Abstract
The unique segregation of homologs, rather than sister chromatids, at the first meiotic division requires in most species the formation of crossovers between homologs by meiotic recombination. Crossovers do not form at random along chromosomes. Rather, their formation is carefully controlled, both at the stage of formation of DNA double-strand breaks (DSBs) that can initiate crossovers and during the repair of these DSBs. We review control of DSB formation and two recently recognized controls of DSB repair: crossover homeostasis and crossover invariance. Crossover homeostasis maintains a constant number of crossovers per cell when the total number of DSBs in a cell is experimentally or stochastically reduced. Crossover invariance maintains a constant crossover density (crossovers per kb of DNA) across much of the genome in spite of strong DSB hotspots in some intervals. These recently uncovered phenomena show that crossover control is even more complex than previously suspected.
Multiple controls of meiotic recombination during gamete formation
The formation of haploid cells (gametes) from diploid precursor cells during meiosis is essential to maintain a constant number of chromosomes from generation to generation in sexually reproducing species. Haploids arise in meiosis because there are two nuclear divisions but only one round of replication. The major problem is to ensure that exactly one copy of each chromosome pair is inherited by each haploid cell. This requires that homologs, or more precisely homologous centromeres, segregate from each other at the first meiotic division and that sister centromeres segregate at the second meiotic division. In most species homolog segregation requires formation of a physical connection between homologs. This connection is detected genetically as a crossover or microscopically as a chiasma (pl., chiasmata). Meiotic recombination also forms new combinations of alleles, thereby speeding the evolution of species. Thus, recombination plays a dual role in meiosis, with both immediate and long-term consequences.
Almost from the time of their discovery a century ago, meiotic crossovers and chiasmata were known to be non-randomly distributed along chromosomes. Crossovers do not occur independently: a crossover in one interval decreases the likelihood of a crossover in a nearby interval, a phenomenon called crossover interference, the first recognized control (Box 1). Crossovers are rare in and around centromeres, because their occurrence there interferes with proper chromosome segregation. Crossovers too far from the centromere (i.e., near the telomere) less effectively direct proper segregation, and in at least some species crossing over is reduced near the telomeres.
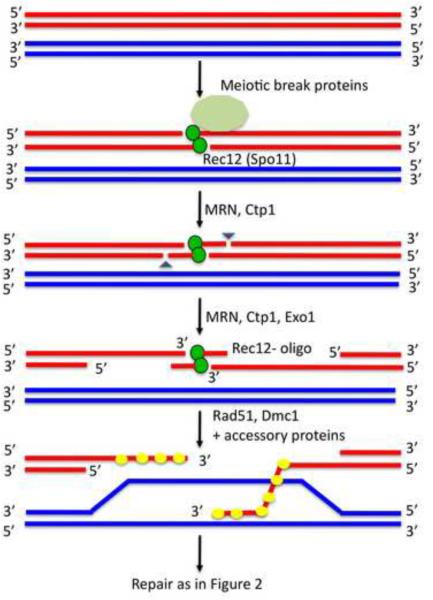
One key to understanding these controls has come from studies of the mechanism of crossing over, which is initiated by the formation of lesions in one of the interacting DNA molecules. Double-strand breaks (DSBs) in DNA can initiate crossing over in two well-studied species, the budding yeast Saccharomyces cerevisiae and the very distantly related fission yeast Schizosaccharomyces pombe, although other lesions, such as single-strand breaks (nicks), have not been excluded (Box 2). DSBs are made by a meiosis-specific topoisomerase-like protein Spo11 (called Rec12 in S. pombe, Table 1) in conjunction with several “meiotic break proteins”, which, like Spo11, are essential for both DSB formation and meiotic recombination (Figure 1) [1, 2]. A Spo11 ortholog appears to be present in all species that undergo meiosis, making it likely that DSBs are important for meiotic recombination in all species.
Table 1.
Proteins involved in meiotic recombination.
| Protein | Function |
|---|---|
| Bas1, Bas2, Rap1 | S. cerevisiae transcription factors that regulate meiotic recombination at some loci. |
| Brc-1 | C. elegans DSB repair and recombination protein, homologous to mammalian BRCA1. |
| Ctp1 | S. pombe protein involved in Rec12-oligonucleotide removal; called Sae2 in S. cerevisiae. |
| Clr4 | S. pombe histone H3 methyltransferase specific for lysine 9. Methylated histone H3-K9 is associated with transcriptionally repressed (heterochromatic) regions. |
| Dmc1 | Meiosis-specific DNA strand-exchange protein; paralog of Rad51. |
| Dpy-28 | C. elegans condensin I complex protein that regulates crossover distribution in meiotic cells. |
| Exo1 | 5' to 3' exonuclease and flap endonuclease involved in DSB resection. |
| Hop1 | S. cerevisiae protein present in axial elements; localization depends on Red1. |
| Him-17 | C. elegans chromatin-associated protein |
| Hsk1 | S. pombe protein kinase needed for meiotic DSB formation. |
| Kle-2 | C. elegans condensin II complex protein that regulates crossover distribution in meiotic cells. |
| Mek1 | S. cerevisiae meiosis-specific protein kinase that functions with Red1 and Hop1. |
| Mer2 | S. cerevisiae protein required for DSB formation; a “meiotic break protein” in Figure 1. |
| MRN | S. pombe Mre11, Rad50, Nbs1 proteins involved in DSB repair. Known as MRX in budding yeast where Xrs2 replaces Nbs1. |
| Pch2 | S. cerevisiae meiotic checkpoint protein that inhibits chromosome segregation when meiotic recombination is delayed or aberrant. |
| Rad51 | DNA strand-exchange protein required for recombination; ortholog of bacterial RecA. |
| Rad54 | ATP-dependent chromatin remodeling factor that stimulates DNA strand-exchange during recombination. |
| Rec8 | Meiosis-specific subunits of sister chromatid cohesin; S. pombe Red 11 is another meiosis-specific subunit. |
| Rec25 | S. pombe linear element protein; functions with Rec10 and Rec27. |
| Red1 | S. cerevisiae protein component of axial elements and the lateral elements of the synaptonemal complex. |
| RTEL-1 | Human DNA helicase; C. elegans homolog has anti-recombination activity. |
| Set1 | S. cerevisiae histone H3 methyltransferase specific for lysine 4; mammalian Prdm9 also has a SET domain and methylates histone H3-K4. Methylated histone H3-K4 is mostly associated with actively transcribed (euchromatic) regions. |
| Sir2 | S. cerevisiae histone deacetylase |
| Smc5, Smc6 | Structural maintenance of chromosomes proteins; present in a complex important for DNA repair. |
| Spo11 | DNA topoisomerase II-like protein that makes meiotic DSBs; called Rec12 in S. pombe and Mei-W68 in Drosophila. |
| Swi5 | Mitotic and meiotic DNA repair protein; an “accessory protein” in Figure 1. |
| Xnd-1 | C. elegans chromatin protein that affects crossover distribution |
| ZMM | S. cerevisiae Zip1, 2, 3, 4, Msh4, 5 and Mer3 proteins in the synaptonemal complex. |
Figure 1. Meiotic recombination initiation in the fission yeast S. pombe.
Programmed DNA double-strand breaks (DSBs) initiated by Rec12 (Spo11 in other species) during meiosis are efficiently repaired by homologous recombination with high fidelity (for simplicity only one chromatid from each homolog is depicted). Rec12 is aided by several meiotic break proteins to localize and form DSBs. In S. cerevisiae the MRX complex (Mre11, Rad50, Xrs2) is required for DNA breakage and repair, whereas in S. pombe MRN (Mre11, Rad50, Nbs1) is needed only for repair. Rec12, covalently linked to the 5' ends of the DSB, is clipped off attached to short oligonucleotides (~15–45 long) by MRN in conjunction with Ctp1 (Sae2 in S. cerevisiae). The 5' end is further resected by Ctp1 or Exo1 in conjunction with MRN, resulting in a free 3' DNA end. Rad51 and Dmc1, along with numerous accessory proteins, bind the ssDNA end and facilitate invasion of an intact duplex DNA with homology to the invading end. Synthesis of DNA from the end uses the invaded DNA as a template for repair. See Figure 2 for further reactions.
At a DSB, the 5' ends are digested away (resected), and the resultant 3'-ended single-stranded (ss) DNA invades an intact duplex at a point of extensive nucleotide sequence identity. Base-pairing between the two interacting DNA molecules forms hybrid DNA (Figure 1). If the hybrid DNA contains one or more mismatches stemming from a genetic difference between the two parents, mismatch correction can produce three copies of one allele and only one of the other, a phenomenon called gene conversion or non-reciprocal recombination, an exception to Mendel's rule of 2:2 inheritance. Gene conversion can also arise from DNA synthesis that replaces the resected DNA. Resolution of the hybrid DNA intermediate can reciprocally recombine alleles flanking the hybrid DNA region to form a crossover. Alternatively, resolution can leave the flanking alleles in the parental configuration to form a non-crossover. Gene conversion can thus be accompanied by either a crossover or a non-crossover, both of which are forms of reciprocal recombination.
The formation of DSBs and their repair provide multiple levels for control of recombination. Gene conversion frequencies vary greatly along chromosomes, and not surprisingly DSBs were first detected at hotspots of gene conversion, sites that convert at a frequency higher than the genome average. Genome-wide studies have shown that DSB formation is far from random in both budding and fission yeasts, and DSB hotspots and gene conversion hotspots appear to be coincident. Since DSB formation occurs after replication, a DSB can be repaired by interaction with its sister chromatid or with either chromatid of the homolog. Until recently, it was assumed that DSB repair in meiosis occurred only with the homolog, because only crossovers between homologs can properly direct homolog segregation (Figure 2). Studies in both yeasts unexpectedly show, however, that DSBs can be repaired with either the sister or one of the two chromatids of the homolog [3, 4]. A further surprise is that resolution to crossover vs. non-crossover can be regulated in response to the total number of DSBs in the cell, as observed in budding yeast [5]. Here, we discuss factors that influence DSB formation and repair, including two recently described phenomena - crossover homeostasis [5] and crossover invariance [6].
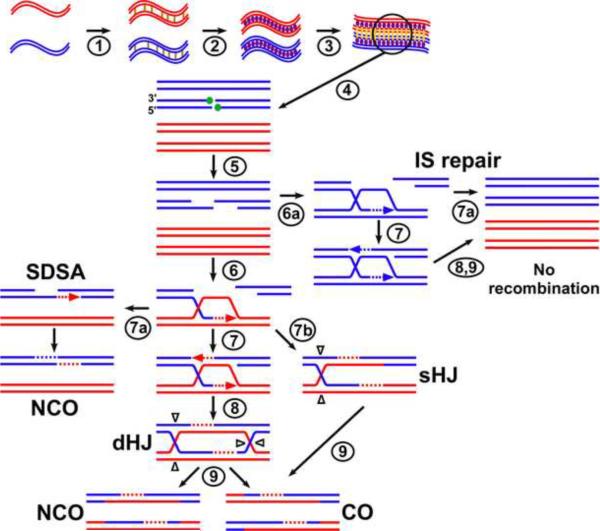
Figure 2. Pathways of meiotic DSB repair.
During meiosis chromosomes are first replicated (step 1) and the identical sister chromatids (red and blue double lines indicate duplex DNA) are linked together by meiosis-specific cohesins (step 1, gold lines) and additional proteins that form axial elements (step 2, purple ovals). Pairing of homologs leads to formation of the synaptonemal complex (step 3, yellow bars); in D. melanogaster, for example, synapsis occurs independently of DSBs, while in S. cerevisiae synapsis would not occur until step 8. Recombination is initiated by programmed DSBs by Rec12 or Spo11 (Figure 1) and numerous partners (step 4, green circles). The now covalently bound Rec12 or Spo11 is removed and the DNA end is resected to create free 3' DNA ends (step 5). The 3' DNA ends invade either the homolog (step 6) or the sister chromatid (step 6a) to create a displacement loop (D-loop), which is extended by DNA synthesis primed by the invading 3' end (see Figure 1 for details). Rad51- or Dmc1-promoted annealing of the other 3' end (“second end capture,” step 7) and ligation of ends forms a double Holliday junction (dHJ; step 8). A single HJ (sHJ; step 7b) is formed if the D-loop is cleaved before the second end anneals. HJ resolution yields a crossover (CO) or non-crossover (NCO), depending on the orientation of cleavage of the HJ(s) (white arrowheads, step 9). If, however, the D-loop is dissociated and the invading end, previously extended by DNA synthesis, anneals with the other DSB end (step 7a), a NCO is formed; this repair is called synthesis-dependent strand annealing (SDSA). Crossover control can act at steps 6, 7, or 9.
Controlling DSB formation
Regulating the timing of DSB formation
In both yeasts, DSBs arise after pre-meiotic DNA replication [7–9]. There are two likely explanations for why this is so. First, sister chromatid cohesion distal to a crossover is essential to generate the tension needed for accurate chromosome segregation; sister cohesion is possible only after replication, as is crossing over between just two chromatids (Figure 2). Second, restricting DNA breaks until after DNA replication avoids errors that could arise by replicating across unrepaired breaks. So how are DSBs restricted to occur after replication?
Early data indicated that local DNA replication is a prerequisite for DNA breakage. In budding yeast when meiotic replication is inhibited by hydroxyurea treatment or by the absence of cyclins, DSBs are not observed [7, 10]. When replication timing is selectively delayed on a chromosome arm with inactive origins, DSBs appear later in that chromosomal region [7]. Later data, however, showed that when replication origin-firing is prevented in S. pombe or S. cerevisiae using pre-replicative complex mutants, meiotic DSBs are formed at wild-type levels, implying that DNA replication per se is not essential for DSB formation [11, 9].
Currently, the solution to this puzzle is unclear. Perhaps most DSBs depend on sister chromatid axis components and DNA replication, whereas others are independent. Alternatively, meiotic checkpoints may inhibit DSB formation when activated by stalled replication forks. If replication origins do not fire at all, these checkpoints are not activated and DSBs form normally (reviewed in [12, 13]). Further experiments are needed to test these scenarios.
DSBs may be formed post-replication because Spo11 or Rec12 and their partner proteins (Figure 1) are present only during this time. Break timing may also be regulated by replication fork-associated proteins that functionally modify these partner proteins [13]. For example, phosphorylation of the budding yeast Spo11 partner protein Mer2 [14–16] regulates the interaction of Mer2 with the DSB-forming complex. Phosphorylated Mer2 in turn recruits other break proteins to break sites. If the cyclin-activated protein kinases were active only at the replication fork, this would, in part, explain the temporal control of meiotic DNA breakage [13]. A similar control may also exist in S. pombe. The protein kinase Hsk1 is required for meiotic DNA breakage [17] and for binding of Rec12 to the mbs1 DSB hotspot [15]. Hsk1 may phosphorylate a Rec12 partner protein.
Chromosomal region-specific requirements for chromosome axis and linear element proteins
Three classes of proteins differentially affect, across the genome, meiotic breakage and recombination: cohesins, condensins, and axial or linear element proteins. Cohesins are required for sister chromatid cohesion following replication, and condensins are required for compacting the chromatin and changing chromosome architecture to allow accurate chromosome segregation during cell division [18, 19]. Linear element or axial element proteins aid interactions between homologous chromosomes during meiosis [20, 21]. In S. pombe the absence of meiosis-specific cohesin subunits Rec8 and Rec11 strongly reduces meiotic DSB frequency and recombination in some chromosomal regions but much less so in others [22]. Absence of linear element protein Rec25 reduces intragenic recombination less than 2-fold at ura1 but 135-fold at ade6 [23]. Similar differential reductions are observed in S. cerevisiae lacking the axial element protein Red1 [24]. Why some regions are more dependent on these proteins than others remains a mystery.
In C. elegans condensins affect meiotic crossover distribution in specific regions of the genome [25]. Condensin I mutations (dpy-28) increase crossovers on the right end of the X chromosome, while condensin II mutations (kle-2) increase crossovers on the left end. In dpy-28 mutants the crossover distribution is correlated with the Rad51 focus-distribution along the X chromosome, indicating that break distribution is affected in these mutants. Mutations in both dpy-28 and kle-2 alter the chromosome axis structure, indicating a role for chromosome structure in determining the position and frequency of meiotic breakage.
Requirements for transcription factors, local DNA sequence, and genomic context for DSB formation
Some hotspots require transcription factor-binding at a distinct local sequence for activity. One of the most thoroughly studied examples is the M26 hotspot in S. pombe, created by a single base-pair mutation in ade6 [26]. 5'-ATGACGT-3' or a closely related sequence is necessary for M26 hotspot activity (i.e., high frequency gene conversion) and DSB-formation specifically during meiosis [27–29]. The transcription factor Atf1-Pcr1, which binds this sequence, is essential for hotspot activity [30, 28]. Similarly, Rts2 and Php2, 3, 5 transcription factors activate their cognate sequences as hotspots when created in the ade6 gene [31].
The chromosomal context of M26 is crucial for its hotspot activity in cells. When 3–6 kb DNA fragments with ade6-M26 centrally located are transplaced to distant chromosomal loci or onto a multi-copy plasmid, M26 is in most but not all cases inactive [32, 33]. These results show that DNA more than 1.5 kb from M26 can influence its activity. Optimal binding of Atf1-Pcr1 to naked DNA implicates an18 bp consensus sequence, and the M26 hotspot requires appropriate base-pairs spread over at least 14 of these bp for full activity in cells [34]. When the consensus sequence was used to identify other potential break sites in the genome, 10 of 15 sites tested showed DSB hotspots within 1 kb of the M26 sequence, and in the one case tested this sequence is required for hotspot activity [29]. Variable DSB frequency among these 10 sites implies additional, as-yet-unidentified factors that influence the intensity of M26 hotspot activity.
Chromosomal context also affects budding yeast DSB hotspots. Insertion of foreign DNA can create a DSB hotspot at the insertion site while decreasing frequency of breakage at a site farther away [35, 36]. Deletions in the promoter region of ARG4 can reduce, enhance or have no effect on gene conversion in ARG4 [37]. Insertions of ARG4 into hot and cold regions show chromosomal context-dependence of DSB propensity paralleling that of the native region [38].
It seems that transcription per se is not required for hotspot activity, since transcriptional strength does not correlate with hotspot activity. The budding yeast HIS4 hotspot requires the transcription factors Bas1, Bas2 and Rap1 [39, 40], but reduction of transcription by deleting part of the promoter does not affect hotspot activity [39]. At both the HIS4 hotspot and the M26 hotspot in fission yeast no correlation has been observed between hotspot activity and transcript levels [39, 30].
Although individual transcription factor-dependent hotspots clearly exist, transcription factor-binding sites are not good indicators of DSBs on a genome-wide level. Early genome-wide analysis in budding yeast showed that of 20 DSB hotspots within intergenic regions, 13 are located between the 5' ends of two divergent genes, implying that a majority of intergenic hotspots require either transcription factor binding or divergent transcription for hotspot activity [41]. However, recent fine-scale mapping of oligonucleotides covalently linked to Spo11 (Figure 1) shows that transcription factor-binding sites have an equal probability of lying in a hotspot or not; indeed, some binding sites seem to obstruct Spo11 action [42]. Binding of transcription factors to their cognate sites may modify chromatin architecture and allow break proteins to act in some cases but not others.
Histone modifications affect meiotic breakage
Some chromosomal features, such as local structure and histone modifications, are correlated with break formation. In budding yeast meiotic DSB sites are associated with sensitivity to micrococcal nuclease (MNase) and DNase1, a defining feature of “open” chromatin [43, 44]. In fission yeast, nucleosome phasing and MNase sensitivity are altered by M26 [45]. In mice, the Eb recombination hotspot is DNase1-hypersensitive, but the Lmp2 hotspot is not [46].
Post-translational modifications of N-terminal tails of histones are associated with changes in the “openness” of chromatin. Thus, it is no surprise that chromatin modifying proteins affect meiotic DNA breakage, but the effects are complex. In budding yeast, absence of the histone deacetylase Sir2 affects meiotic breakage at ~12% of all genes, with increases at some sites and decreases at others [47]. Similarly, absence of Set1, which methylates histone H3-K4, a modification mostly associated with actively transcribed regions, reduces break formation at several hotspots [48]. Analysis of ssDNA that accumulates adjacent to DSBs in dmc1 mutants shows that >80% of DSB hotspots genome-wide are dependent on Set1, but ~7% are repressed by Set1 [49]. set1 mutants have delayed replication [48], which complicates the interpretation, since as noted above DSB formation is coupled to replication.
Histone modifications regulate DSB formation and meiotic recombination in S. pombe as well. The M26 hotspot has hyper-acetylated histones H3 and H4, both marks of transcription activation [50]. Absence of the histone acetyltransferase Gcn5 and the chromatin remodeler Snf22 completely removes the chromatin remodeling associated with M26 and reduces meiotic recombination at the M26 hotspot.
Chromatin modifications have also been implicated in break formation in C. elegans. Partial deletion of him-17, encoding a chromatin-associated protein, reduces the level of H3-K9 methylation and causes almost complete absence of Rad51 DSB-repair foci in the meiotic cells in which recombination would normally occur [51]. Similarly, mutation in xnd-1, encoding a chromatin-binding factor, reduces meiotic Rad51 foci on the X chromosome and increases H2A-K5-Ac in meiotic cells [52].
A role for histone modifications in regulating meiotic breakage has been implicated in mammals. In mice, H3-K4 trimethylation and H4 acetylation are enriched at the Psmb9 and Hlx1 recombination hotspots [53]. The histone H3-K4 methyl transferase Prdm9 binds a degenerate 13-mer motif found in about 40% of human linkage disequilibrium-defined hotspots and also in the mouse Psmb9 and Hlx1 hotspots [54, 55]. Alleles of Prdm9 correlate with hotspot activity, suggesting that Prdm9 binds to a hotspot and activates it.
Centromeres are transcriptionally and recombinationally silent regions of the genome. In fission yeast RNAi-mediated methylation of histone H3-K9 by Clr4 represses transcription at the centromeres [56]. Disrupting genes that encode RNAi factors or Clr4 dramatically increases meiotic DSBs at and around centromere 3 (cen3) and increases recombination between genetic markers flanking cen3 ~100-fold [57]. Interestingly, disruption of some factors that increase transcription in the fission yeast centromeres does not affect meiotic recombination in cen3, suggesting that overlapping as well as distinct histone modifications dictate the accessibility of a region to transcription and recombination functions [57].
The studies mentioned above substantiate the correlations between meiotic DSB formation and histone modifications, chromosomal context, transcription factors and their cognate sites, and chromatin remodeling in numerous species. However, a clear and universal picture is not observed. Multiple factors affect DNA breakage variably, depending on the chromosomal site and the species studied. At some sites the transcription complex may localize to “open” promoters and recruit meiotic break proteins, while at other sites the transcription complex may occlude them. Modified chromatin may differentially control the accessibility of the transcription and the meiotic recombination machinery. In addition meiotic DSB proteins may recognize and bind to specific combinations of modified histones or to a “recruiting factor” that recognizes modified histones. Elucidating the role(s) of these factors in DSB-formation awaits further study.
Controlling DSB repair
There are more DSBs formed per meiosis, as measured by Rad51 DSB-repair foci, than crossovers in many organisms, including mice (~10:1) and Arabidopsis thaliana (~15:1) [58, 59]. A genome-wide study in S. cerevisiae estimated that about 60% of DSBs are repaired to generate crossovers (COs), with the remainder repaired as non-crossovers (NCOs) (Figure 2) [60], though some NCOs could be lost via mismatch correction and thus underestimated. The frequency of gene conversions (GCs) with an associated CO varies in different organisms, from an average of ~35% in S. cerevisiae to as high as 80% in some regions of the S. pombe genome [61, 62]. The restriction of COs is exceptionally strong in C. elegans: only one CO forms per homolog pair even when two chromosomes, each of which normally has one crossover, are fused end-to-end [63], a result of strong chromosome-wide crossover interference (Box 1).
It isn't as simple as one DSB–one crossover; so, what factors control how frequently a CO is formed during DSB repair? Crossover control during DSB repair can be viewed as two alternatives at each of two steps: whether repair is with the sister or with a homolog chromatid (Figure 2, steps 6 and 6a), and whether a CO or a NCO forms during repair (Figure 2, steps 7a and 9). Genetically observable COs and NCOs [i.e., GCs] can be formed only by repair with a homolog chromatid as the template, as the sister chromatid is identical and repair with it would be genetically silent (Figure 2, step 6a). Repair with the homolog can result in either a CO or NCO by means discussed below.
Crossover vs. non-crossover repair of DSBs
The step of recombination at which COs are differentiated from NCOs has been the focus of substantial research. The canonical model of DSB repair proposes that COs and NCOs arise from alternative resolution of Holliday junctions (HJs), i.e., at essentially the last step of DSB repair [64]. Enzymatic cleavage of one pair of strands generates a CO, and cleavage of the other a NCO (Figure 2, step 9). Electron microscopy and gel electrophoresis provided evidence for HJs in S. cerevisiae, in this case mostly, but not exclusively, double HJs (Figure 2, step 8) [65, 66], as well as mostly single HJs in S. pombe (Figure 2, step 7b) [3].
There is, however, growing evidence against such “late” control of CO vs. NCO formation during meiosis. In S. cerevisiae heteroduplex DNA (hDNA) generated during NCO formation does not show the expected symmetric structure predicted from double HJ resolution [67–69]. In the absence Zip1, 2, 3, and 4, Mer3, or Msh4, 5 (“ZMM” proteins, which act before HJ formation) (Figure 2, steps 3 and 6) COs and HJ intermediates are substantially reduced but NCOs are unaffected [70]. This result has been interpreted as differentiation of COs and NCOs before HJ resolution. Furthermore, hDNA from NCOs was detected earlier (at the same time as HJs) than hDNA from COs [71]. In strains deficient for the meiosis-specific transcription factor Ndt80, unresolved HJs accumulate and CO hDNA is reduced, but NCO hDNA is not reduced. It was proposed that NCOs are formed in a different, HJ-independent pathway via synthesis-dependent strand annealing (SDSA, Figure 2, step 7a), and that most or all HJs give rise only to COs. In support of this view, a temporal analysis of single-end invasions (Figure 2, step 6) suggested that the CO vs. NCO designation happens earlier than HJ resolution [72]. In S. pombe, mutants lacking the HJ-resolvase Mus81-Eme1 accumulate HJs, and COs are virtually eliminated, but gene conversions (NCOs) form with wild-type frequency [73, 74, 3].
To date, DNA intermediates specific to SDSA repair have not been detected, possibly due to their short half-life or high instability. A genetic assay for a class of NCOs most easily explained by SDSA, however, provided evidence that S. cerevisiae uses SDSA during meiosis [75]. The structure of NCOs and COs formed during mitotic gap repair is also most consistent with NCOs being formed via SDSA, and COs via HJ resolution [76].
Although the results discussed above are consistent with COs and NCOs arising from different intermediates, they are also compatible with induction of a factor, perhaps Ndt80-dependent in S. cerevisiae, that directs HJ resolution to COs only late in meiosis. In the absence of this factor and HJ resolution, intermediates may be diverted into NCOs exclusively, perhaps via SDSA.
Homolog vs. sister chromatid for DSB repair
A DSB can be repaired using either the homolog or the sister chromatid as template, but what determines partner choice is unclear (Figure 2, step 6 and 6a). The most thorough examination of what influences the choice between interhomolog (IH) and intersister (IS) DNA repair has been done in S. cerevisiae. During meiosis in this yeast there is a strong preference for IH repair, at least by assay of HJs at one (artificial) hotspot via 2-D gel electrophoresis – IH HJs outnumber IS HJs 5 to 1 [77]. This homolog preference is dependent on Red1, Hop1, Mek1, and Rec8 [78–80]. In the absence of these chromosome axis-associated proteins, IH HJs and COs are reduced; IS HJs are increased in mek1 mutants. Thus, IH repair is actively promoted by several proteins; IS repair may be actively suppressed.
In spite of these observations and conclusions, a recent study indicated that IS repair can be frequent during S. cerevisiae meiosis. This study used homologous chromosomes hemizygous for large (3.5 or 90 kb) deletions, and therefore DSBs on the intact homolog could be repaired only with the sister chromatid; the observed, high-level DSBs were efficiently repaired [4]. Additional observations suggest that a substantial fraction of DSBs on chromosomes with an intact homolog are also frequently repaired with the sister. IS repair may have been previously underestimated by measuring only HJ formation; a majority of IS repair could be via SDSA, or IS HJs may be less stable or actively destabilized.
Sister chromatid repair has also been implicated at S. cerevisiae centromeres, where recombination is reduced more than DSBs, relative to the genome average [81]. But unlike the rest of the genome, crossover homeostasis, described below, cannot account for this observation, as NCO and CO levels are equally suppressed; NCOs would be expected to increase in response to the DSB repair that does not proceed towards CO formation. In a zip1 mutant, centromeric recombination is not suppressed, but DSBs are not increased, suggesting that Zip1 suppresses IH repair at the centromere in favor of IS repair [81]. The genome-wide CO reduction in an msh4 mutant – in which DSBs are not reduced – is not accompanied by an increase in NCOs, suggesting IS repair as well [60]. Evidence of homolog-independent, and presumably intersister, DSB repair has been observed in smc5, smc6, and brc1 mutants of C. elegans [82, 83]. IS repair during meiosis appears to be more prevalent than previously estimated and may partly account for Rad51 foci greatly outnumbering COs in some species.
Crossover homeostasis
The maintenance of a constant level of COs by adjusting the number of NCOs when DSB levels change has been termed crossover homeostasis, based on results from a study of S. cerevisiae non-null spo11 mutants in which DSB formation was decreased [5]. In spite of DSBs being reduced to 80, 30, and 20% of wild type, the level of COs remained constant at numerous genetic loci, but NCOs were reduced in parallel with DSB reduction. Thus, COs were maintained at the expense of NCOs (Figure 3a). Genome-wide microarray analysis of CO distribution in S. cerevisiae meiotic tetrads supports crossover homeostasis in wild type as well [81, 60]. COs and NCOs are not correlated, as a nearly constant level of COs is observed from cell to cell, while NCOs vary in parallel with DSBs per cell.
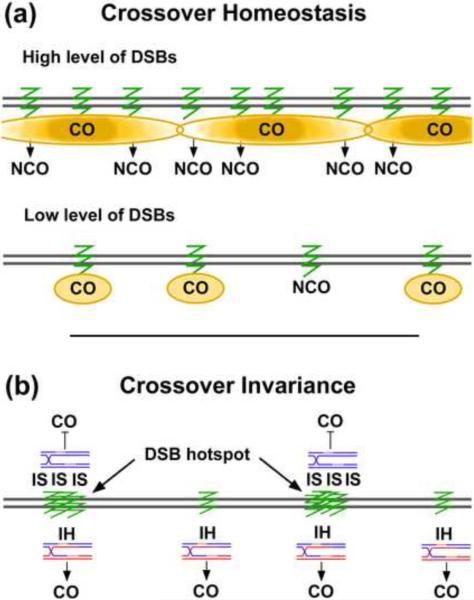
Figure 3. Two mechanisms to distribute crossovers across chromosomes.
(a) Crossover homeostasis observed in S. cerevisiae [5, 81, 60]. A crossover (CO) generated at one DSB (green zig-zag) inhibits nearby DSBs from generating another CO, a phenomenon known as crossover interference (COI; Box 1) (represented by yellow clouds; deeper color representing greater interference). Instead, these adjacent DSBs are repaired as non-crossovers (NCOs). The amount of DSBs may vary from cell to cell in meiosis: one with abundant DSBs (top diagram) has more NCOs than one with few DSBs (lower diagram), but the overall level of COs remains constant. Homeostasis is thought to arise from the same mechanism as COI and to occur in other species. (b) A different mechanism of crossover control, crossover invariance, observed in S. pombe [6]. S. pombe has intense DSB hotspots that are widely space across the genome but a nearly constant level of COs per kb of DNA. In other words, a genetic interval with a DSB hotspot has about the same frequency of COs as one of similar size without a DSB hotspot. At these DSB hotspots, intersister (IS) repair is more frequent than interhomolog (IH) repair, but away from hotspots DSB repair is mostly or all IH. Since IS repair does not yield genetically observable COs, the amount of COs from hotspots is roughly equal to the COs generated away from hotspots, resulting in the observed crossover invariance.
Crossover homeostasis is proposed to be part of the mechanism of crossover interference (Box 1). S. cerevisiaezip2 and zip4 mutants display reduced COs genome-wide, and there is a significant increase in the correlation between COs and NCOs [81]. Both homeostasis and interference are impaired, supporting a link between the two. The meiosis checkpoint protein Pch2 helps organize the meiotic axis components during synaptonemal complex formation and localizes to putative CO sites [84]. In the absence of Pch2, interference is reduced, gene conversions are elevated, and crossovers increase on large chromosomes (though not on smaller ones), but DSBs do not increase, suggesting the extra COs come from NCO or intersister events [84, 85]. While pch2 mutants have no defect in spore viability alone, when combined with reduced DSBs in the spo11 mutations noted above, spore viability is strongly reduced as the DSBs are reduced, due to chromosome missegregation events [84, 85]. Pch2 therefore appears necessary to establish interference and crossover homeostasis, suppressing excess COs in abundant DSB situations and ensuring proper CO distribution when DSBs are limiting, perhaps by balancing COs and NCOs and preventing excess IS repair. Organisms without interference, S. pombe for example, are predicted not to have homeostasis [5]; this prediction has yet to be tested.
How crossover homeostasis is maintained is still unclear, but it may be enforced during DSB repair. In C. elegans, the protein Rtel-1 was recently inferred to influence DSB repair towards NCOs, presumably via the SDSA pathway [86]. Purified human RTEL-1 disassembles D-loops, preferentially those with a 3' end invasion thought to be intermediates in DSB repair (Figure 2, step 6) [87, 86]. COs are increased in C. elegans rtel-1 mutants, and increasing the number of DSBs by a dpy-28 mutation [25] or ionizing irradiation further increases COs [86]. Interference was also impaired, as multiple COs per chromosome were observed, consistent with a role for Rtel-1 in crossover control by disrupting single-end invasions and preventing HJ and CO formation, sending repair down the SDSA pathway. Conversely, there might be proteins that push repair towards HJ and CO formation. D-loops formed by human DMC1, but not those formed by human RAD51, are resistant to dissociation by RAD54 [88]. This resistance may allow DMC1-promoted recombination intermediates to proceed to HJs and COs, but RAD51-promoted intermediates to SDSA and NCOs. Alternatively, homeostasis could be enforced early during the formation of DSBs.
Crossover invariance
The number of crossovers per unit of DNA in S. pombe is nearly uniform across much of the genome, even though the DSB distribution is highly variable [89, 90]. A recent study [6] explained this seeming discrepancy by partner choice differing at hotspots and in DSB-cold regions. Two DSB hotspots, the wild-type mbs1 and the M26-like single base-pair mutation ade6-3049, were analyzed for HJ formation. At each hotspot IS HJs outnumber IH HJs ~4 to 1. These data provide direct evidence for IS repair being more frequent than IH repair, opposite of the result in S. cerevisiae. Both IS and IH HJs are completely dependent on Rad51 but independent of Dmc1, as is recombination at each hotspot. In contrast, HJs and recombination in regions distant from DSB hotspots are highly dependent on Dmc1 and Swi5, a mediator for both Dmc1 and Rad51 that is required only for IH HJ formation at hotspots. Dmc1 is also necessary specifically for IH DSB repair and recombination in S. cerevisiae [91, 78]. Deletion of the mbs1 hotspot reduces COs less than DSBs, showing that substantial COs can be generated in DSB-cold regions, and these COs are strongly Dmc1-dependent. Although IS and IH HJs in DSB-cold regions were not directly measured due to their low levels, the dependence on Swi5 and Dmc1 implies that most HJs distant from DSB hotspots are IH (Figure 3b).
Genetic recombination data agree with this notion. Genetic intervals with weak DSB hotspots have about as many COs as comparable-size intervals with intense DSB hotspots. This phenomenon was termed crossover invariance and reflects a type of crossover control not previously reported: DSB hotspots primarily use IS repair and hence produce few COs per DSB, while regions with few DSBs predominantly use IH repair and may contribute as many COs as regions with DSB hotspots (Figure 3b). This feature keeps the level of crossing over nearly uniform across the genome, although for markers closely flanking mbs1 this DSB hotspot is clearly a crossover hotspot [62].
Some organisms go to great lengths to promote IH repair and COs, so why is this IH preference not seen at DSB hotspots S. pombe? Non-disjunction of homologous chromosomes has serious consequences for all organisms, and IH connections are important to establish proper segregation, but not universally. In the fruit fly Drosophila melanogaster males do not recombine, but their chromosomes still segregate properly; single achiasmate chromosome pairs also segregate properly in females, though recombination is still vital for proper female meiosis [92]. Similarly, homolog segregation is nonrandom in S. pombe mutants lacking Rec12 (Spo11 homolog), properly occurring 63% of the time by a secondary system dependent on the dynein motor; spore viability is reduced to only ~20% of wild type in rec12 null mutants [93]. Additionally, S. pombe has only three chromosomes that are relatively large (4 – 6 Mb) and have 10 – 20 COs each. Therefore, enforcement of a complicated IH-promoting system for crossing over isn't necessary. Rather, maintaining uniform COs across the genome increases the likelihood of advantageous haplotype rearrangements, and crossover invariance limits excess COs from forming at the same position, involving the same genes, generation after generation.
Concluding remarks and future perspectives
Multiple proteins are required to deliberately break DNA at distinct sites (hotspots) during meiosis. Hotspot determination is complex, involving multiple factors that activate Spo11 – break proteins and their modification, chromatin state, transcription factors, and chromosomal structural components. Although some hotspots are activated by transcription factors, it is not clear how wide-spread this control is, nor is it known how transcription factors act in DSB formation. The same is true for the other factors. Particularly puzzling is how chromosomal axis proteins, such as Rec8, impart their specificity on some but not other chromosomal regions. Correlating the binding of these factors with hotspots genome-wide should be informative.
Though crossover homeostasis and invariance can be seen as two distinct methods of achieving control via DSB repair, our understanding of both processes is still in its infancy. Whether homeostasis occurs in species other than S. cerevisiae and perhaps C. elegans, and invariance in species other than S. pombe, is unknown. These controls may arise from similar (or the same) mechanisms, since both maintain constancy of COs. IS repair of DSBs, as observed in S. pombe, S. cerevisiae, and C. elegans, could be as effective as NCO formation in preventing COs from occurring too close or too frequently in the same location. Indeed, the 50% association of COs with GCs in S. cerevisiae would allow DSBs to be reduced by only 2-fold while maintaining homeostasis, if NCO:CO were the only option; nevertheless, DSBs can be reduced by 5-fold and homeostasis is maintained [5]. Changing IS repair to IH repair [4] is another possible source of COs when DSBs are reduced. It will be interesting to see the effect of impeding IS repair on CO and NCO distribution and homeostasis; NCOs are expected to increase to prevent extra COs. Whether NCOs arise by SDSA would be substantiated by finding DNA intermediates specific to SDSA, thereby lending physical reality to this genetic concept. A genome-wide map of COs and NCOs in S. pombe meiotic tetrads using the methods established in S. cerevisiae might reveal homeostasis in addition to invariance in that species and could illuminate their relation. Measuring in S. pombe the effect of fewer DSBs via hypomorphic Rec12 alleles on both the CO:NCO ratio and the IS:IH ratio at DSB hotspots could further help determine the relation. Ultimately, one would like to examine the DSB sites themselves to determine if they are designated, perhaps by chromatin modifications, for CO or NCO, and for IS or IH, repair prior to DSB formation. Finally, it will be exciting to see what layers of crossover control still remain to be discovered.
Box 1. Crossover interference.
If the formation of COs during meiosis were random, a CO at one location would not affect the formation of a CO elsewhere in the genome. In most organisms studied, however, a CO decreases the likelihood that another forms nearby on the same chromosome. The mechanism of this phenomenon, called crossover interference, is not understood, but several models have been proposed. The polymerization model [94] suggests that, after a CO is initiated, an inhibitor is bi-directionally polymerized outward from the CO and prevents any sites nearby from also forming a CO. The mechanical stress model [95] is based on the chromosome being subjected to physical stress during meiosis; an initial DSB (or CO) relieves this stress and forms a CO, but the resulting lack of stress prevents other COs from forming nearby. The counting model [96] posits that after the first CO is formed a fixed number of NCOs must form along the chromosome before another CO is formed, though the observation of crossover homeostasis provides evidence against this model [5]. Recently, COs have been confirmed to interfere not just with other COs but with NCOs as well [60].
Interference can be measured quantitatively via the coefficient of coincidence, S = RD/(R1*R2), where R1 and R2 are the frequencies of crossovers in two adjacent genetic intervals and RD is the frequency of double crossovers. Interference is then calculated as I = 1 − S; therefore, S < 1 indicates interference present, while S > 1 indicates negative interference.
Studies in S. cerevisiae indicate that not all COs are subject to interference [97]. The majority of COs are Msh4-Msh5-dependent and are subject to interference, whereas Mus81-Mms4-dependent COs are not. In A. thaliana and M. musculus most COs are Msh4-Msh5-dependent and manifest interference; in C. elegans all COs are Msh4-Msh5-dependent, and each bivalent forms only one CO (complete interference). On the other end of the spectrum, S. pombe has no CO interference; nearly all COs are dependent on Mus81-Eme1 [73, 74, 3].
Box 2. Detecting meiotic DSBs.
In both budding and fission yeast DSBs are formed by a highly conserved, meiosis-specific protein Spo11 (Rec12 in S. pombe). Like a DNA topoisomerase, this protein becomes covalently linked to the DNA, but several “meiotic break proteins” are also essential (Figure 1). In these yeasts, DSBs are directly observed by Southern blot hybridization of meiotic DNA and are non-uniformly distributed across the genome – there are both “hotspots,” with frequent DSBs, and “coldspots” or cold regions, with infrequent breaks. Examined carefully, hotspots are seen to be clusters of breaks generally spanning ~100–200 bp in S. cerevisiae [42] and up to 4 kb in S. pombe [90]. In mice, labeling of broken DNA ends using terminal deoxynucleotidyl transferase (TdT) revealed a DSB hotspot, although these could be single- rather than double-strand breaks [98]. To map DSBs genome-wide, meiotically broken DNA is enriched either by purifying ssDNA, naked or bound by Dmc1 or a single-strand binding protein, that accumulates on both sides of a DSB [99, 100] or by immunoprecipitation of Spo11 or Rec12 covalently bound to DSB ends [41, 90, 42] and hybridizing this DNA to whole genome microarrays. High-throughout sequencing of Spo11-bound oligonucleotides (Figure 1) has provided a nucleotide-level resolution of break sites [42]. The coincidence of break sites determined directly by Southern blot analyses and by the microarray and sequencing methods indicates that the latter two methods detect DSBs.
Since DSBs are thought to be prerequisites for recombination, DSB hotspots are inferred from exceptionally high local frequencies of gene conversion and crossovers or from linkage disequilibrium, taken to reflect historical recombination at high frequency between haplotype blocks, genetic markers between which there is little recombination during the evolution of a population [64, 101]. DSBs are also often inferred by fluorescence microscopy of meiotic recombination proteins, such as Rad51 or phosphorylated histone H2AX, that are recruited to break sites, but these low-resolution methods do not reveal hotspots. TdT labeling of DSB ends with fluorescently marked nucleotides (TUNEL staining) confirmed Rad51 foci as break sites in C. elegans [25].
Acknowledgements
We thank Michael Lichten, anonymous reviewers, and Joël Savard for helpful comments. Research in our laboratory is supported by grants GM031693 and GM032194 from the National Institutes of Health of the United States of America.
Glossary
- Meiotic recombination
Exchange of genetic information or DNA between homologous chromosomes during meiosis
- Double-strand break (DSB)
A DNA molecule with both strands broken at the same position or closely spaced positions
- Crossover
Recombination that results in the reciprocal exchange of genetic markers flanking the site of DNA interaction
- Non-crossover
Recombination that results in local genetic exchange without exchange of flanking genetic markers
- Gene conversion
Recombination that results in non-reciprocal transfer of genetic information from one DNA duplex to another; can occur by correction of base mismatches in heteroduplex DNA or by local DNA synthesis at the site of DNA interaction
- Hotspots
Chromosomal sites with higher than genome average frequency of DSBs or of recombination
- Hybrid DNA
A double-stranded DNA molecule with one strand from each of two interacting DNA duplexes
- Heteroduplex DNA
Hybrid DNA with one or more base differences in the two interacting DNA duplexes
- Synthesis-dependent single-strand annealing (SDSA)
A mechanism for DSB repair without crossing over but with potential gene conversion (see Figure 2, step 7a)
- Meiotic checkpoint
A set of molecular pathways that control and regulate meiotic nuclear division progression
- Haplotype blocks
A set of genetic alleles that are transmitted together through meiosis
- Cohesins
Proteins required for holding sister chromatids together (cohesion). Rec8 is a meiosis-specific cohesin subunit
- Condensins
Proteins involved in chromosome compaction during cell division
- Linear elements
S. pombe nuclear structures replacing the SC and containing Rec10 (which has limited homology to S. cerevisiae Red1), Rec25, and Rec27
- Synaptonemal complex (SC)
A proteinaceous structure formed during meiosis that aids chromosome pairing and recombination. The SC is made up of lateral elements connected by central regions. Each lateral element arises from an axial element, formed prior to SC formation along sister chromatids
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keeney S. Mechanism and control of meiotic recombination initiation. Current Topic Development Biology. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 2.Cromie GA, Smith GR. Meiotic recombination in Schizosaccharomyces pombe: A paradigm for genetic and molecular analysis. In: Egel R, Lankenau D-H, editors. Recombination and meiosis: Models, means, and evolution. Springer-Verlag: 2008. pp. 195–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cromie GA, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldfarb T, Lichten M. Frequent and efficient use of the sister chromatid for DNA double-strand break repair during budding yeast meiosis. PLoS Biol. 2010;8:e1000520. doi: 10.1371/journal.pbio.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martini E, et al. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyppa RW, Smith GR. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell. 2010;142:243–255. doi: 10.1016/j.cell.2010.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borde V, et al. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 8.Cervantes MD, et al. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 9.Hochwagen A, et al. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–873. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Smith KN, et al. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Current Biology. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 11.Murakami H, Nurse P. Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nature. 2001;28:290–293. doi: 10.1038/90142. [DOI] [PubMed] [Google Scholar]
- 12.Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol. 2006;16:R217–228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Murakami H, Keeney S. Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev. 2008;22:286–292. doi: 10.1101/gad.1642308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson KA, et al. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasanuma H, et al. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 2008;22:398–410. doi: 10.1101/gad.1626608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan L, et al. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes & development. 2008;22:386–397. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogino K, et al. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 2006;23:8131–8136. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legagneux V, et al. Multiple roles of Condensins: a complex story. Biol. Cell. 2004;96:201–213. doi: 10.1016/j.biolcel.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Peters JM, et al. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 20.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 21.Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- 22.Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis L, et al. Rec25 and Rec27, novel components of meiotic linear elements, link cohesin to DNA breakage and recombination in fission yeast. Current Biology. 2008;18:849–854. doi: 10.1016/j.cub.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szankasi P, et al. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe: Wild-type and mutant alleles including the recombination hotspot allele ade6-M26. J. Mol. Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 27.Schuchert P, et al. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. Embo J. 1991;10:2157–2163. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox ME, et al. A family of CRE-related DNA sequences with meiotic recombination hotspot activity in Schizosaccharomyces pombe. Genetics. 2000;156:59–68. doi: 10.1093/genetics/156.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steiner WW, Smith GR. Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol Cell Biol. 2005;25:9054–9062. doi: 10.1128/MCB.25.20.9054-9062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kon N, et al. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 1997;94:13756–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steiner WW, et al. Important characteristics of sequence-specific recombination hotspots in Schizosaccharomyces pombe. Genetics. 2011;187:385–396. doi: 10.1534/genetics.110.124636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponticelli AS, Smith GR. Chromosomal context dependence of a eukaryotic recombinational hot spot. Proc. Natl. Acad. Sci. USA. 1992;89:227–231. doi: 10.1073/pnas.89.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virgin JB, et al. Active and inactive transplacement of the M26 recombination hotspot in Schizosaccharomyces pombe. Genetics. 1995;141:33–48. doi: 10.1093/genetics/141.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner WW, Smith GR. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2005;169:1973–1983. doi: 10.1534/genetics.104.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. Embo J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultes NP, Szostak JW. A poly(dA.dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borde V, et al. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4832–4842. doi: 10.1128/mcb.19.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White MA, et al. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Current Genetics. 1992;21:109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- 40.White MA, et al. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerton JL, et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan J, et al. A Hierarchical Combination of Factors Shapes the Genome-wide Topography of Yeast Meiotic Recombination Initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohta K, et al. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu TC, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 45.Mizuno K, et al. The meiotic recombination hot spot created by the single-base substitution ade6-M26 results in remodeling of chromatin structure in fission yeast. Genes and Development. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- 46.Mizuno K, et al. Molecular analysis of a recombinational hotspot adjacent to Lmp2 gene in the mouse MHC: fine location and chromatin structure. Mamm. Genome. 1996;7:490–496. doi: 10.1007/s003359900149. [DOI] [PubMed] [Google Scholar]
- 47.Mieczkowski PA, et al. Loss of a histone deacetylase dramatically alters the genomic distribution of Spo11p-catalyzed DNA breaks in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3955–3960. doi: 10.1073/pnas.0700412104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sollier J, et al. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. Embo J. 2004;23:1957–1967. doi: 10.1038/sj.emboj.7600204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borde V, et al. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. Embo J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada T, et al. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 52.Wagner CR, et al. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature. 2010;467:839–843. doi: 10.1038/nature09429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buard J, et al. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. Embo J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myers S, et al. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 55.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 57.Ellermeier C, et al. RNAi and heterochromatin repress centromeric meiotic recombination. Proc. Natl. Acad. Sci. USA. 2010;107:8701–8705. doi: 10.1073/pnas.0914160107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moens PB, et al. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci. 2002;115:1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- 59.Mercier R, et al. Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3,whereas the other one is not. Curr Biol. 2005;15:692–701. doi: 10.1016/j.cub.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 60.Mancera E, et al. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fogel S, et al. Meiotic gene conversion: a signal of the basic recombination event in yeast. Cold Spring Harbor Symp. Quant. Biol. 1979;43:1325–1341. doi: 10.1101/sqb.1979.043.01.152. [DOI] [PubMed] [Google Scholar]
- 62.Cromie GA, et al. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 64.Szostak JW, et al. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 65.Bell LR, Byers B. Homologous association of chromosomal DNA during yeast meiosis. Cold Spring Harbor Symp. Quant. Biol. 1982;47:829–840. doi: 10.1101/sqb.1983.047.01.095. [DOI] [PubMed] [Google Scholar]
- 66.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 67.Porter SE, et al. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics. 1993;134:5–19. doi: 10.1093/genetics/134.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilbertson LA, Stahl FW. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1996;144:27–41. doi: 10.1093/genetics/144.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Birmingham EC, et al. Testing predictions of the double-strand break repair model relating to crossing over in Mammalian cells. Genetics. 2004;168:1539–1555. doi: 10.1534/genetics.104.029215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borner GV, et al. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 71.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 72.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 73.Osman F, et al. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 74.Smith GR, et al. Fission yeast Mus81***•Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McMahill MS, et al. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitchel K, et al. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: implications for recombination. Mol Cell. 2010;38:211–222. doi: 10.1016/j.molcel.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 78.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 79.Niu H, et al. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim KP, et al. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen SY, et al. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15:401–415. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adamo A, et al. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008;9:287–292. doi: 10.1038/sj.embor.7401167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bickel JS, et al. Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet. 2010;6:e1001028. doi: 10.1371/journal.pgen.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joshi N, et al. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genet. 2009;5:e1000557. doi: 10.1371/journal.pgen.1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zanders S, Alani E. The pch2Delta mutation in baker's yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genet. 2009;5:e1000571. doi: 10.1371/journal.pgen.1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Youds JL, et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barber LJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–271. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bugreev DV, et al. The resistance of DMC1 D-loops to dissociation may account for the DMC1 requirement in meiosis. Nat Struct Mol Biol. 2011;18:56–60. doi: 10.1038/nsmb.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Young JA, et al. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 90.Cromie GA, et al. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLoS Genetics. 2007;3:e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bishop DK, et al. DMC1: A meiosis-specific homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 92.Orr-Weaver TL. Meiosis in Drosophila: seeing is believing. Proc. Natl. Acad. Sci. USA. 1995;92:10443–10449. doi: 10.1073/pnas.92.23.10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Davis L, Smith GR. Dynein promotes achiasmate segregation in Schizosaccharomyces pombe. Genetics. 2005;170:581–590. doi: 10.1534/genetics.104.040253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.King JS, Mortimer RK. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 1990;126:1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kleckner N, et al. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Foss E, et al. Chiasma interference as a function of genetic distance. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Qin J, et al. Mouse strains with an active H2-Ea meiotic recombination hot spot exhibit increased levels of H2-Ea-specific DNA breaks in testicular germ cells. Mol Cell Biol. 2004;24:1655–1666. doi: 10.1128/MCB.24.4.1655-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blitzblau HG, et al. Mapping of meiotic single-stranded DNA reveals double-stranded-break hotspots near centromeres and telomeres. Curr Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- 100.Buhler C, et al. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Arnheim N, et al. Mammalian meiotic recombination hot spots. Annu. Rev. Genet. 2007;41:369–399. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]