Abstract
Hematopoietic cell transplantation (HCT) is now a curative option for certain categories of patients with hematological malignancies and other life-threatening illnesses. Technical and supportive care has resulted in survival rates that exceed 70% for those who survive the first two years after HSCT. However, long-term survivors carry a high burden of morbidity, including endocrinopathies, musculoskeletal disorders, cardiopulmonary compromise, and subsequent malignancies. Understanding the etiologic pathways that lead to specific post-HCT morbidities is critical to developing targeted prevention and intervention strategies. Understanding the molecular underpinnings associated with graft vs. host disease (GvHD), organ toxicity, relapse, opportunistic infection and other long-term complications now recognized as health care concerns will have significant impact on translational research aimed at developing novel targeted therapies for controlling chronic GvHD, facilitating tolerance and immune reconstitution, reducing risk of relapse and secondary malignancies, minimizing chronic metabolic disorders and improving quality of life. However, several methodological challenges exist in achieving these goals; these issues are discussed in detail in this article.
Introduction
Progress in hematopoietic cell transplantation (HCT) over the last 30–40 years has resulted from improvements in several areas, notably better supportive care and less toxic conditioning regimens. It is for this reason that HCT is being increasingly offered as a curative option for patients with hematological malignancies and other life-threatening illnesses. Advances in how HCT is performed and how patients are supported in the immediate post-HCT period, have resulted in survival rates that exceed 70% for those who survive the first two years after HCT.(1–3) However, full restoration of health does not necessarily accompany cure or control of the underlying disease. Endocrinopathies, musculoskeletal disorders, cardiopulmonary compromise, and subsequent malignancies are well-described in HCT survivors.(4–13) In fact, the cumulative incidence of a chronic health condition among HCT survivors is reported to be 59% (95% CI, 56%-62%) at 10 years after HCT; for severe/life-threatening conditions or death due to chronic health conditions, the 10-year cumulative incidence approached 35% (95%CI, 32%-39%).(14) Furthermore, HCT survivors with chronic graft versus host disease (GvHD) are 5 times as likely to develop severe/life threatening conditions. Understanding the burden of morbidity carried long-term by the HCT survivors is important to the healthcare providers and policy makers in identifying and procuring resources for the long-term care of these patients; it is important to the researchers in identifying etiologic pathways that lead to the morbidity; and finally it is important to the HCT survivors in helping them make informed decisions regarding the quality of life concerns long-term after HCT.
Understanding the etiology and pathogenesis of long-term complications in survivors of HCT performed in childhood is important for a variety of reasons. It is well-established that exposure to genotoxic agents during the earlier years of life has a significant impact on the structure and function of the developing organs. Examples of this abound in the children treated with conventional therapy, and include the higher risk of anthracycline-related cardiotoxicity, of radiation-related cognitive impairment, and of radiation-related second malignancies when the exposure occurs at a younger age. However, large-scale studies attempting a comprehensive understanding of the etiology and pathogenesis of long-term complications after HCT performed in childhood are lacking. There are several methodological challenges that have precluded the conduct of such studies, the first and foremost is the fact that large cohorts of patients need to be assembled to address these issues. The following sections describe the methodological challenges in the conduct of such studies, drawing upon studies conducted in adult populations as examples of how such studies can be accomplished.
Magnitude of risk of chronic medical conditions after HCT
In order to describe the magnitude of risk of chronic medical conditions after HCT with precision, it is critical to assemble large cohorts of children who have undergone HCT, with near complete follow-up. The emphasis on large cohorts is due to the fact that HCT survivors represent a heterogeneous population; therefore, understanding the magnitude of risk in homogeneous sub-populations necessitates the assembly of a large cohort. Most of the large studies have utilized registry data. Registry data is limited by the passive reporting of the outcome of interest, inability to validate the outcome, and incomplete follow-up of the patients, thus resulting in imprecise measurements of risk. These limitations can be overcome by the establishment of a consortium of institutions dedicated to the study of late effects and with available resources to ensure complete and accurate follow-up of the patients under consideration.
Comprehensive exposure assessment
Studies focusing on children treated with conventional therapy have established the role of therapeutic exposures as the primary etiologic factor in the development of long-term complications quite conclusively. This is exemplified by the dose-dependent association between anthracyclines and congestive heart failure; radiation and secondary breast cancer or brain tumors or thyroid cancer. It would therefore follow, that adverse events after HCT would be due in part to the pre-HCT therapeutic exposures, HCT-related conditioning, and post-HCT exposures (chemotherapy, radiation or immunosuppressive therapy). However, for the most part, previous reports describing post-HCT complications have failed to take into account the pre-HCT therapeutic exposures, thus increasing the risk of creating imperfect associations between the HCT process and the outcome of interest, when the true association was in part related to the pre-HCT exposures. Obtaining pre-HCT exposures can be challenging for a large cohort of HCT survivors, and one way of overcoming this challenge is to conduct a nested case-control study within the cohort – with abstraction of detailed pre-HCT therapeutic exposure limited to the cases and matched controls.
Validation of outcomes of interest
When assessing outcomes of interest, it is critical that methodologies be established upfront as to how these outcomes will be identified and whether there is a process in place for validating the outcome. Under ideal circumstances, there would be stringent criteria established up front to validate the outcome of interest. Typically, this would include a pathology report (with cytogenetics where appropriate) for second malignancies, and echocardiographic evidence of cardiomyopathy (with established cut offs in ejection fraction and fractional shortening for defining cardiac dysfunction). If the outcome of interest is elicited by self-report, it should be stated clearly in the report, along with the attendant limitations.
Develop clear relation between exposure and outcome
Utilization of the methodology described above will allow the establishment of a clear relation between exposure and outcome, and will also allow a clear delineation of the independent contribution of pre-HCT vs. HCT-related vs. post-HCT exposures. Having such a relationship then allows the exploration of the inter-individual variability in risk, given constant exposure. Such inter-individual variability is clearly observed in patients treated with anthracyclines, where doses exceeding 1000 mg/m2 are tolerated by some, while low doses (<150 mg/m2) result in congestive heart failure in the others. Identifying those at highest risk upfront becomes critical in determining the best therapy for the individual patient, maximizing the chance of cure, while minimizing the long-term toxicity – the basis for “personalized medicine”. In addition, understanding the pathogenetic mechanism underlying the development of an outcome of interest, given a therapeutic exposure would allow for the development of novel therapeutic interventions, allowing for early detection and potential reversal of the process.
Advances in genetics research has also been a significant contributing factor in defining the human major histocompatibility system (HLA) and high impact translational research that has guided donor selection and refined through the development of DNA-based tools for high resolution genotyping robust criteria for the optimal selection of HLA matched unrelated donors.(15, 16) However, despite precise matching for HLA, graft-versus-host (GVHD) disease, opportunistic infection and other complications remain significant obstacles to safety and overall success. Genetic polymorphism is responsible not only for HLA mismatching between donor and recipient, but also for the mismatching of a potentially large number of minor histocompatibility antigens encoded by genes located across the genome.(17, 18) Genetic variants encoding minor histocompatibility antigens include single nucleotide polymorphisms (SNPs) responsible for amino acid substitutions in cellular proteins that can function as minor histocompatibility antigens,(18) or deletions in genes that abrogate protein production and thereby alter donor and recipient disparity for minor histocompatibility antigens as illustrated in a recent paper by McCarroll et al.(19) In addition to genetic polymorphism causing recipient and donor disparity, certain polymorphisms can also affect gene function. Many SNPs have been identified, both intergenic and in nearby promoter regions, that modify gene function by changing expression levels, modify functional amino acid substitutions, or cause alternative splicing of gene transcripts all of which may alter gene function.(20)
In order to develop a deeper understanding of the molecular underpinnings of therapy-related long-term complications, it becomes important to collect appropriate biospecimens from the patients who do and do not develop the outcome. Ideally, this should be in the form of blood, collected to allow subsequent extraction of DNA and RNA, as well as establish lymphoblastoid cell lines. In patients undergoing allogeneic HCT, the blood should be collected prior to HCT, in order to reflect the DNA of the host. Study of GvHD ideally would require the collection of paired DNA from the host and donor. There are two approaches to the study of genetic variation in disease: (1) candidate gene studies based on the selection of a limited number of tag SNPs for analyzing specific genes and pathways; and (2) genome-wide association studies (GWAS) using DNA arrays capable of detecting a million or more SNPs.
Both approaches have attendant strengths and limitations. Candidate gene studies are both complementary and additive to genome-wide studies. Table 1 compares the characteristics of both strategies. A GWAS approach offers the ability to study complex pathways, allowing for an assessment of the action/ interaction of many genes; it also allows for new genes to be identified. GWAS has gained significant favor, partly due to the fact that several studies that have utilized a candidate gene approach have failed replication. However, a GWAS approach requires a large sample size in order to account for false discovery. In addition, there is a need for a replication cohort so that the genes identified in the discovery set can be validated in the test set.
Table 1.
Comparison of Candidate Gene and Genome-Wide Study Approaches
| Candidate Gene Studies | Genome-Wide Studies |
|---|---|
| Rational selection of genes for study increases biological plausibility of findings, however novel genes will not be identified. | Analysis of whole genome allows identification of genes not previously thought to be important for the outcome in question. |
| Limited number of genes reduces the possibility of false discovery. | Large number of analyses means a high risk of false discovery. |
| Expense reduced if candidate genes selected prove to be relevant to outcome. | Increased expense associated with following up the large number of positive findings. |
| Moderate sample size may be sufficient | Very large sample sizes are needed because of the large number of comparisons being made. |
The GWAS approach does not have an a priori hypothesis, and is considered to be hypothesis-generating and more suited for complex disorders where clear etiologic lead is not established. However, this is not true for many post-HCT outcomes, where for the most part there is a clearly established etiologic association between the exposure and outcome (e.g., radiation and subsequent malignancies). In such cases, an argument could be made for a comprehensively selected (and biologically plausible) list of genes identified along the path of the action of radiation on the target organ. As mentioned earlier, GWAS is limited by the need for a large sample size due to issues related to multiple testing – this is less of an issue with a candidate gene approach. Finally, gene-gene interactions may require prohibitively large samples in a GWAS setting, but are logistically feasible when conducting a candidate gene study.
Candidate gene polymorphisms and late effects after HCT
Genetic polymorphisms can influence outcome of transplant by modifying the function of the gene, and many studies have looked at genetic variants that modify expression of key cytokines, a plausible mechanism for modification of outcome. The first studies commonly explored one variant, and many included relatively small numbers of heterogeneous cases. Many (perhaps most) of the positive findings of these studies have not been replicated in later studies. Difficulties in replication may indicate that the original findings were simply statistical chance, that the finding is true, but specific to particular populations or transplant strategies, or that there may be gene-gene interactions that modify observations. In addition to modifying gene function, polymorphisms may serve as minor antigens. There are several examples of studies that have utilized the candidate gene approach in order to understand molecular underpinnings of disease. These examples are illustrated here.
Analysis of GvHD
Several candidate gene studies have generated data showing that genetic variation can affect the risk of GvHD. More than 26 publications have appeared between 1998 to 2009 reporting associations of donor or recipient genotypes with the risk of acute GvHD for 30 candidate SNPs in 16 genes including: CD31, CTLA4, FAS, HSPA1L, IL1α, IL1β, IL2, IL23R, IL6, IL10,(21) IL10RB,(22) MADCAM1, MTHFR, NOD2, TGFβ1, TNF, TNFRII and VEGFα. Previous candidate gene studies however have been generally limited to relatively small study populations of a few hundred patients and donors, and results have not been consistent. Nevertheless, the resulting publications have indelibly impacted the research agenda and demonstrated the need for more comprehensive and robust approaches to identifying specific genetic variation associated with transplant outcome. Clinical risk assessment models have advanced significantly in the last few years, especially in the identification and proper weighing of relevant clinical variables, but the “genetic effect” remains largely undefined and unmeasured.
Analysis of opportunistic infection and AFO
In addition to acute GvHD, candidate gene studies have been used to productively explore genetic risk factors in other clinically important complications of HCT. The Seattle group has used the candidate gene approach in an analysis of gram negative bacteremia which identified variation in the LBP gene associated with risk of gram negative bacteria and death,(23) fungal disease implicating variation in the TLR4 gene with risk of invasive aspergillosis,(24) and air flow obstruction (AFO) disease of the lungs.(25) AFO, also known as bronchiolitis obliterans syndrome, is a serious and often fatal complication of chronic GvHD that compromises pulmonary function and quality of life.(26–28) Fifteen candidate genes involved in the innate immunity pathway were examined in a discovery cohort of 363 patient-donor pairs. Significant associations were found in multivariate models for two SNPs in the BPI gene of the patient (SNP 33065, p=0.038; and SNP 36045, p=0.025); and this association was confirmed in an independent validation cohort, with 9 BPI SNP-defined risk haplotypes identified (p-values 0.013 to 0.043).(25)
Candidate gene studies exploring genetic associations with complications and late effects of transplantation have been reported for more than 15 years, and yet the findings have yet to be included in clinical practice. Technology to perform genotyping on a large scale has advanced rapidly, and in common with candidate gene studies, has perhaps surpassed our ability to interpret results and incorporate them into clinical practice. The challenge of understanding the impact of polymorphisms on outcomes is illustrated by the complexities of the genetic polymorphisms in the gene CD31.
Behar and colleagues reported a case-control study in 1996, showing that disparity between recipient and donor for a polymorphism (SNP 125) in CD31 increased the risk of GvHD (Table 2).(29) A larger study published in the same year showed no effect of the variant.(30) Later, studies performed in Japan and elsewhere, suggested that SNP 125 was in linkage equilibrium with other SNPs (SNP 563/670) and that these linked variants were driving the association,(31, 32) although a study of unrelated donor cord blood transplants showed no effect.(33) A more recent British study found that disparity for CD31 polymorphisms did not associate with outcomes, but that genotype for SNP 125 was associated with risk of GvHD.(34) These authors suggest that the variant CD31 genotype had an effect on T-cell function. These papers, which include 15 years of work, offer no clarity regarding the clinical importance or mechanism of action of variants in a single gene, illustrating the complexity of translation of laboratory findings into clinical practice.
Table 2.
Studies Reporting the Association of CD31 Polymorphisms with GVHD Risk
| Ref | Stem Cell Source | # | Conclusion |
|---|---|---|---|
| Behar et al, NEJM, 1996 | Sibling marrow, Stanford, US | 46 | CD31 SNP125 is an important minor antigen |
| Nichols et al, Blood 1996 | Sibling marrow, US Multi-center | 301 | CD31 SNP 125 is NOT an important minor antigen |
| Maruya et al. Blood, 1998 | Sibling marrow, Japan | 118 | CD31 MIGHT be an important minor antigen due to SNPs 563/670 |
| Grumet et al, BBMT, 2001 | Sibling marrow, Stanford, US | 120 | CD31 IS a minor antigen and polymorphism at SNPs 563/670 is more important than SNP 125 |
| Rocha et al, Blood, 2002 | Sibling marrow, France | 107 | CD 31 polymorphism at SNPs 563/670 is a minor antigen |
| Kogler et al. Transplantation, 2002 | Unrelated donor cord blood | 115 | CD31 polymorphism at SNP125 has no impact on clinical outcomes |
| Balduini et al, Br. J Haem, 2001 | Sibling marrow, Italy | 150 | CD 31 polymorphism at SNP 563 is a minor antigen |
| Goodman et al. Bone Marrow Transplantation, 2005 | Sibling marrow, British | 85 | No effect of CD31 SNPsl25 or 563/670 as a minor antigen, but CD 125 genotype correlated with GVHD risk suggesting a functional effect. |
References (for Table 2)
Balduini CL, Francesco F, Noris P et al. Donor-recipient Incompatibility at CD31-codon 563 is a major risk factor for acute graft-versus-host disease after allogeneic bone marrow transplantation from a human leucocyte antigen-matched donor. Br J Haematol 2001; 114: 951.
Behar E, Chao NJ, Hiraki DD et al. Polymorphism of adhesion molecule CD 31 and it’s role in graft versus host disease. N Engl J Med 1996;334: 286–91.
Goodman RS, Ewing J, Evans PC, Donor CD31 genotype and its association with acute graft-versus-host disease in HLA identical sibling stem cell transplantation. Bone Marrow Transpl. 2005, 36, 151–156.
Grumet FC, Hiraki DD, Brown BW et al. CD31 Mismatching Affects Marrow Transplantation Outcome. Biol. Blood and Marrow Transplant. 7:503–512 (2001).
Kogler G, Middleton PG, Wilke, M et al. Recipient cytokine genotypes for TNF∝ and IL 10 and the minor histocompatibility antigens HY and CD31 codon 125 are not associated with occurrence or severity of acute GVHD in unrelated donor cord blood transplantation. Transplantation, 2002 74, 1167–1175.
Maruya E, Saji H, Seki S et al. Evidence That CD31, CD49b, and CD62L Are Immunodominant Minor Transplants Histocompatibility Antigens in HLA Identical Sibling Bone Marrow. Blood, 1998 92: 2169–2176.
Nichols WC, Antin, JH, Lunetta KL et al. Polymorphism of CD31 is not a significant risk factor for acute graft versus host disease. Blood, 1996 88: 4429–4434.
Ostrovsky O, Shimoni A, Rand A et al. Genetic variations in the heparanase gene (HPSE) associate with increased risk of GVHD following allogeneic stem cell transplantation: effect of discrepancy between recipients and donors. Blood, 2010, 115: 2319–2328.
Rocha V, Franco RF, Porcher R et al. Host defense and inflammatory gene polymorphisms are associated with outcomes after HLA-identical sibling bone marrow transplantation,. Blood, 2002 100: 3908–3918.
The utility of genetic variants that predict risk of complications may allow selection of an optimal donor. However, it should be recognized that in some patients there are limited donor choices, and it is highly unlikely that studies will identify variants that would be more important than the dominant effect of HLA-matching, limiting the clinical usefulness of alternative strategies for donor selection.
Genetic association studies do offer the opportunity to explore the biology of transplant complications, and may allow identification of novel drug targets. For example, recent work by Ostrovsky et al has shown an association between heparanase genotype and GvHD, identifying this molecule as a potential target for GvHD therapy.(35)
Single gene disorders, such as Fanconi anemia and dyskeratosis congenita also have an important impact on outcome of transplantation, with an attributable risk much greater than that likely to be associated with a population polymorphism. Recognition of these disorders prior to transplantation is plays a critical role in avoidance of specific toxic exposures (radiation, busulfan) and modification of dosing. When the characteristic physical abnormalities are present diagnoses are usually made, but can be overlooked in patients presenting late, particularly in adult centers unfamiliar with these disorders. The availability of telomere testing has improved the recognition of dyskeratosis congenita, as physical manifestations may appear after marrow failure and the need for transplantation.
In summary, candidate gene studies require prior knowledge (or at least a hypothesis) regarding which pathways are important. The key strength of a genome-wide approach is the ability to make new discoveries. It should be recognized, however, that positive findings in a genome-wide study become candidates for replication in a candidate gene study.
Genome-wide association studies (GWAS)
Over the last decade, there has been extraordinary progress in the development of methods and tools for the characterization of the human genome. The recent completion of the human genome map(36, 37) and the development of dense SNP marker maps of the genome,(38, 39) as well as development of massively parallel genotyping technologies,(40–42) have made it possible to screen genes in an unbiased manner for polymorphisms that correlate with phenotype, disease status and relevant quantitative traits. Consideration of the entire genome in an unbiased fashion permits the discovery of genetic determinants, genes and pathways that would have never been considered otherwise.
Estimation of statistical power for genetic association studies
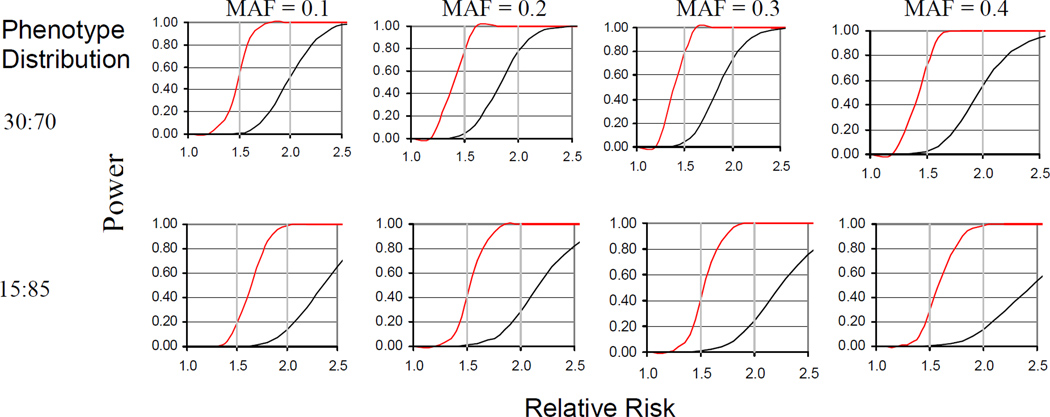
The statistical power question has been mostly ignored in previous candidate gene studies and the use of inadequately powered study populations has undoubtedly been a critical issue leading to many false positive results. Statistical power is an even greater issue for GWAS when the number of genetic factors tested for association approaches one million or more SNPs. An example of the study sample size needed to adequately power a GWAS is illustrated by the estimated statistical power to be gained by increasing the sample size from 1500 to the more than 5000 transplant pairs (Figure 1). Given a phenotype with a 30:70 distribution, such as chronic GvHD or grades 2–4 acute GvHD, a sample size of 1500 provides 60 to 80% power to observe a 2.0 relative risk for SNPs with a minor allele frequency (MAF) of 0.10 to 0.40, whereas a sample size of 5000 is necessary to detect a 1.5 relative risk, after the necessary correction for multiple comparisons. For a phenotype with a 15:85 distribution, such as grade 3–4 acute GvHD, a sample size of 1500 provides 60–80% power to observe a 2.4 to 2.5 relative risk for SNPs with MAF of 0.10 to 0.30, whereas a sample size of 5000 provides similar power to observe a 1.6 to 1.7 relative risk. Relative risks in the range of 1.5 to 1.7 are consistent with values previously reported by Lee et al for the effect size of HLA mismatching and mortality estimated in a multi-center NMDP study of 3,857 unrelated donor HCT cases.(15)
Figure 1.
Improvement in statistical power by increasing the sample size from 1500 samples (black lines) to 5000 samples (red lines). Horizontal gray lines indicate relative risks of 1.5 and 2. 0. Panels show results for SNPs with minor allele frequencies (MAF) ranging between 0.10 and 0.40.
GWAS in HCT
Two preliminary publications have appeared for GWAS of HCT, one by the Japanese group lead by Ogawa(43) and one summarizing preliminary GWAS results for the first 1500 patient-donor pairs from the Seattle HCT cohort.(44) Overall the Seattle results demonstrate that genes and pathways associated with clinically significant HCT outcomes can be identified, however the size of the initial GWAS cohort was not sufficiently large and lacked statistical power for measuring effect sizes with magnitudes of risk <2.0. Seattle investigators are currently expanding this single center GWAS-HCT project to include a total of 5,000 HCT donor-recipient pairs anticipating that data generated will provide important information for assessing risk, patient counseling and treatment planning, and provide insight into the basic mechansims responsible for transplant complications and rationale for new targeted therapies.
Planning future genetic studies
Key issues that need to be addressed include (1) study design beginning with a precise definition of the endpoints or phenotypes, (2) identification of an appropriate and adequately sized study population together with a reliable plan for collecting and maintaining high quality DNA, and (3) selection of an appropriate approach or platform for genotyping.
Study design
If the intent is to analyze a single endpoint, e.g. chronic GVHD, a case-control study design would be the most efficient; however a cohort design would be more appropriate for the study of complex or multiple phenotypes. Furthermore, consideration should be given to issues related to survival bias when designing prevalent case-control studies, especially where the endpoint is associated with a high lethality rate. Study design must include rigorous power estimations to determine the number of subjects necessary to meet statistical objectives.
Study population
Registration of transplant patients for a cohort would ideally begin prior to transplant to assure complete ascertainment of the population at risk and collection of DNA samples from both patient and donor. Prospective enrollment has the advantage of minimizing sampling bias, obtaining appropriate consent for genetic studies and facilitating when feasible the establishment of a biorepository for long-term sustainable maintenance of cells and/or DNA.
Genetic analysis
A candidate gene approach should be guided by a specific hypothesis and relevant preliminary data, whereas a genome-wide approach is necessary for comprehensive discovery analysis. If limited to a few genetic variants, the candidate gene approach has the advantage of requiring a relatively smaller study population and being less costly compared to a genome-wide discovery approach (a few dollars per SNP for genotyping ~1000 samples, multiplied by the number of SNPs), whereas a GWAS requires a much larger number of study samples and is more costly ($200–400/sample for ~1 million SNPs). Customized arrays or “ImmunoChip” are also available that identify a few thousand SNPs many of which have been previously shown to represent markers for association with immune-mediated diseases. The NIH maintains two related GWAS databases, one known as the case control GWAS database (https://gwas.lifesciencedb.jp/cgi-bin/gwasdb/gwas_top.cgi) that catalogs a growing number of SNPs associated with different diseases including cardiovascular and metabolic disorders, and a second known as dbGaP (http://www.ncbi.nlm.nih.gov/gap) that maintains detailed genotype and phenotype data for NIH supported GWAS projects.
Summary
The discovery of markers and functional variants associated with HCT outcomes will have significant implications for future translational research aimed at improving risk assessment and directing mechanistic research. Identification of new and informative genetic markers for HCT complications will have utility in the clinic for further strengthening objective pretransplant risk assessment and patient counseling, and serving as rationale tools for clinical management and treatment planning. The discovery of previously unrecognized functional variants defining genes and pathways associated with GvHD, organ toxicity, relapse, opportunistic infection and other long-term complications now recognized as emerging health care concerns will have significant impact on translational research aimed at developing novel targeted therapies for controlling chronic GvHD, facilitating tolerance and immune reconstitution, reducing risk of relapse and secondary malignancies, minimizing chronic metabolic disorders and improving quality of life.
Acknowledgements
Funding for this work was made possible in part by the following National Institute of Health grants: 1R13CA159788-01 (MP, KSB), U01HL069254 (MP), R01 CA078938 [SB], P01 CA 30206 [SJF], P01 AI33484 (JH), R01 AI094260 (JH) and R01 AI105914 (JH). The views expressed in this manuscript do not reflect the official policies of the Department of Health and Human Services; nor does mention by trade names, commercial practices, or organizations imply endorsement by the U.S. Government. Further support was provided by a generous grant from the St. Baldrick’s Foundation, a Leukemia Lymphoma Society grant (2192) (SB), and the Lance Armstrong Foundation, as well as the following pharmaceutical companies: Genzyme, Otsuka America Pharmaceutical, Inc., and Sigma-Tau Pharmaceuticals, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of those that provided funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bhatia S, Francisco L, Carter A, et al. Late mortality after allogeneic hematopoietic cell transplantation and functional status of long-term survivors: report from the Bone Marrow Transplant Survivor Study. Blood. 2007;110:3784–3792. doi: 10.1182/blood-2007-03-082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatia S, Robison LL, Francisco L, et al. Late mortality in survivors of autologous hematopoietic-cell transplantation: report from the Bone Marrow Transplant Survivor Study. Blood. 2005;105:4215–4222. doi: 10.1182/blood-2005-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Socie G, Stone JV, Wingard JR, et al. Long-term survival and late deaths after allogeneic bone marrow transplantation. Late Effects Working Committee of the International Bone Marrow Transplant Registry. N Engl J Med. 1999;341:14–21. doi: 10.1056/NEJM199907013410103. [DOI] [PubMed] [Google Scholar]
- 4.Armenian SH, Sun CL, Francisco L, et al. Late congestive heart failure after hematopoietic cell transplantation. J Clin Oncol. 2008;26:5537–5543. doi: 10.1200/JCO.2008.17.7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker KS, Ness KK, Steinberger J, et al. Diabetes, hypertension, and cardiovascular events in survivors of hematopoietic cell transplantation: a report from the Bone Marrow Transplant Survivor Study. Blood. 2007;109:1765–1772. doi: 10.1182/blood-2006-05-022335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan BM, Shalet SM. Endocrine late effects after bone marrow transplant. Br J Haematol. 2002;118:58–66. doi: 10.1046/j.1365-2141.2002.03527.x. [DOI] [PubMed] [Google Scholar]
- 7.Choi M, Sun CL, Kurian S, et al. Incidence and predictors of delayed chronic kidney disease in long-term survivors of hematopoietic cell transplantation. Cancer. 2008;113:1580–1587. doi: 10.1002/cncr.23773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser CJ, Bhatia S, Ness K, et al. Impact of chronic graft-versus-host disease on the health status of hematopoietic cell transplantation survivors: a report from the Bone Marrow Transplant Survivor Study. Blood. 2006;108:2867–2873. doi: 10.1182/blood-2006-02-003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leiper AD. Non-endocrine late complications of bone marrow transplantation in childhood: part II. Br J Haematol. 2002;118:23–43. doi: 10.1046/j.1365-2141.2002.03471.x. [DOI] [PubMed] [Google Scholar]
- 10.Majhail NS, Ness KK, Burns LJ, et al. Late effects in survivors of Hodgkin and non-Hodgkin lymphoma treated with autologous hematopoietic cell transplantation: a report from the bone marrow transplant survivor study. Biol Blood Marrow Transplant. 2007;13:1153–1159. doi: 10.1016/j.bbmt.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizzo JD, Curtis RE, Socie G, et al. Solid cancers after allogeneic hematopoietic cell transplantation. Blood. 2009;113:1175–1183. doi: 10.1182/blood-2008-05-158782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Socie G, Salooja N, Cohen A, et al. Nonmalignant late effects after allogeneic stem cell transplantation. Blood. 2003;101:3373–3385. doi: 10.1182/blood-2002-07-2231. [DOI] [PubMed] [Google Scholar]
- 13.Wingard JR, Vogelsang GB, Deeg HJ. Stem cell transplantation: supportive care and long-term complications. Hematology Am Soc Hematol Educ Program. 2002:422–444. doi: 10.1182/asheducation-2002.1.422. [DOI] [PubMed] [Google Scholar]
- 14.Sun CL, Francisco L, Kawashima T, et al. Prevalence and predictors of chronic health conditions after hematopoietic cell transplantation: a report from the Bone Marrow Transplant Survivor Study. Blood. 2010;116:3129–3139. doi: 10.1182/blood-2009-06-229369. quiz 3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 16.Hansen JA, Petersdorf EW, Lin MT, et al. Genetics of allogeneic hematopoietic cell transplantation. Role of HLA matching, functional variation in immune response genes. Immunol Res. 2008;41:56–78. doi: 10.1007/s12026-007-0043-x. [DOI] [PubMed] [Google Scholar]
- 17.Perreault C, Decary F, Brochu S, Gyger M, Belanger R, Roy D. Minor histocompatibility antigens. Blood. 1990;76:1269–1280. [PubMed] [Google Scholar]
- 18.Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference Volume I; Seattle, WA: International Histocompatibility Working Group Press; 2006. [Google Scholar]
- 19.McCarroll SA, Bradner JE, Turpeinen H, et al. Donor-recipient mismatch for common gene deletion polymorphisms in graft-versus-host disease. Nat Genet. 2009;41:1341–1344. doi: 10.1038/ng.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen JA, Chien JW, Warren EH, Zhao LP, Martin PJ. Defining genetic risk for graft-versus-host disease and mortality following allogeneic hematopoietic stem cell transplantation. Curr Opin Hematol. 2010;17:483–492. doi: 10.1097/MOH.0b013e32833eb770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin MT, Storer B, Martin PJ, et al. Relation of an interleukin-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. N Engl J Med. 2003;349:2201–2210. doi: 10.1056/NEJMoa022060. [DOI] [PubMed] [Google Scholar]
- 22.Lin MT, Storer B, Martin PJ, et al. Genetic variation in the IL-10 pathway modulates severity of acute graft-versus-host disease following hematopoietic cell transplantation: synergism between IL-10 genotype of patient and IL-10 receptor beta genotype of donor. Blood. 2005;106:3995–4001. doi: 10.1182/blood-2004-11-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chien JW, Boeckh MJ, Hansen JA, Clark JG. Lipopolysaccharide binding protein promoter variants influence the risk for Gram-negative bacteremia and mortality after allogeneic hematopoietic cell transplantation. Blood. 2008;111:2462–2469. doi: 10.1182/blood-2007-09-101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochud PY, Chien JW, Marr KA, et al. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med. 2008;359:1766–1777. doi: 10.1056/NEJMoa0802629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chien JW, Zhao LP, Hansen JA, Fan WH, Parimon T, Clark JG. Genetic variation in bactericidal/permeability-increasing protein influences the risk of developing rapid airflow decline after hematopoietic cell transplantation. Blood. 2006;107:2200–2207. doi: 10.1182/blood-2005-06-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrow D, Buskard N, Hill RS, Vickars L, Churg A. Bronchiolitis obliterans complicating bone marrow transplantation. Chest. 1985;87:828–830. doi: 10.1378/chest.87.6.828. [DOI] [PubMed] [Google Scholar]
- 27.Epler GR, Colby TV. The spectrum of bronchiolitis obliterans. Chest. 1983;83:161–162. doi: 10.1378/chest.83.2.161. [DOI] [PubMed] [Google Scholar]
- 28.Johnson FL, Stokes DC, Ruggiero M, Dalla-Pozza L, Callihan TR. Chronic obstructive airways disease after bone marrow transplantation. J Pediatr. 1984;105:370–376. doi: 10.1016/s0022-3476(84)80007-4. [DOI] [PubMed] [Google Scholar]
- 29.Behar E, Chao NJ, Hiraki DD, et al. Polymorphism of adhesion molecule CD31 and its role in acute graft-versus-host disease. N Engl J Med. 1996;334:286–291. doi: 10.1056/NEJM199602013340502. [DOI] [PubMed] [Google Scholar]
- 30.Nichols WC, Antin JH, Lunetta KL, et al. Polymorphism of adhesion molecule CD31 is not a significant risk factor for graft-versus-host disease. Blood. 1996;88:4429–4434. [PubMed] [Google Scholar]
- 31.Maruya E, Saji H, Seki S, et al. Evidence that CD31, CD49b, and CD62L are immunodominant minor histocompatibility antigens in HLA identical sibling bone marrow transplants. Blood. 1998;92:2169–2176. [PubMed] [Google Scholar]
- 32.Grumet FC, Hiraki DD, Brown BWM, et al. CD31 mismatching affects marrow transplantation outcome. Biol Blood Marrow Transplant. 2001;7:503–512. doi: 10.1053/bbmt.2001.v7.pm11669217. [DOI] [PubMed] [Google Scholar]
- 33.Kogler G, Middleton PG, Wilke M, et al. Recipient cytokine genotypes for TNF-alpha and IL-10 and the minor histocompatibility antigens HY and CD31 codon 125 are not associated with occurrence or severity of acute GVHD in unrelated cord blood transplantation: a retrospective analysis. Transplantation. 2002;74:1167–1175. doi: 10.1097/00007890-200210270-00019. [DOI] [PubMed] [Google Scholar]
- 34.Goodman RS, Ewing J, Evans PC, et al. Donor CD31 genotype and its association with acute graft-versus-host disease in HLA identical sibling stem cell transplantation. Bone Marrow Transplant. 2005;36:151–156. doi: 10.1038/sj.bmt.1705013. [DOI] [PubMed] [Google Scholar]
- 35.Ostrovsky O, Shimoni A, Rand A, Vlodavsky I, Nagler A. Genetic variations in the heparanase gene (HPSE) associate with increased risk of GVHD following allogeneic stem cell transplantation: effect of discrepancy between recipients and donors. Blood. 2010;115:2319–2328. doi: 10.1182/blood-2009-08-236455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 37.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 38.Miller RD, Phillips MS, Jo I, et al. High-density single-nucleotide polymorphism maps of the human genome. Genomics. 2005;86:117–126. doi: 10.1016/j.ygeno.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzaki H, Dong S, Loi H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 41.Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 42.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa S, Matsubara A, Onizuka M, et al. Exploration of the genetic basis of GVHD by genetic association studies. Biol Blood Marrow Transplant. 2009;15:39–41. doi: 10.1016/j.bbmt.2008.11.020. [DOI] [PubMed] [Google Scholar]
- 44.Chien JW, Zhao LP, Storer B, et al. Improving hematopoietic cell transplant outcomes in a new era of genomic research. Biol Blood Marrow Transplant. 2009;15:42–45. doi: 10.1016/j.bbmt.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]