Abstract
The C4'-oxidized abasic site (C4-AP) forms two types of interstrand cross-links with the adjacent nucleotides in DNA. Previous experiments revealed that dG does not react with the lesion and that formation of one type of cross-link is catalyzed by the opposing dA. Iso-guanosine•dC and 2-aminopurine•dT base pairs were used to determine why dG does not cross-link with C4-AP despite its well known reactivity with other bis-electrophiles. 7-Deaza-2'-deoxyadenosine was used to probe the role of the nucleotide opposite C4-AP in the catalysis of interstrand cross-link formation.
Keywords: DNA damage, interstrand cross-link, abasic sites, modified oligonucleotides, modified nucleotides
1. Introduction
Abasic sites (AP) and related oxidized abasic lesions (e.g. C4-AP, DOB) are common forms of DNA damage.1,2 While toxic in their own right, recent studies have shown these lesions can react with their surroundings to form even more deleterious types of damage.3–6 This includes interstrand cross-links (ICLs), which are potent blocks to replication and transcription.7 Accurate ICL repair is essential to genomic integrity and how this occurs is an active area of research.8–11 ICLs are produced by a variety of cytotoxic damaging agents, as well as ionizing radiation. Therapeutic agents whose cytotoxicity is linked to ICL formation include the nitrogen mustards, mitomycin C, cis-platin, and the enediyne C1027.12–15 A variety of bis-electrophiles, such as acrolein, and others that are produced endogenously also alkylate opposing strands of duplex DNA to produce ICLs.16,17 Recently, an adduct of 2'-deoxycytidine was identified in cellular DNA that was consistent with interstrand (and/or intrastrand) cross-link formation between it and the oxidized abasic site resulting from hydrogen atom abstraction from the C4'-position of DNA (C4-AP).18 C4-AP is produced by a variety of oxidizing agents, including bleomycin, the enediynes, and ionizing radiation.19–22 This oxidized abasic lesion was known to be mutagenic and has been shown to irreversibly inhibit DNA polymerase β, but until recently had not been suggested to form DNA interstrand cross-links.18,23–25
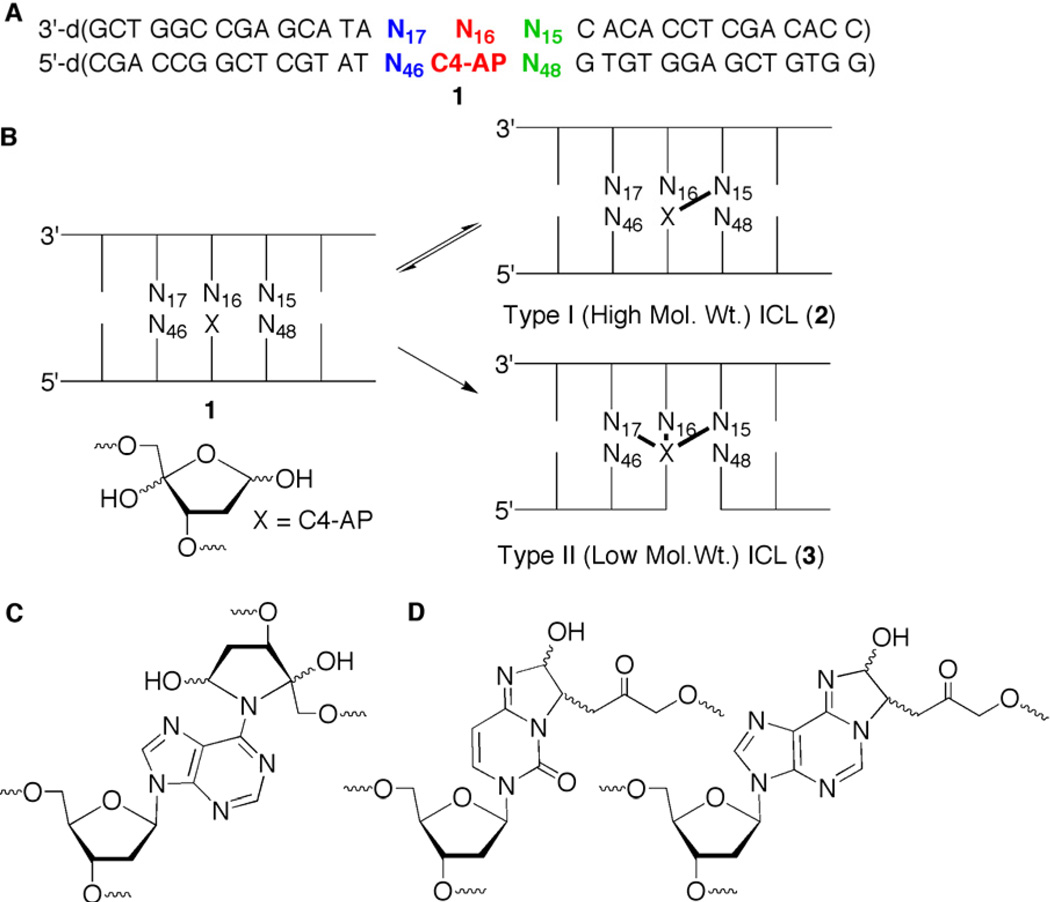
C4-AP was unequivocally shown to form two types of ICLs with dA and one with dC by using chemically synthesized 31 base pair oligonucleotide substrates (1, Figure 1A) containing the lesion at a defined site.4,26 Both strands were intact in Type I ICLs (high molecular weight, 2, Figures 1B, C), which were only observed with dA in duplexes containing this nucleotide at position 15. β-Elimination from C4-AP gave rise to Type II ICLs (low molecular weight, 3, Figures 1B, D) with dA and dC at multiple positions (N14–N17) in the vicinity of the abasic site. Type I ICLs (2) formed reversibly, and reverted to C4-AP in several hours, but the Type II ICLs (3) were stable.26 Type II ICLs are transformed into double strand breaks during nucleotide excision repair, indicating that a simple oxidized abasic lesion can give rise to the most deleterious form of DNA damage.27
Figure 1.
DNA interstrand cross-link formation from C4-AP. A) General sequence (1) used in cross-linking experiments. B) Cartoon illustrating the formation of two types of cross-links from C4-AP. C) Detailed structure of the Type I ICL formed between C4-AP and dA. D) Detailed structure of the Type II ICL formed between C4-AP and dC or dA.
Although the alkylating agents such as acrolein typically react with dG, cross-linking was not detected between C4-AP and this nucleotide regardless of its position in the duplex relative to the oxidized abasic lesion. Another unusual feature of C4-AP cross-link formation was that the DNA in the vicinity of the abasic site catalyzed the reaction. These aspects of this chemical process are explored further in this contribution using nonnative nucleotides as probes and by taking advantage of our ability to use chemical synthesis to prepare any desired DNA sequence containing C4-AP.
2. Results and Discussion
2.1 Substrate design
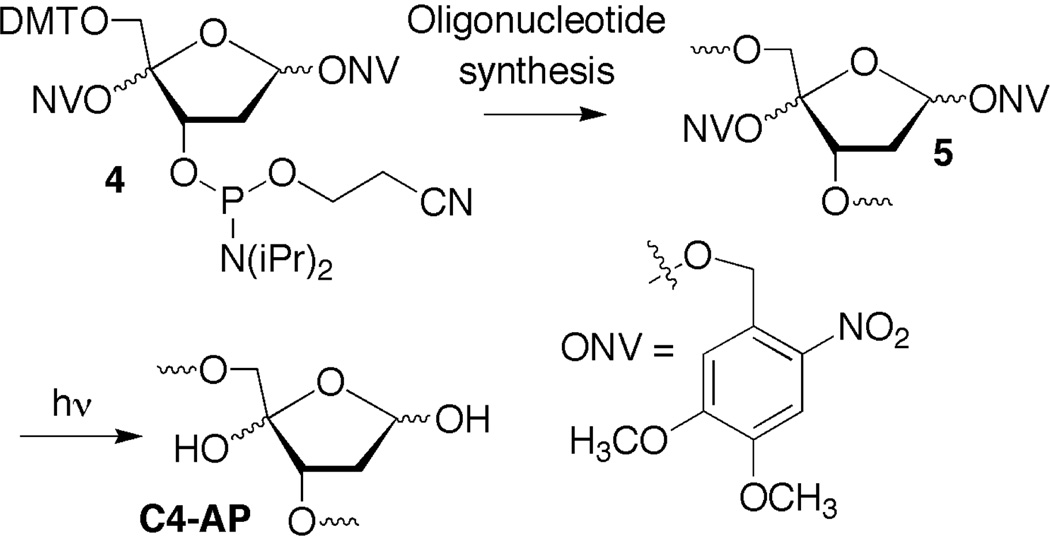
The C4'-oxidized abasic site (C4-AP) is produced by a variety of DNA oxidizing agents, but in greatest yield by bleomycin under O2 deficient conditions. However, these agents typically do not produce C4-AP at a single site in substrates longer than several nucleotides. Consequently, we and other groups have developed methods for independently producing C4-AP from stable precursors in chemically synthesized oligonucleotides.4,28–31 In this study, C4-AP was generated on an as needed basis from the photolabile precursor (5), which was incorporated into oligonucleotides from phosphoramidite 4 during solid-phase DNA synthesis (Scheme 1).4 The general sequence in 1 is part of a restriction fragment from pBR322, which possesses a hot-spot for cleavage by bleomycin.4
Scheme 1.
Generation of C4-AP in synthetic oligonucleotides.
2.2 Probing the absence of C4-AP cross-linking with dG
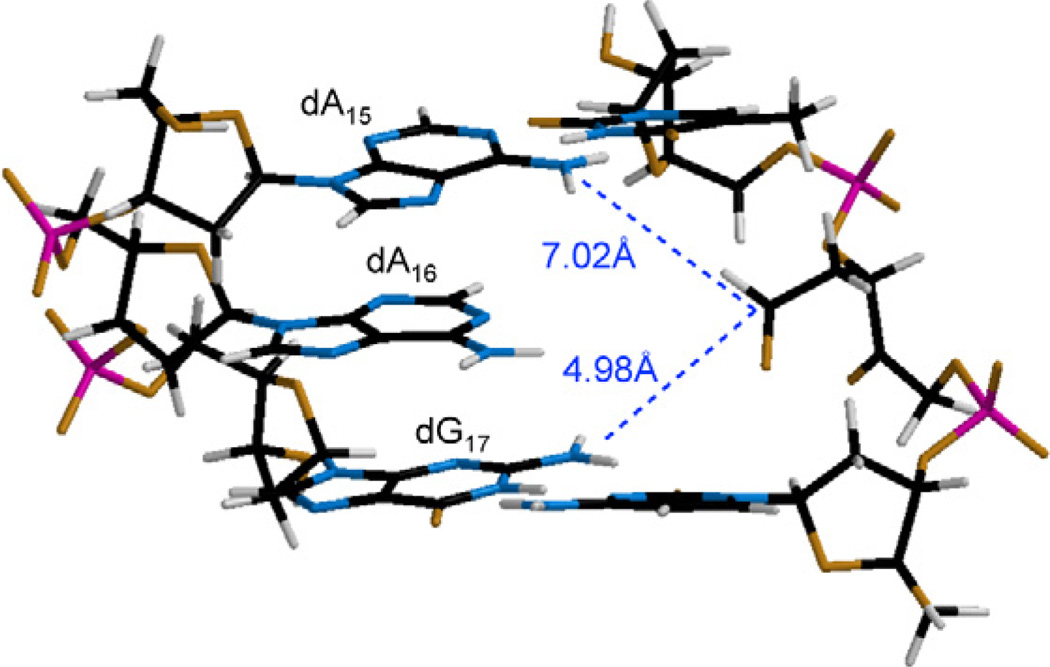
Diffusible bis-electrophiles such as acrolein, 4-hydroxynon-2-enal and but-2-ene-dial react readily with dG.16,17,32 An ICL between dG and AP was also trapped under reductive conditions.3 Although proximity influences C4-AP reactivity, it alone does not explain the lack of cross-linking between C4-AP and dG. Molecular models suggested that the N2-amino group of dG at position 17 (dG17) is ~2 Å closer to the lesion's reactive C1-aldehyde than is the N6-amine of dA at position 15 (dA15, Figure 2).26 However, interstrand cross-links occurred exclusively with the more distant N6 of dA15. Similarly, dG does not form ICLs with dioxobutane, or the bis-electrophile derived from furan oxidation, which are also 1,4-dicarbonyl molecules.5,33,34
Figure 2.
Molecular model of C4-AP showing proximity of potential nucleophiles created using Spartan®.
2.2.1 Cross-linking in duplexes containing iso-guanosine
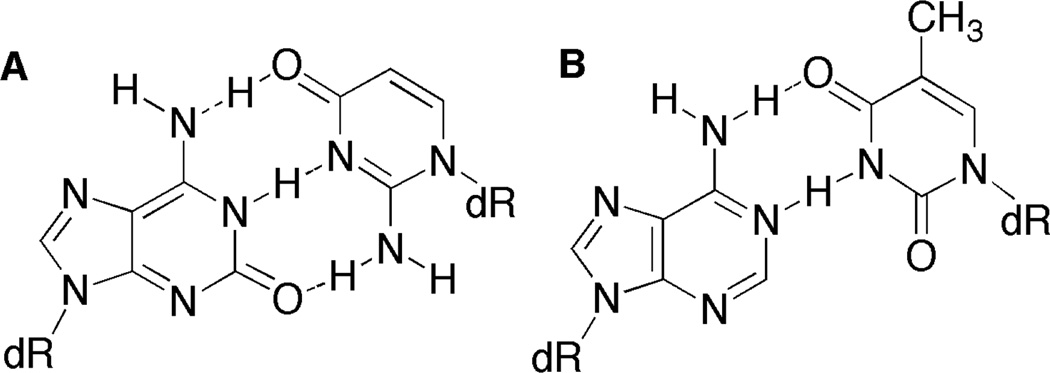
The exocyclic amine and carbonyl group in guanosine and iso-guanosine form comparable hydrogen bonds to dC and iso-guanosine (iG), respectively, but with opposite orientation with respect to the double helix. Introducing an iG:iC base pair positions the amine of iG in the major groove in approximately the same position as the N6-amine of dA (Figure 3), which forms Type I and II cross-links with C4-AP (Table 1). Iso-Guanosine was introduced into 1 at position 17 (1l, 1m) and position 15 (1c, 1d, 1g, 1h) in 1. Type I ICLs were not observed in any of the duplexes. This is consistent with previous studies on duplexes containing C4-AP and dG at the corresponding positions (1a, 1b, 1e, 1f, 1i, 1j).26 Type II ICLs formed when iG was incorporated at N17 (1l, 1m), albeit not with iso-guanosine. The relatively higher ICL yield when C4-AP was opposite dA (1l) compared to dT (1m) was consistent with the previous reported ICL catalysis by the purine nucleotide opposite the lesion (1i, 1j).4,26
Figure 3.
Base pairing of A) iso-guanosine with iso-cytosine and B) dA with dT showing similar positioning of exocyclic amines on iso-guanosine and dA in the major groove.
Table 1.
The effect of iso-guanosine and dG:dT mispairing on Type II on (3) interstrand cross-link formation.
 | |||
|---|---|---|---|
| Substrate |  |
%Yield 3a | ICL Position |
| 1ab | 5.1 ± 0.4 | A16, C17, (0.9:1) | |
| 1bb | 0.7 ± 0.1 | C17 | |
| 1c | 7.2 ± 1.1 | iG15 | |
| 1d | 2.0 ± 0.1 | C17, iG15, (2.3:1) | |
| 1eb | 28.8 ± 0.8 | A16, C17, (1.3:1) | |
| 1fb | 1.6 ± 0.3 | C17 | |
| 1g | 18.4 ± 0.5 | iG15 | |
| 1h | 3.7 ± 0.3 | iG15 | |
| 1ib | 49.3 ± 1.6 | A16 | |
| 1jb | 0.3 ± 0.1 | Not determined | |
| 1k | 36.5 ± 1.5 | A16, C14 (1.1) | |
| 1l | 33.2 ± 2.1 | A16, C14, (1.3:1) | |
| 1m | 0.9 ± 0.1 | C14 | |
Yields are an average of at lease 3 replicates.
Data take from ref. 26.
The lack of reactivity with iG when it is present at dN17 is not surprising, given the positioning of the exocyclic amine. For comparison, ICLs do not form with dA17 either. iG is better positioned to react with C4-AP when it is incorporated at dN15 (1c, 1d, 1g, 1h) because its exocyclic amine is in a position comparable to the N6-amino group of dA15, where cross-links do form. Type II ICLs (3) are produced in each of these duplexes and iG15 is involved in all cases (Table 1). In contrast, no reaction is observed with dG15 in comparable duplexes (1a, 1b, 1e, 1f). Type II ICL (3) yields increase when iG15 is mispaired with dT (1g versus 1c, 1h versus 1d) and all cross-linking occurs with the nonnative nucleotide. These data suggest that iG accessibility improves its reactivity. However, we do not know whether this is due to greater access to its Watson-Crick face in the mispair and/or greater dynamic flexibility overall.
The increased iG reactivity when part of a mispair (1g, 1h) led us to examine the reactivity of dG17 when it too is opposite dT (1k). Type II ICL (3) formation was similar to that when a dC:dG base pair was present at this position (1i). The similarity included the absence of any reactivity at dG17. However, dT46:dG17 containing DNA (1k) also yielded Type I ICLs (2, 6.8 ± 0.1 %). Moreover, cross-linking occurred at dG17. (Cross-linking sites were determined using hydroxyl radical cleavage.) This is the only sequence of the more than 30 examined in which a Type I ICL was observed between C4-AP and a native nucleotide other than dA This observation is also consistent with the idea that the lack of reactivity previously observed with dG is partially attributable to relative inaccessibility to its nucleophilic functional groups, perhaps due to stronger base pairing with dC than that observed in a dA:dT base pair.26
2.2.2 Cross-linking in duplexes containing 2-aminopurine: thymidine base pairs
Cross-linking by iG15 and dT46:dG17 led us to introduce 2-aminopurine (2AP) at the comparable locations (1p, 1q, 1s, Table 2). 2AP17 positions the exocyclic amine in the vicinity where the N2-amino group of dG17 resides. Type I and II cross-links form in duplexes containing 2AP17 when C4-AP is opposite dA or dT (1p, 1q, Table 2). As in other instances involving comparable duplexes containing dA17 instead of 2AP17 (e.g. 1n, 1o), ICL yields are higher when C4-AP is opposite dA (1p) than dT (1q). The subtleties of local duplex structure are evidenced by the distribution of cross-links. Type II cross-links (3) form at dC14 and dA16 rather than at 2AP17 when C4-AP is opposite dA (1p), even though the overall yield is more than 20%. This reactivity is consistent with an otherwise identical duplex containing dA17 (1n, Table 2). In contrast, a low yield of Type II ICL at 2AP17 is produced when dT is opposite C4-AP (1o) even though none form at dA17 when 1o reacts.
Table 2.
The effect of 2-aminopurine on interstrand cross-link formation.
 | |||
|---|---|---|---|
| Substrate |  |
% Yield 2 (Pos.)a | % Yield 3 (Pos.)a |
| 1nb | Not observed | 21.0 ± 0.6 (A16) | |
| 1ob | Not observed | 0.7 ± 0.1 (ND) | |
| 1p | 14.8 ± 1.2 (2AP17) | 20.5 ± 0.2 (A16, C14, 1.2:1) | |
| 1q | 2.7 ± 0.4 (2AP17) | 1.3 ± 0.2 (2AP17) | |
| 1rb | 21.3 ± 0.9 (A15) | 7.3 ± 1.0 (A15) | |
| 1s | 3.9 ± 0.1 (ND) | 18.8 ± 1.1 (C17, C14, 1:1) | |
Yields are an average of at lease 3 replicates.
Data take from ref. 26.
The formation of Type I ICLs (2) at 2AP17 from 1p and 1q is most distinctive (Table 2). 2-Aminopurine is the only correctly base paired nucleotide that forms a Type I ICL at position 17. The importance of the positioning 2AP is underscored by the lack of cross-linking with this nucleotide when it is incorporated at position 15 (1s, Table 2). Although no cross-linking involving 2AP15 (1s) is observed, Type I ICLs readily form with dA15 (1r).
2.2.3 Adenine catalysis of Type II cross-link formation in duplexes containing 2-aminopurine or iso-guanosine
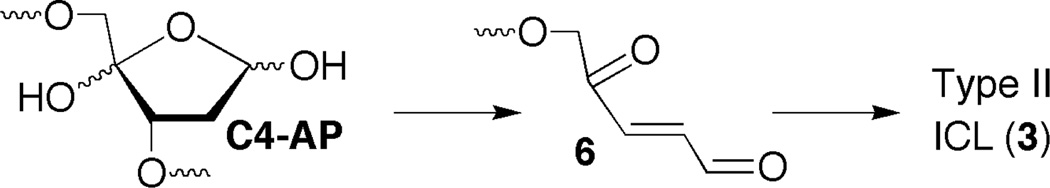
Previous C4-AP cross-linking experiments (e.g. 1b, 1j, 1o, Table 3) showed that the addition of adenine and other purines that contain exocyclic amines increased the yield of Type II ICLs when C4-AP was opposite thymidine.26 The effect of adenine saturates at ~1 mM, consistent with binding of the purine by the C4-AP containing duplex.4 The purines were proposed to increase the rate constant of the rate determining β-elimination of C4-AP to 6 (Scheme 2), which then reacts with one or more nucleotides on the opposite strand. Adenine has a similar effect on Type II ICLs (3) formed from duplexes containing 2AP (1q) or iG (1d, 1h, 1m, Table 3). In instances for which data are available from the reaction of comparable duplexes containing native nucleotides (e.g. 1o versus 1q, 1b versus 1d, and 1j versus 1m), the relative increases in yield of 3 upon addition of adenine are very similar. These data suggest that the formation of Type II ICLs from duplexes containing the nonnative nucleotides, 2AP and iG proceed via the same mechanism through 6 as those containing dA and dG, respectively.
Table 3.
The effect of adenine on Type II (3) interstrand cross-link formation.
 | |||
|---|---|---|---|
| % Yield 3a | |||
| Substrate |  |
No Adenine | 0.1 mM Adenine |
| 1ob | 0.7 ± 0.1 | 4.2 ± 0.8 | |
| 1q | 1.3 ± 0.2 | 5.2 ± 0.8 | |
| 1bb | 0.7 ± 0.1 | 12.2 ± 0.8 | |
| 1d | 2.0 ± 0.1 | 32.2 ± 1.8 | |
| 1jb | 0.3 ± 0.1 | 3.3 ± 0.4 | |
| 1m | 0.9 ± 0.1 | 9.1 ± 0.1 | |
| 1h | 3.7 ± 0.3 | 14.1 ± 1.0 | |
Yields are an average of at lease 3 replicates.
Data take from ref. 26.
Scheme 2.
Type II ICL (3) formation via β-elimination from C4-AP.26
2.2 Probing the catalysis of ICL formation via an opposing dA using 7-deaza-2'-deoxyadenosine (7dA)
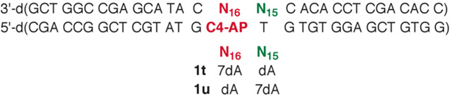
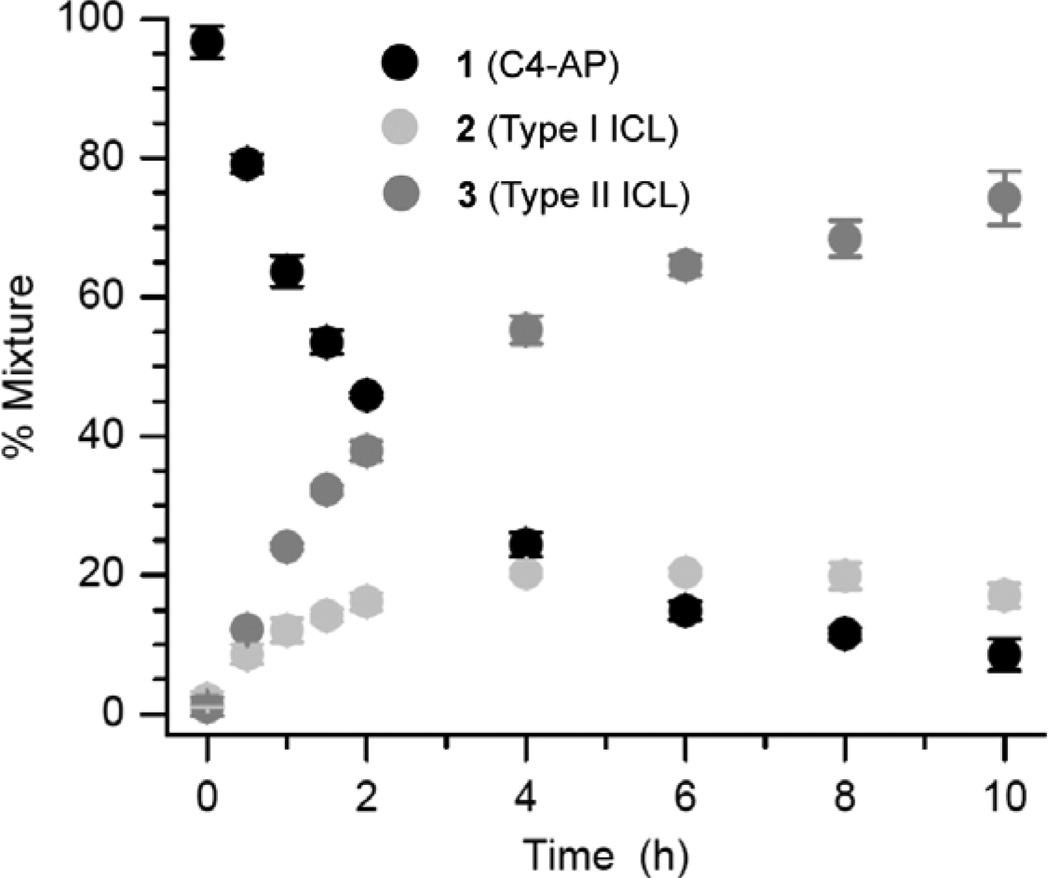
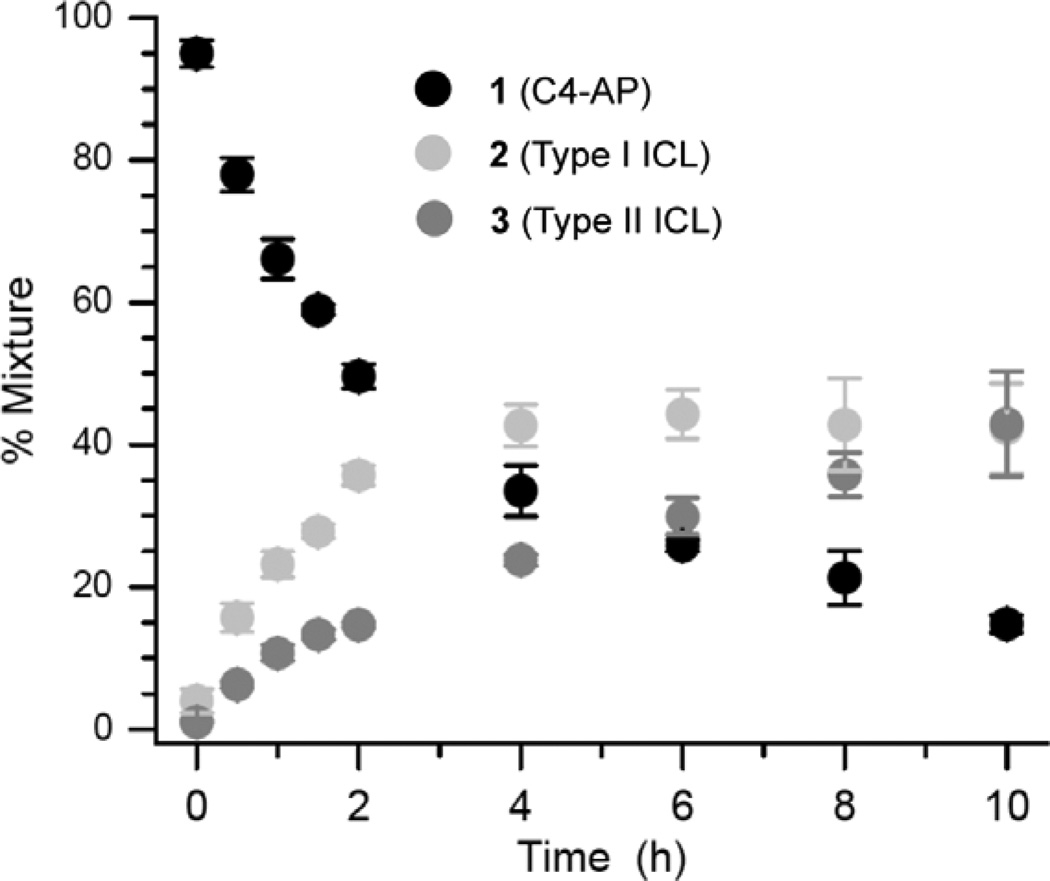
The role of the dA opposing C4-AP as a catalyst for ICL formation was probed further by preparing duplexes containing 7-deaza-2'-deoxyadenosine (7dA). 7dA has a higher pKa (5.2) than dA (3.5).35,36 A greater fraction of 7dA molecules will be protonated at pH 7.2 where the reaction is carried out, suggesting that it would be a more effective acid-base catalyst. Consequently, 7dA was substituted for dA16 in 1r (1t). ICL formation was followed in 1t over the course of 10 h (Figure 4). The amount of Type I ICL (2) reached a maximum in ~4 h and then slowly decreased. This is consistent with the reactions of other duplexes containing C4-AP, and is an indication that 2 forms reversibly.4,26 Type II ICL (3) formed exclusively with dA15 in 1t. Its yield reached more than 70% in 10 h, which is more than twice as great as when dA is opposite C4-AP (1r) in an otherwise identical sequence.
Figure 4.
The effect of 7-deaza-2'-deoxyadenosine (7dA) on cross-link formation when it is opposite C4-AP (1t).
Placing 7dA at position 15 (1u), which is where cross-linking occurs when dA is present (1r, Table 2), also alters ICL formation (Figure 5). The overall rate at which the starting material disappears is comparable to when 7dA is opposite C4-AP (1t, Figure 4). However, the distribution of ICL products is different. The Type I ICL (2, Figure 5) is formed more rapidly and in higher yield (at the expense of 3) than when 7dA is opposite C4-AP. Furthermore, Type II cross-links (3) are distributed over dA16 and dC17, as well as at 7dA15 in 1u.
Figure 5.
The effect of 7-deaza-2'-deoxyadenosine (7dA) on cross-link formation involving C4-AP when it is opposite the 3'-adjacent thymidine (1u).
3. Conclusions
Interstrand cross-link formation involving C4-AP occurs in cells subjected to bleomycin or ionizing radiation, and was detected at low levels in cells not exposed to oxidative stress.18 Chemical studies using duplexes containing C4-AP at specific sites indicated that the formation of cross-links with the surrounding nucleotides was dependent upon the local sequence. However, reaction with dG did not occur even though this nucleotide reacts with a number of diffusible electrophiles. This surprising lack of reactivity with dG was investigated using the nonnative nucleotides iso-guanosine (iG) and 2-aminopurine (2AP). The data indicate that the lack of dG cross-linking to C4-AP is attributable to two factors 1) poor positioning of its N2-amino group when it is opposite the nucleotide bonded to the 3'-phosphate of C4-AP (dG15), and 2) overall reduced reactivity compared to dA and 2AP. The latter is at least partially due to stronger base pairing with dC compared to dA (and 2AP) with dT, as evidenced by the increased cross-linking when dG or iG are mispaired.
Experiments using duplexes containing 7-deaza-2'-deoxyadenosine (7dA) reinforce the hypothesis that the neighboring nucleotides affect ICL formation by catalyzing the elimination of C4-AP. How the nucleotide opposite C4-AP catalyzes ICL formation is uncertain. It may function as an acid or base catalyst, or a nucleophilic one via Schiff-base formation. However, it is unlikely that the opposing nucleotide functions as a catalyst independent of the local sequence. 2'-Deoxycytidine (pKa = 4.2) induces Type II ICL (3) formation less efficiently than dA (pKa = 3.5), suggesting that other properties, such as base stacking may influence reactivity.36
4. Experimental
General Methods
Oligonucleotides were synthesized on an Applied Biosystems Incorporated 394 oligonucleotide synthesizer. Commercially available oligonucleotide synthesis reagents, including the phosphoramidite for 7dA, iG, iC, and 2AP were purchased from Glen Research (Sterling, VA). T4 polynucleotide kinase and terminal deoxynucleotide transferase were obtained from New England Biolabs. γ-32P-ATP and α-32P-cordycepin 5′-triphosphate were purchased from Perkin Elmer. C18-Sep-Pak cartridges were obtained from Waters. Quantification of radiolabeled oligonucleotides was carried out using a Molecular Dynamics Phosphorimager 840 equipped with ImageQuant TL software. All photolyses of oligonucleotides were carried out in clear eppendorf tubes in a Rayonet photoreactor (RPR-100) fitted with 16 lamps having an output maximum at 350 nm. Syntheses of DNA substrates containing 5 were carried out as previously reported.4
General procedure for ICL formation in C4-AP containing duplexes
DNA (200 nM) in PBS buffer (100 mM NaCl and 10 mM sodium phosphate, pH 7.2) was photolyzed for 30 min at room temperature. One part DNA was then diluted to 100 nM with one part PBS buffer or a 200 mM solution of adenine in PBS buffer. These reactions were typically incubated at 37 °C for 10 h. For time course experiments, a 3 µL aliquot was removed every 2 h. Aliquots were mixed with an equal volume of formamide loading buffer and the DNA was resolved using 20% denaturing polyacrylamide gel electrophoresis (PAGE).
Determination of ICL location using the hydroxyl-radical cleavage reaction
Duplex DNA constructed of a C4-AP containing strand and a 3′-labeled complement was incubated at 37°C overnight in PBS. Note: low yielding ICL reactions were incubated for as long as three days. The reaction was concentrated and purified by 20% analytical (0.4 mm thickness) PAGE. The bands corresponding to the ICL products were carefully cut from the gel, crushed, and eluted in 500 µL of water for 1 h at room temperature with periodic vortexing. In some cases, Type II ICLs (3) were evident as multiple resolvable bands and were isolated independently. The solution was filtered using a 10 mL Polyprep Column (Biorad) and desalted using a C18 column (100 mg Sep-Pak). After concentration, a portion of the DNA (30,000–60,000 cpm was dissolved in 8 µL of water and mixed with 10 µL of the 2 × oxidation buffer (20 mM NaCl, 20 mM sodium phosphate, pH 7.2, 2 mM sodium ascorbate, and 1 mM hydrogen peroxide). To initiate the reaction, 2 µL of the Fe•EDTA solution (1 mM EDTA and 0.5 mM Fe(NH4)2(SO4)2•6H2O) was added, and the reaction was allowed to proceed for no longer than 3 min. The reaction was quenched with 10 µL of 100 mM thiourea and concentrated. The DNA was then suspended in formamide loading buffer and separated by 20% PAGE.
Acknowledgments
We are grateful for support of this research from the National Institute of General Medical Sciences (GM-063028).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Gates KS. Chem. Res. Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatgilialoglu C. Wiley Ser. React. Intermed. Chem. Biol. 2009;2:99–133. [Google Scholar]
- 3.Dutta S, Chowdhury G, Gates KS. J. Am. Chem. Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sczepanski JT, Jacobs AC, Greenberg MM. J. Am. Chem. Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 5.Guan L, Greenberg MM. J. Am. Chem. Soc. 2009;131:15225–15231. doi: 10.1021/ja9061695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Proc. Natl. Acad. Sci. U. S. A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumari A, Minko IG, Harbut MB, Finkel SE, Goodman MF, Lloyd RS. J. Biol. Chem. 2008;283:27433–27437. doi: 10.1074/jbc.M801237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumaresan KR, Sridharan DM, McMahon LW, Lambert MW. Biochemistry. 2007;46:14359–14368. doi: 10.1021/bi7015958. [DOI] [PubMed] [Google Scholar]
- 9.Smeaton MB, Hlavin EM, McGregor Mason T, Noronha AM, Wilds CJ, Miller PS. Biochemistry. 2008;47:9920–9930. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Räschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Schärer OD, Walter JC. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panasci L, Xu ZY, Bello V, Aloyz R. Anticancer Drugs. 2002;13:211–220. doi: 10.1097/00001813-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Zietlow L, Smith LA, Bessho M, Bessho T. Biochemistry. 2009;48:11817–11824. doi: 10.1021/bi9015346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy DR, Lu J, Shen B, Beerman TA. Proc. Nat. Acad. Sci. USA. 2007;104:17632–17637. doi: 10.1073/pnas.0708274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kothandapani A, Dangeti VSMN, Brown AR, Banze LA, Wang X-H, Sobol RW, Patrick SM. J. Biol. Chem. 2011;286:14564–14574. doi: 10.1074/jbc.M111.225375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozekov ID, Turesky RJ, Alas GR, Harris CM, Harris TM, Rizzo CJ. Chem. Res. Toxicol. 2010;23:1701–1713. doi: 10.1021/tx100179g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stone MP, Cho Y-J, Huang H, Kim H-Y, Kozekov ID, Kozekova A, Wang H, Minko IG, Lloyd RS, Harris TM, Rizzo CJ. Acc. Chem. Res. 2008;41:793–804. doi: 10.1021/ar700246x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regulus P, Duroux B, Bayle P-A, Favier A, Cadet J, Ravanat J-L. Proc. Nat. Acad. Sci. USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama H, Xu C, Murugesan N, Hecht SM. J. Am. Chem. Soc. 1985;107:4104–4105. [Google Scholar]
- 20.Rabow LE, Stubbe J, Kozarich JW. J. Am. Chem. Soc. 1990;112:3196–3203. [Google Scholar]
- 21.Xi Z, Goldberg IH. In: Comprehensive Natural Products Chemistry. Kool ET, editor. Vol. 7. Amsterdam: Elsevier; 1999. pp. 553–592. [Google Scholar]
- 22.von Sonntag C. The Chemical Basis of Radiation Biology. London: Taylor & Francis; 1987. [Google Scholar]
- 23.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Biochemistry. 2004;43:13621–13627. doi: 10.1021/bi048337r. [DOI] [PubMed] [Google Scholar]
- 24.Guan L, Greenberg MM. J. Am. Chem. Soc. 2010;132:5004–5005. doi: 10.1021/ja101372c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guan L-R, Bebenek K, Kunkel TA, Greenberg MM. Biochemistry. 2010;49:9904–9910. doi: 10.1021/bi101533a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. J. Am. Chem. Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sczepanski JT, Jacobs AC, Van Houten B, Greenberg MM. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Gil JM, Greenberg MM. Angew. Chem. Int. Ed. 2003;42:5882–5885. doi: 10.1002/anie.200352102. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Weledji YN, Greenberg MM. J. Org. Chem. 2004;69:6100–6104. doi: 10.1021/jo049033d. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Stubbe J. Biochemistry. 2004;43:5278–5286. doi: 10.1021/bi0495376. [DOI] [PubMed] [Google Scholar]
- 31.Aso M, Usui K, Fukuda M, Kakihara Y, Goromau T, Suemune H. Org. Lett. 2006;8:3183–3186. doi: 10.1021/ol060987v. [DOI] [PubMed] [Google Scholar]
- 32.Chen B, Bohnert T, Zhou X, Dedon PC. Chem. Res. Toxicol. 2004;17:1406–1413. doi: 10.1021/tx049818e. [DOI] [PubMed] [Google Scholar]
- 33.Op de Beeck M, Madder A. J. Am. Chem. Soc. 2011;133:796–807. doi: 10.1021/ja1048169. [DOI] [PubMed] [Google Scholar]
- 34.Stevens K, Claeys DD, Catak S, Figaroli S, Hocek M, Tromp JM, Schürch S, Van Speybroeck V, Madder A. Chem. - Eur. J. 2011;17:6940–6953. doi: 10.1002/chem.201100067. [DOI] [PubMed] [Google Scholar]
- 35.Suydam T, Levandoski SD, Strobel SA. Biochemistry. 2010;49:3723–3732. doi: 10.1021/bi100234v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saenger W. Principles of Nucleic Acid Structure. New York: Springer Verlag; 1983. [Google Scholar]