Abstract
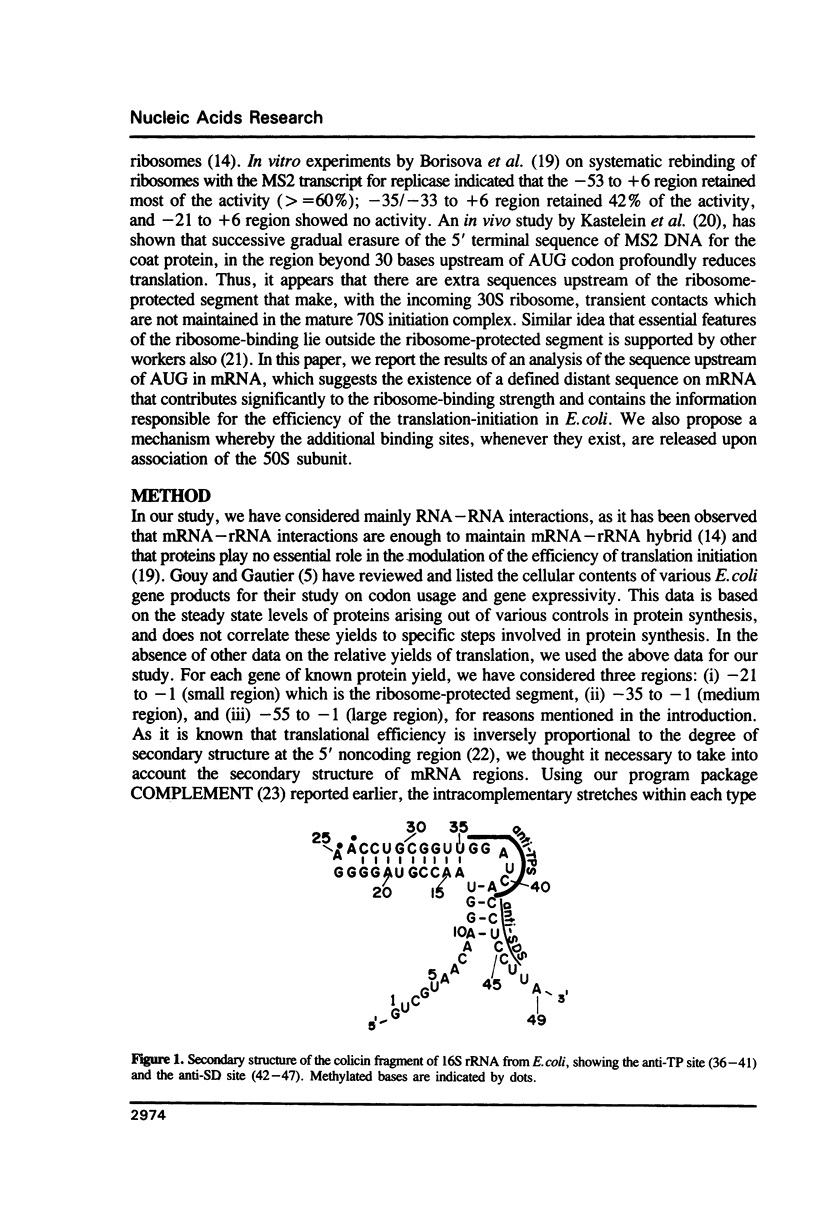
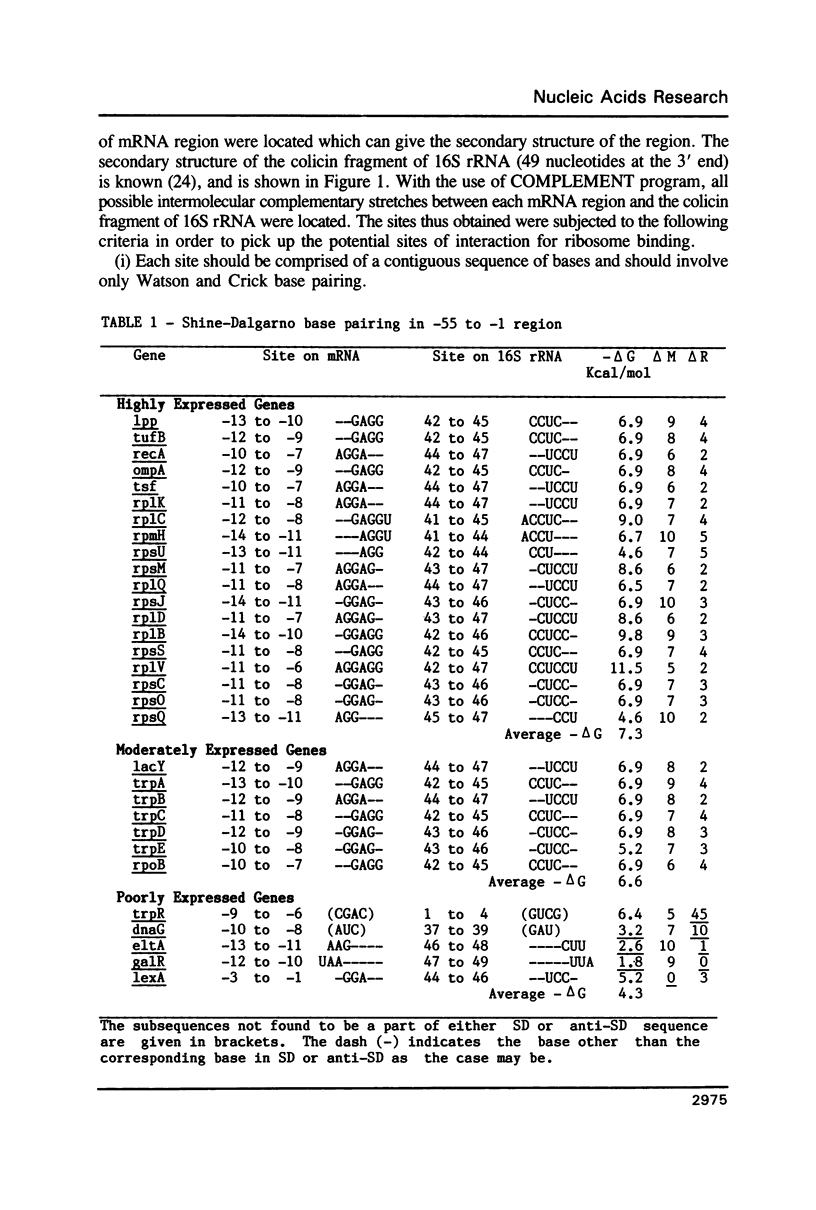
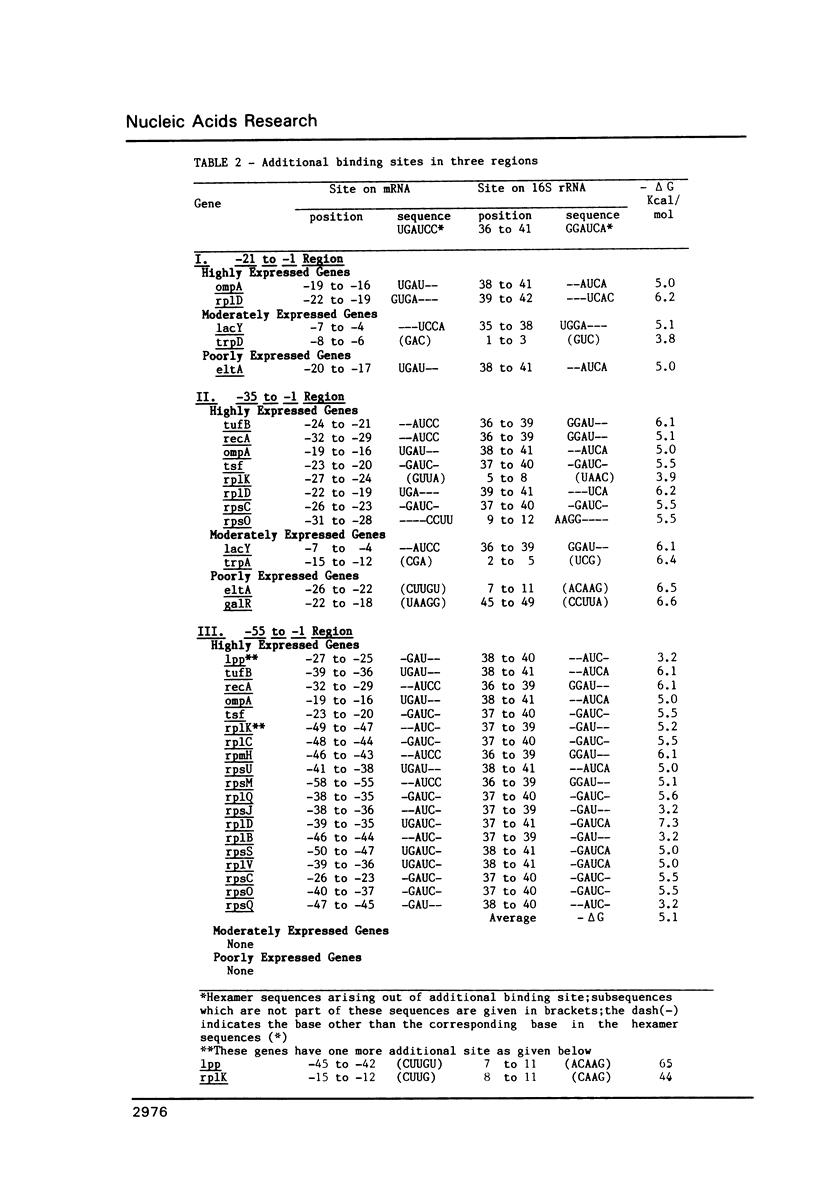
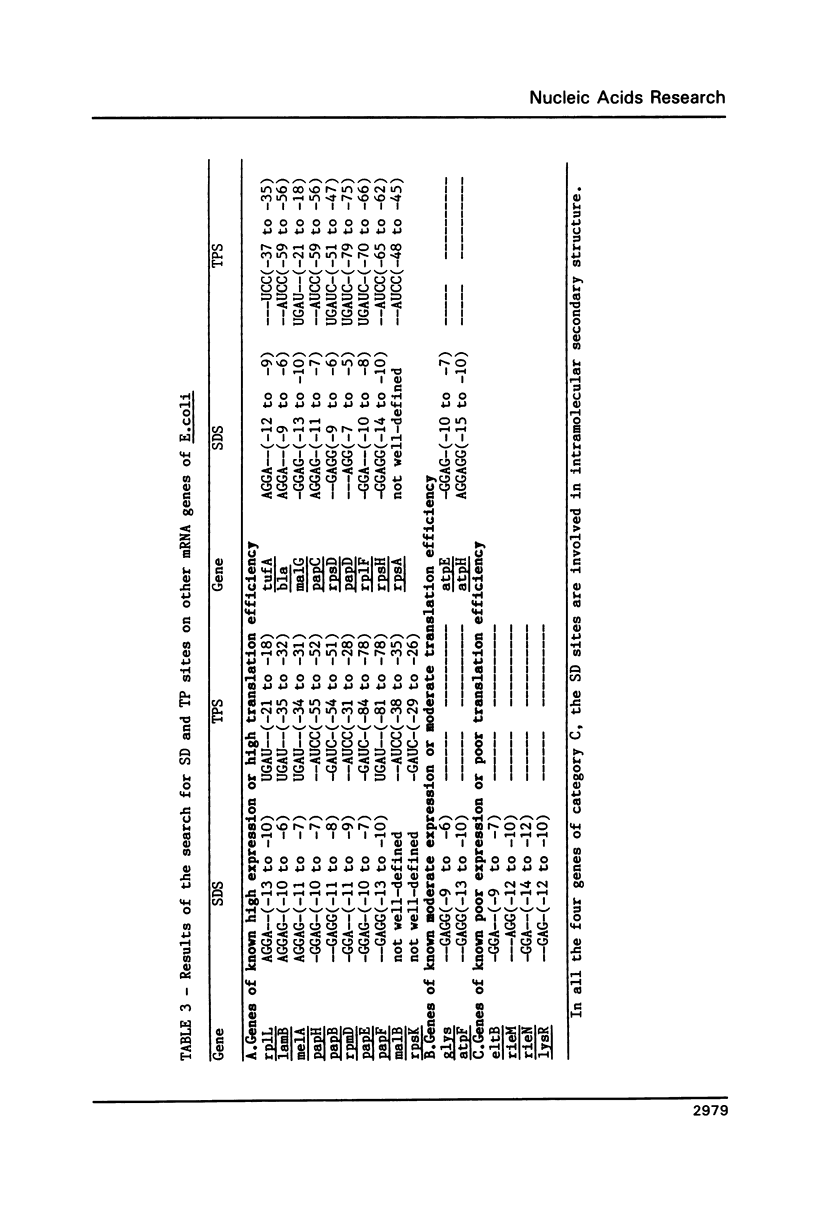
For various genes of E. coli, three regions (-55 to -1; -35 to -1; -21 to -1) 5' to AUG codon on mRNA were searched for sites of interaction with colicin fragment of 16S rRNA. The detailed sequence comparison points out that apart from Shine-Dalgarno base pairing, an additional ribosome-binding site, a subsequence of 5'-UGAUCC-3' invariably exists in mRNA for highly expressed genes. Poorly expressed genes appear to be controlled by only Shine-Dalgarno base pairing. The analysis indicates that in the initiator region, the -55 to -1 region contains the signal which decides the efficiency of the translation-initiation. The site on 16S rRNA, 5'-GGAUCA-3' at position 1529, that can base pair to the above site, has a recognition site on 23S rRNA at position 2390. In the light of the conserved nature and accessibility of these sites, it is proposed that the site on 16S rRNA plays a bifunctional role--initially it binds to mRNA from highly expressed genes to form a stable 30S initiation complex, and upon association with 50S subunit it exchanges base pairing with 23S rRNA, thus leaving the site on mRNA free.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backendorf C., Overbeek G. P., Van Boom J. H., Van Der Marel G., Veeneman G., Van Duin J. Role of 16-S RNA in ribosome messenger recognition. Eur J Biochem. 1980 Sep;110(2):599–604. doi: 10.1111/j.1432-1033.1980.tb04904.x. [DOI] [PubMed] [Google Scholar]
- Bergmann J. E., Lodish H. F. A kinetic model of protein synthesis. Application to hemoglobin synthesis and translational control. J Biol Chem. 1979 Dec 10;254(23):11927–11937. [PubMed] [Google Scholar]
- Borisova G. P., Volkova T. M., Berzin V., Rosenthal G., Gren E. J. The regulatory region of MS2 phage RNA replicase cistron. IV. Functional activity of specific MS2 RNA fragments in formation of the 70 S initiation complex of protein biosynthesis. Nucleic Acids Res. 1979;6(5):1761–1774. doi: 10.1093/nar/6.5.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma D. P., Nag B., Tewari D. S. Association of 16S and 23S ribosomal RNAs to form a bimolecular complex. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4875–4878. doi: 10.1073/pnas.80.16.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Curry K. A., Tomich C. S. Effect of ribosome binding site on gene expression in Escherichia coli. DNA. 1988 Apr;7(3):173–179. doi: 10.1089/dna.1988.7.173. [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Christensen A., Garrett R. A. Higher order structure in the 3'-minor domain of small subunit ribosomal RNAs from a gram negative bacterium, a gram positive bacterium and a eukaryote. J Mol Biol. 1983 Sep 5;169(1):249–279. doi: 10.1016/s0022-2836(83)80183-1. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen D., Iserentant D., Derom C., Fiers W. Systematic alteration of the nucleotide sequence preceding the translation initiation codon and the effects on bacterial expression of the cloned SV40 small-t antigen gene. Gene. 1982 Jan;17(1):55–63. doi: 10.1016/0378-1119(82)90100-7. [DOI] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gren E. J. Recognition of messenger RNA during translational initiation in Escherichia coli. Biochimie. 1984 Jan;66(1):1–29. doi: 10.1016/0300-9084(84)90188-3. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Débarbouillé M., Schwartz M. A role for mRNA secondary structure in the control of translation initiation. Nature. 1982 Feb 18;295(5850):616–618. doi: 10.1038/295616a0. [DOI] [PubMed] [Google Scholar]
- Herr W., Chapman N. M., Noller H. F. Mechanism of ribosomal subunit association: discrimination of specific sites in 16 S RNA essential for association activity. J Mol Biol. 1979 Jun 5;130(4):433–449. doi: 10.1016/0022-2836(79)90433-9. [DOI] [PubMed] [Google Scholar]
- Herr W., Noller H. F. Nucleotide sequences of accessible regions of 23S RNA in 50S ribosomal subunits. Biochemistry. 1978 Jan 24;17(2):307–315. doi: 10.1021/bi00595a018. [DOI] [PubMed] [Google Scholar]
- Herr W., Noller H. F. Protection of specific sites in 23 S and 5 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J Mol Biol. 1979 Jun 5;130(4):421–432. doi: 10.1016/0022-2836(79)90432-7. [DOI] [PubMed] [Google Scholar]
- Heus H. A., van Knippenberg P. H. The 3' terminal colicin fragment of Escherichia coli 16S ribosomal RNA. Conformational details revealed by enzymic and chemical probing. J Biomol Struct Dyn. 1988 Feb;5(4):951–963. doi: 10.1080/07391102.1988.10506437. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Jansone I., Berzin V., Gribanov V., Gren E. J. The regulatory region of MS2 phage RNA replicase cistron. III. Characterization of fragments resulting from S1 nuclease digestion. Nucleic Acids Res. 1979;6(5):1747–1760. doi: 10.1093/nar/6.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay E., Seth A. K., Jay G. Specific binding of a chemically synthesized prokaryotic ribosome recognition site. Prospect for molecular cloning and expression of eukaryotic genes. J Biol Chem. 1980 May 10;255(9):3809–3812. [PubMed] [Google Scholar]
- Kastelein R. A., Berkhout B., Overbeek G. P., van Duin J. Effect of the sequences upstream from the ribosome-binding site on the yield of protein from the cloned gene for phage MS2 coat protein. Gene. 1983 Sep;23(3):245–254. doi: 10.1016/0378-1119(83)90015-x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljenström H., von Heijne G. Translation rate modification by preferential codon usage: intragenic position effects. J Theor Biol. 1987 Jan 7;124(1):43–55. doi: 10.1016/s0022-5193(87)80251-5. [DOI] [PubMed] [Google Scholar]
- Maitra U., Stringer E. A., Chaudhuri A. Initiation factors in protein biosynthesis. Annu Rev Biochem. 1982;51:869–900. doi: 10.1146/annurev.bi.51.070182.004253. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Schairer H. U., Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985 Feb;4(2):519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. The involvement of mRNA secondary structure in protein synthesis. Biochem Cell Biol. 1987 Jun;65(6):576–581. doi: 10.1139/o87-074. [DOI] [PubMed] [Google Scholar]
- Poldermans B., Van Buul C. P., Van Knippenberg P. H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3' end of 16 S ribosomal RNA of Escherichia coli. II. The effect of the absence of the methyl groups on initiation of protein biosynthesis. J Biol Chem. 1979 Sep 25;254(18):9090–9093. [PubMed] [Google Scholar]
- Ryoji M., Berland R., Kaji A. Reinitiation of translation from the triplet next to the amber termination codon in the absence of ribosome-releasing factor. Proc Natl Acad Sci U S A. 1981 Oct;78(10):5973–5977. doi: 10.1073/pnas.78.10.5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Blundell M., Kennell D. Translation and mRNA decay. Mol Gen Genet. 1978 Apr 6;160(2):121–129. doi: 10.1007/BF00267473. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpaer E. G. Constraints on codon context in Escherichia coli genes. Their possible role in modulating the efficiency of translation. J Mol Biol. 1986 Apr 20;188(4):555–564. doi: 10.1016/s0022-2836(86)80005-5. [DOI] [PubMed] [Google Scholar]
- Steitz J. A., Jakes K. How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4734–4738. doi: 10.1073/pnas.72.12.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanaraj T. A., Kolaskar A. S., Pandit M. W. Prediction of the recognition sites on 16S and 23S rRNAs from E. coli for the formation of 16S-23S rRNA complex. J Biomol Struct Dyn. 1988 Dec;6(3):587–592. doi: 10.1080/07391102.1988.10506509. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Hearst J. E. Structure-function relations in E. coli 16S RNA. Cell. 1983 May;33(1):19–24. doi: 10.1016/0092-8674(83)90330-6. [DOI] [PubMed] [Google Scholar]
- Van Duin J., Kurland C. G., Dondon J., Grunberg-Mangago M., Branlant C., Ebel J. P. New aspects of the IF3-ribosome interaction. FEBS Lett. 1976 Feb 15;62(2):111–114. doi: 10.1016/0014-5793(76)80030-0. [DOI] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]



