fainaro R. In vivo delivery of small interfering RNA to tumors and their vasculature by novel dendritic nanocarriers. FASEB J. 2010;24(9):3122–3134. doi: 10.1096/fj.09-149641. [DOI] [PubMed] [Google Scholar]
- 23.Leng Q, Scaria P, Ioffe OB, Woodle M, Mixson AJ. A branched histidine/lysine peptide, H2K4b, in complex with plasmids encoding antitumor proteins inhibits tumor xenografts. J Gene Med. 2006;8(12):1407–1415. doi: 10.1002/jgm.982. [DOI] [PubMed] [Google Scholar]
- 24.Felding-Habermann B, O’Toole TE, Smith JW, Fransvea E, Ruggeri ZM, Ginsberg MH, et al. Integrin activation controls metastasis in human breast cancer. Proc Natl Acad Sci U S A. 2001;98(4):1853–1858. doi: 10.1073/pnas.98.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34(1):322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104(39):15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23(4):457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 28.Kim JY, Choung S, Lee EJ, Kim YJ, Choi YC. Immune activation by siRNA/liposome complexes in mice is sequence- independent: lack of a role for Toll-like receptor 3 signaling. Mol Cells. 2007;24(2):247–254. [PubMed] [Google Scholar]
- 29.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11(3):263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 30.Zamanian-Daryoush M, Marques JT, Gantier MP, Behlke MA, John M, Rayman P, et al. Determinants of cytokine induction by small interfering RNA in human peripheral blood mononuclear cells. J Interferon Cytokine Res. 2008;28(4):221–233. doi: 10.1089/jir.2007.0090. [DOI] [PubMed] [Google Scholar]
- 31.Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2′-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther. 2007;15(9):1663–1669. doi: 10.1038/sj.mt.6300240. [DOI] [PubMed] [Google Scholar]
- 32.Buyens K, Meyer M, Wagner E, Demeester J, De Smedt SC, Sanders NN. Monitoring the disassembly of siRNA polyplexes in serum is crucial for predicting their biological efficacy. J Control Release. 2010;141(1):38–41. doi: 10.1016/j.jconrel.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 33.Schraa AJ, Kok RJ, Berendsen AD, Moorlag HE, Bos EJ, Meijer DK, et al. Endothelial cells internalize and degrade RGD-modified proteins developed for tumor vasculature targeting. J Control Release. 2002;83(2):241–251. doi: 10.1016/s0168-3659(02)00206-7. [DOI] [PubMed] [Google Scholar]
- 34.Boturyn D, Coll JL, Garanger E, Favrot MC, Dumy P. Template assembled cyclopeptides as multimeric system for integrin targeting and endocytosis. J Am Chem Soc. 2004;126(18):5730–5739. doi: 10.1021/ja049926n. [DOI] [PubMed] [Google Scholar]
- 35.Kim WJ, Yockman JW, Lee M, Jeong JH, Kim YH, Kim SW. Soluble Flt-1 gene delivery using PEI-g-PEG-RGD conjugate for anti-angiogenesis. J Control Release. 2005;106(1–2):224–234. doi: 10.1016/j.jconrel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 36.Kim WJ, Yockman JW, Jeong JH, Christensen LV, Lee M, Kim YH, et al. Anti-angiogenic inhibition of tumor growth by systemic delivery of PEI-g-PEG-RGD/pCMV-sFlt-1 complexes in tumor-bearing mice. J Control Release. 2006;114(3):381–388. doi: 10.1016/j.jconrel.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Chen YC, Tseng YC, Mozumdar S, Huang L. Biodegradable calcium phosphate nanoparticle with lipid coating for systemic siRNA delivery. J Control Release. 2010;142(3):416–421. doi: 10.1016/j.jconrel.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3(5):579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 39.Hood JD, Bednarski M, Frausto R, Guccione S, Reisfeld RA, Xiang R, et al. Tumor regression by targeted gene delivery to the neovasculature. Science. 2002;296(5577):2404–2407. doi: 10.1126/science.1070200. [DOI] [PubMed] [Google Scholar]
- 40.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279(5349):377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 41.Lee JH, Lee K, Moon SH, Lee Y, Park TG, Cheon J. All-in-one target-cell-specific magnetic nanoparticles for simultaneous molecular imaging and siRNA delivery. Angew Chem Int Ed Engl. 2009;48(23):4174–4179. doi: 10.1002/anie.200805998. [DOI] [PubMed] [Google Scholar]
- 42.Yoshimoto M, Ogawa K, Washiyama K, Shikano N, Mori H, Amano R, et al. alpha(v)beta(3) Integrin-targeting radionuclide therapy and imaging with monomeric RGD peptide. Int J Cancer. 2008;123(3):709–715. doi: 10.1002/ijc.23575. [DOI] [PubMed] [Google Scholar]
- 43.Lee ER, Marshall J, Siegel CS, Jiang C, Yew NS, Nichols MR, et al. Detailed analysis of structures and formulations of cationic lipids for efficient gene transfer to the lung. Hum Gene Ther. 1996;7(14):1701–1717. doi: 10.1089/hum.1996.7.14-1701. [DOI] [PubMed] [Google Scholar]
- 44.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Solodin I, Brown CS, Bruno MS, Chow CY, Jang EH, Debs RJ, et al. A novel series of amphiphilic imidazolinium compounds for in vitro and in vivo gene delivery. Biochemistry. 1995;34(41):13537–13544. doi: 10.1021/bi00041a033. [DOI] [PubMed] [Google Scholar]
- 46.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;16(6):510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]