Abstract
Par proteins establish discrete intracellular spatial domains to polarize many different cell types. In the single-cell embryo of the nematode worm Caenorhabditis elegans, the segregation of Par proteins is crucial for proper division and cell fate specification. Actomyosin-based cortical flows drive the initial formation of anterior and posterior Par domains, but cortical actin is not required for the maintenance of these domains. Here we develop a model of interactions between the Par proteins that includes both mutual inhibition and PAR-3 oligomerization. We show that this model gives rise to a bistable switch mechanism, allowing the Par proteins to occupy distinct anterior and posterior domains seen in the early C. elegans embryo, independent of dynamics or asymmetries in the actin cortex. The model predicts a sharp loss of cortical Par protein asymmetries during gradual depletion of the Par protein PAR-6, and we confirm this prediction experimentally. Together, these results suggest both mutual inhibition and PAR-3 oligomerization are sufficient to maintain distinct Par protein domains in the early C. elegans embryo.
Introduction
Polarization of single cells involves the spatial segregation of proteins, lipids, and other molecules. Cell polarization is crucial for a range of biological processes including embryonic development, directed motility, and epithelial cell function, and is observed in a wide variety of organisms (1–3). Many instances of cell polarization rely on a core module of conserved proteins consisting of the Par proteins PAR-1, PAR-2, PAR-3, and PAR-6, the atypical protein kinase aPKC, and the tumor-suppressor protein LGL (4). A key feature of this module is that its members become asymmetrically distributed during cell polarization, and these asymmetries are essential for the elaboration of a polarized state. Thus, a key challenge is to understand how Par protein asymmetries are established and maintained during polarization.
The one-cell embryo (zygote) of the nematode worm Caenorhabditis elegans has emerged as an important model system for studying Par protein dynamics during intracellular polarization (5). Polarization of the zygote occurs ∼20 min after fertilization and involves two distinct phases: establishment and maintenance (6). Just before polarity establishment, the proteins PAR-1, PAR-2, and LGL (which segregate to the posterior pole in the polarized embryo) are cytoplasmic, while the proteins PAR-3, PAR-6, and aPKC (which segregate to the anterior pole) are enriched at the interface between the plasma membrane (PM) and the cell cortex (a thin layer just beneath the membrane that is enriched in filamentous actin and the contractile protein nonmuscle-myosin II) (7,8).
During polarity establishment, a transient sperm-derived cue triggers actomyosin-based cortical flows that transport the anterior Par proteins toward the future anterior pole, while the posterior Par proteins and LGL become enriched in a complementary posterior domain (5,9,10). These complementary distributions are then maintained, in the absence of the sperm cue, for ∼10 min as the cell prepares for first division (6).
Photokinetic studies suggest that both anterior and posterior Par proteins exchange freely between cortex and cytoplasm and can diffuse within the plane of the membrane (1,11). The anterior Par proteins PAR-3, PAR-6, and aPKC can bind one another to form a trimeric complex (12–18). Structure/localization studies in worms (19), flies (20), and mammalian cells (reviewed in Goldstein and Macara (3)) suggest that recruitment of PAR-3 to the cortex/PM involves multiple domains that bind to distinct targets including phosphoinositide lipids, other proteins and possibly F-actin (19,20). In addition, recruitment requires an N-terminal oligomerization domain that mediates self-association of PAR-3 and can assemble filaments when expressed as a purified fragment in vitro (19,21–23). Together, these data suggest that PAR-3 binds the cortex/PM as a multivalent oligomer. PAR-1, PAR-2, and LGL likewise associate reversibly with the cortex/PM, although the details of this association are less well understood.
The likely basis for complementarity of Par protein distributions lies in mutually antagonistic interactions between anterior and posterior Par proteins (24). aPKC phosphorylates and promotes the dissociation of PAR-1 (25–27), PAR-2 (28), and LGL (29,30); in Drosophila and mammalian cells, PAR-1 phosphorylates and promotes PAR-5-dependent dissociation of PAR-3 at residues that are conserved in C. elegans (31,32). Recent studies in C. elegans suggest that PAR-2 and LGL can also act redundantly to prevent association of PAR-6/aPKC, although the molecular mechanisms remain poorly understood (18,30). An emerging view is that actin-independent maintenance of Par asymmetries involves a continuous balance of local exchange among cytoplasmic and cortex/PM pools, diffusion and local protein/protein interactions (1), but how globally stable asymmetries emerge from these local kinetics is unclear.
Reaction-diffusion models have provided a useful tool for studying the dynamics of intracellular polarization, mainly in the context of cell motility. These models have revealed a number of potential mechanisms, including spatial bistability driven by nonlinearities in the underlying biochemical kinetics (33–35), and Turing instabilities that do not require such nonlinearities for pattern formation (36,37). See Suzuki et al. (17) and Iglesias and Devreotes (38) for some excellent reviews. To date, one previous theoretical study has specifically addressed Par protein segregation in the early C. elegans embryo (39). By coupling reaction-diffusion models to simple representations of actomyosin contractility and cortical flow, Tostevin and Howard (39) showed that mutual phosphorylation, plus enhanced anterior Par protein binding to a polarized actomyosin cortex and feedback control of cortical contractility and elasticity by anterior Par proteins, would be sufficient to form stable complementary Par protein domains. Interestingly, while asymmetrical contractility and cortical flow are essential for polarity establishment, inhibiting myosin activity (40) or disrupting cortical actin (1,41) during the maintenance phase does not abolish complementarity, but results only in small effects on the boundary position between the Par domains. This suggests that actin-independent mechanisms are sufficient to dynamically stabilize complementary Par domains once they have formed.
Here we use an approach analogous to that of Tostevin and Howard (39) to investigate under what conditions Par proteins can maintain distinct domains in the absence of polarized actomyosin, as observed in previous experimental studies (1,41). We show that the simplest form of mutual inhibition in which anterior and posterior Par proteins mutually promote one another's dissociation through mass action kinetics is insufficient to stabilize complementary Par domains in the absence of actomyosin, but that initial asymmetries can be maintained when PAR-3 oligomerization is included. We show that PAR-3 oligomerization endows the system with bistability; accordingly, our model predicts a sharp threshold for the loss of spatial asymmetries as the levels of anterior (or posterior) Par proteins are reduced, and we verify this prediction experimentally using RNA interference.
The Mathematical Model
Model assumptions
Based on the discussion above, we make the following assumptions about the Par proteins and how they interact:
Assumption 1. The Par proteins can be grouped into two modules based on their localization in the polarized embryo: the anterior Par module (denoted ParA), consisting of PAR-3, PAR-6, and aPKC; and the posterior Par module (denoted ParP), consisting of PAR-1, PAR-2, and LGL.
Assumption 2. ParA can homodimerize.
Assumption 3. The cytoplasmic concentrations of ParA and ParP are at quasi-steady state (we relax this assumption below).
Assumption 4. All interactions (dimerization, cortical association, and dissociation) follow first-order or mass-action kinetics.
Assumption 5. All cortical forms of ParA are capable of catalyzing cortical dissociation of ParP, and vice versa.
Assumption 6. ParA and ParP each modify the other to produce a rapidly dissociating form.
Assumptions 1–3 simplify the structure of the mathematical model. This is not an arbitrary grouping; in many cell types, PAR-3/PAR-6/aPKC and PAR-1/LGL localize to complementary spatial domains (PAR-2 is unique to C. elegans). By making these assumptions, we are ignoring the underlying dynamics of the individual Par proteins and their complex formation but render the model more general. We assume ParA can, at most, dimerize, although higher-order structures are likely.
Assumption 4, that all interactions use mass-action and linear kinetics, represents the simplest model formulation consistent with currently available experimental data. Data available as of this writing suggests that the Par proteins interact directly with one another and do not form intermediate complexes or act indirectly through diffusible signals. Thus, there is no empirical justification for using sigmoidal functions or other nonlinear functions to describe Par protein interactions. Assumptions 5 and 6 are derived from the earlier discussion. A simplified schematic summarizing the interactions incorporated into the mathematical model is shown in Fig. 1.
Figure 1.
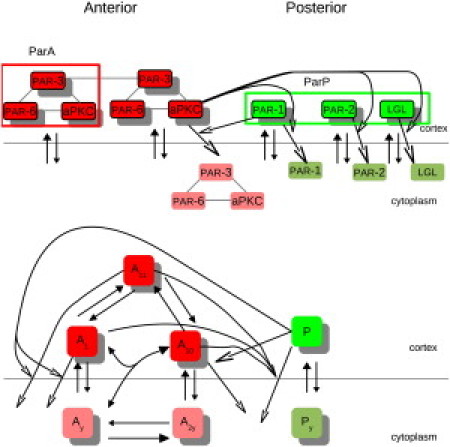
(Upper) Schematic of experimentally determined interactions between the anterior Par proteins (PAR-3, PAR-6, and aPKC) and the posterior Par proteins (PAR-1, PAR-2, and LGL) and (lower) simplified interactions used in the mathematical model. The anterior Par proteins form a complex (12–17) that can dimerize (22,23) and bind to the cortex (7). aPKC phosphorylates and promotes dissociation of PAR-1, PAR-2, and LGL (25–29). PAR-1 can phosphorylate PAR-3 to promote dissociation of anterior Par proteins (30–32). PAR-2 and LGL promote dissociation of anterior Par proteins through mechanisms that are not well understood. For the mathematical model, we consider the anterior and posterior Par proteins as modules, and allow the anterior Par proteins to dimerize. (Solid arrowheads) Cortical loss by regulated dissociation. (Open arrowheads) Cortical loss by conversion. We assume the cytoplasmic Par proteins do not interact with each other. (Lighter color) Phosphorylated forms.
Variables
The model keeps track of the following quantities:
A1, cortical monomeric ParA;
A10, cortical dimeric ParA (singly bound);
A11, cortical dimeric ParA (doubly bound);
P, cortical ParP.
That is, A10 is a ParA dimer with only one molecule bound to the cortex while A11 is a ParA dimer with both molecules bound to the cortex. Fig. 1 (top) shows A11, a ParA dimer with both molecules bound. We assume neither part of the dimer is modified in A10 and A11.
We assume the following quantities are at quasi-steady state and so are included in the model but do not vary over time:
Ay, cytoplasmic ParA monomers;
A2y, cytoplasmic ParA dimers;
Py, cytoplasmic ParP.
Model equations
According to Assumption 3, the cytoplasmic Par proteins are at quasi-steady state, and modified Par proteins that dissociate from the cortex are absorbed into the cytoplasmic pool. Consequently, we neglect the dynamics of the modified and cytoplasmic Par proteins. In addition, we assume the cytoplasmic forms of the Par proteins do not interact with each other, and focus on interactions and dynamics of the cortical Par proteins. The dynamics of the cortically bound unmodified Par proteins can be described as
| (1a) |
| (1b) |
| (1c) |
| (1d) |
Equation parameters and their meaning are listed in Table S1 in the Supporting Material.
Parameter values and nondimensional equations
Previous experimental studies of the Par network have focused on identifying key molecular activities and functional interactions. Although recent photokinetic studies have measured diffusivities and exchange rates for a subset of the Par proteins (1,11), many of the kinetic parameters in our model remain unknown. Thus, we focus here on asking whether qualitative-systems-level features relevant to cell polarization, such as bistability and dynamic stabilization of complementary domains, are possible, given empirically constrained assumptions about the underlying molecular interactions. Accordingly, we began our analysis with a random search of model parameter space, seeking parameter values for which the model yields stable complementarity of Par domains in the absence of diffusion and a quasistable anterior/posterior (AP) boundary in the presence of diffusion (see Fig. S1 in the Supporting Material).
For our parameter searches and further analysis of the solutions, we nondimensionalized the model equations, allowing us to analyze the dynamics in terms of ratios of kinetic parameters and focus on relative rather than absolute rates. We use the following scalings,
and introduce the dimensionless parameters, αy and ρy, that represent the nondimensional cytoplasmic concentrations of ParA and ParP, respectively. After nondimensionalizing, we have
| (2a) |
| (2b) |
| (2c) |
| (2d) |
In addition, we exploited several factors to constrain values of model parameters. First, by using simple kinetic reasoning, we assumed that the rate of ParA dimer association (kA11on) is higher than ParA monomer association (kAon). Second, we used recently reported values for cortical exchange rates and diffusivities of PAR-6 and PAR-2 to set the values for corresponding model parameters, i.e., kAoff = 5.4 × 10−3 s−1; kPoff = 7.3 × 10−3 s−1; DA = 0.28 μm2/s; and DP 0.15 μm2/s (1,11). Thus we find the timescale is τ ≈ 185 s, the scaled diffusivities are D1 ≈ 0.008, D2 ≈ 0.004, and the scaled off-rates are 1 for ParA and β8 ≈ 1 for ParP. Finally, we chose β7 = 1 so that the level of ParP is roughly 1 in the absence of ParA, and we require the maximal levels of total ParA to be <3 (i.e., we required cortical levels of ParA and ParP to be roughly similar). With these constraints on the parameter values, ∼1 in 384 parameter sets of the full model (expressions in Eq. 2) yielded a bistable solution. This implies that, on average, ∼37% of the values chosen independently for each of the six free parameters yields a bistable solution (0.376 ≈ 1/384), suggesting that bistability is a robust property of the model. All other parameters we allowed to range over two orders of magnitude. A cam diagram summarizing the random parameter search results is shown in Fig. S1. The value-ranges used in the random searches and the values used to analyze particular solutions in detail are given in Table S2.
Discretization and numerical implementation
The equations were discretized using the Forward Time Centered Space (42) numerical analysis method and coded using the C programming language. The time and space step sizes used in the simulations were chosen sufficiently small to ensure numerical stability. Parameter space searches were performed using custom software written in Java (http://www.java.com/en/).
Experimental Materials and Methods
Strains
Worms were handled as previously described in Brenner (43). The transgenic strain JH1902 [Ppie−1:RFP:PAR-6; Ppie−1:GFP:PAR-2C56S] (28) was used in the experiments described here.
RNA-mediated interference
RNAi feeding experiments were performed as previously described in Timmons et al. (44) using a feeding strain directed against par-6 (45). For par-6(RNAi), early adult worms were placed on prepared plates and kept at 25°C. Embryos were dissected from the adult worms and examined at 30-min intervals after the beginning of RNAi feeding.
Microscopy
Gravid worms were dissected in egg salts on 22 mm2 coverslips which were then inverted onto a 2% agarose pad and sealed with Vaseline (Unilever, Rotterdam, The Netherlands) (45). A single midplane image was obtained using an RT wide-field epifluorescence microscope (DeltaVision, www.appliedprecision.com) at 20°C through a Plan Apo 60× 1.4 oil immersion lens. A single image was acquired at the maintenance phase to prevent photo bleaching. Each image was rotated to position the anterior pole to the left. The fluorescence level around the circumference of the embryo was found using the ImageJ plug-in “straighten” (http://rsb.info.nih.gov/ij/). The circumference of the cell, including the eggshell, was traced by hand, then the “straighten” plug-in transformed the chosen section into a one-dimensional plot of the average fluorescence of the circumference. The eggshell is included in the section of interest because it autofluoresces in the red channel and in some regions is indistinguishable from the cortex of the embryo. Embryos were deemed polarized if there was >20% difference in average fluorescence between the anterior and posterior poles. The position of the Par protein boundary was determined qualitatively during image acquisition using the SoftWorx (www.appliedprecision.com) software's built-in measurement tool.
Results
Anterior Par protein oligomerization may help maintain distinct Par domains independently of asymmetries in the actomyosin cortex
We began by asking under what conditions the Par proteins could maintain distinct domains, independently of any asymmetries or dynamics of the actin cortex. We first considered the case in which ParA and ParP interact only by promoting each other's dissociation, and ParA is not capable of dimerizing. As a result, we can write the expressions in Eq. 2 as
| (3a) |
| (3b) |
As shown in Fig. 2 (top row), this version of the model cannot maintain an initially asymmetric distribution of anterior and posterior Par proteins.
Figure 2.

Time course of Par protein dynamics alone as determined by the expressions in Eq. 3 (top row) and Eq. 2 (bottom row), independent of interactions with a polarized actin cortex. When Par proteins interact only by mutual promotion of dissociation, there is a loss of any initial asymmetry (top row), while Par proteins are capable of maintaining distinct domains when the anterior Par proteins can also dimerize (bottom row). In both cases, ParA (left) has an initial distribution that is high at the anterior pole (0 egg length) and low at the posterior pole (1 egg length). A reciprocal initial distribution is assumed for ParP (right). Note the timescale in this and all subsequent figures is given in dimensional units.
We can understand why this is so by considering the expressions in Eq. 3 without the spatial diffusion term, which yields two coupled ordinary differential equations with a single steady state, as shown in the phase plane diagram (seen later in Fig. 4, left). Indeed, if we impose biologically reasonable constraints that the parameter values are strictly positive and that A1 and P are greater than or equal to zero, then this steady state is always unique, regardless of the choice of parameter values. We conclude that cross-inhibition in which anterior and posterior Par proteins promote one another's dissociation through mass-action kinetics is insufficient to explain dynamic stabilization of complementary Par domains.
Figure 4.
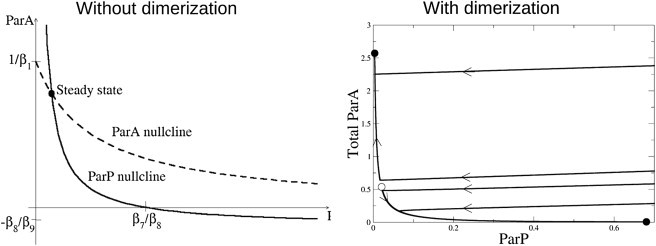
Phase plane plots of temporal Par protein dynamics (neglecting diffusion). (Left) When the Par proteins interact only by mutual promotion of dissociation (expressions in Eq. 3), the nullclines indicate a single steady state, suggesting that regardless of initial conditions, ParA and ParP will coexist at levels determined by their mutual interaction strengths. (Right) When ParA can dimerize (expressions in Eq. 2), the phase plane indicates a bistable solution: ParA and ParP can assume distinct stable steady states corresponding to the protein distributions seen at the anterior and posterior poles of the embryo. Several sample trajectories that tend to the steady states are shown, with direction of evolution over time (arrows). (Solid circles) Stable steady states. (Open circles) Unstable steady states.
In contrast, when we allowed ParA to dimerize and sampled kinetic parameters randomly (see above for details), we could now find many parameter sets for which the model equations (see expressions in Eq. 2) stabilize complementary domains in the absence of diffusion (Fig. 2, bottom row). When we allowed ParA and ParP to diffuse at rates comparable to those measured by Goehring et al. (1), the boundary between the anterior and posterior domains tended to drift toward one or the other pole, ultimately leading to a loss of polarity. The magnitude and direction of drift depended on the relative strengths of mutual inhibition (β4 and β9) and diffusivity (D1 and D2) (Fig. 3). However, for ∼1 out of 622 parameter sets that yielded stable complementarity without diffusion (roughly 1/239,000 of all parameters sampled; 17% of each parameter's independently sampled values), the total drift over 600s was less than a few percent egg length, which is comparable to the drift seen during the maintenance phase in the absence of cortical actin (1).
Figure 3.

The boundary between the Par protein domains can drift toward either the anterior or posterior pole, depending on the relative strength of inhibition (β4 and β9) or the relative speeds of diffusion (D1 and D2). The boundary drifts toward the posterior pole for D1 or β4 smaller than their default values (left column) and toward the anterior pole for values higher than their default (right column). The boundary is metastable for intermediate values (middle column) and held for a large number of time steps. The values for β4 shown here still give rise to bistability in the space-free version of the expressions in Eq. 2. Similar results are found when varying D2 and β9.
Again, we can understand the behavior of the model by considering the expressions in Eq. 2 in the absence of the spatial diffusion term. As shown in Fig. 4 (right), the model can exhibit bistability. That is, each patch of the model cortex can stably assume one of two steady states: high ParA/low ParP, or low ParA/high ParP. Together, these results suggest that ParA dimerization plus cross-promoted dissociation of ParA and ParP can give rise to bistable dynamics. Bistability is sufficient to maintain complementarity of Par domains on short timescales, but maintenance of a stable boundary position on longer timescales requires balancing anterior and posterior Par protein dynamics or including additional mechanisms to buffer against drift.
Par protein dynamics predict loss of distinct domains as cytoplasmic Par proteins are depleted
Our results show that ParA dimerization plus cross-inhibition is sufficient to stabilize complementary Par domains, but we cannot formally exclude other possibilities. For example, Tostevin and Howard (39) showed that mutual inhibition (with mass-action kinetics) plus enhanced recruitment of ParA to anteriorly enriched actomyosin and a self-limiting actomyosin contraction could also stabilize Par domains. While recent work suggests that actomyosin asymmetries are not required for the stable maintenance of Par domains (1,41), it is possible that other binding sites for ParA might be enriched during maintenance, which could substitute for the actomyosin used in the Tostevin and Howard model.
As a first step toward distinguishing these two models experimentally, we examined their predicted responses to gradual depletion of either anterior or posterior Par proteins. The cross-inhibition plus ParA dimerization model predicts a sharp threshold in the level of either ParA or ParP, below which asymmetry is abruptly lost (shown for ParA in Fig. 5, left). The model also predicts that bistability is maintained when levels of ParA and ParP are simultaneously decreased (Fig. 5, right). This is consistent with the observation that reducing levels of anterior Par proteins can rescue partial loss of function in posterior Par proteins and vice versa (46). In contrast, the cross-inhibition plus asymmetric recruitment model of Tostevin and Howard predicts a gradual loss of asymmetry as ParA is depleted, and no loss of asymmetry or change in the size of the Par protein domains as ParP is depleted (Fig. 6).
Figure 5.

Bifurcation diagram (left) and two-parameter continuation diagram (right) for the full space-free (no diffusion) Par protein model when ParA is capable of dimerizing (expressions in Eq. 2). The bifurcation diagram displays the steady-state behavior of the model, as one parameter—here the level of cytoplasmic ParA—is varied. The hysteresis curve suggests the model will lose bistability as the level of cytoplasmic ParA is depleted. The two-parameter continuation diagram indicates the steady-state behavior of the model as two parameters, the levels of cytoplasmic ParA and ParP, are varied. The wedge-shaped region in this diagram demonstrates there is a region of bistability as either or both cytoplasmic pools are depleted. Variations in the cytoplasmic levels that cause the system to exit the region of bistability will result in a loss of bistability, while variations that allow the system to remain in the bistable region will maintain the capacity to adopt distinct steady states.
Figure 6.

Simulations of the model proposed by Tostevin and Howard (39). This model takes into account actomyosin dynamics, mutual inhibition of Par proteins, and binding of ParA to (polarized) actomyosin but not dimerization of ParA. Depleting the total amount of anterior Par protein in the system by lowering the initial condition results in a gradual loss of polarity rather than the sharp loss for the model with bistable dynamics. Shown here is the amount of membrane-associated anterior Par proteins (Am, top row) and the membrane-associated posterior Par proteins (Pm, bottom row). Depleting the total amount of posterior Par proteins produces the same profiles shown here for the corresponding amounts of total anterior Par proteins (not shown).
Anterior Par protein depletion in vivo leads to loss of polarity
To test experimentally the prediction that polarity loss occurs abruptly below a threshold level of ParA, we examined the response of maintenance phase C. elegans embryos to decreasing levels of PAR-6 produced by RNA interference (47,48). PAR-6 is required for cortical enrichment of aPKC (7), which mediates cortical dissociation of the posterior Par proteins, so that depletion of PAR-6 mimics loss of ParA in either model. We synchronized young adult worms engineered to express mCHERRY-tagged PAR-6 and GFP-tagged PAR-2, then placed these worms on plates containing bacteria that synthesize double-stranded RNA that targets PAR-6. We then examined maintenance-phase embryos dissected from the treated worms at 30-min intervals (Fig. 7). Significantly, we observed a sharp transition from a polarized to unpolarized state at ∼5 h of RNAi exposure. This result was consistent across many embryos and multiple RNAi experiments (Fig. 8), supporting the hypothesis that bistability underlies the maintenance of Par protein asymmetries.
Figure 7.
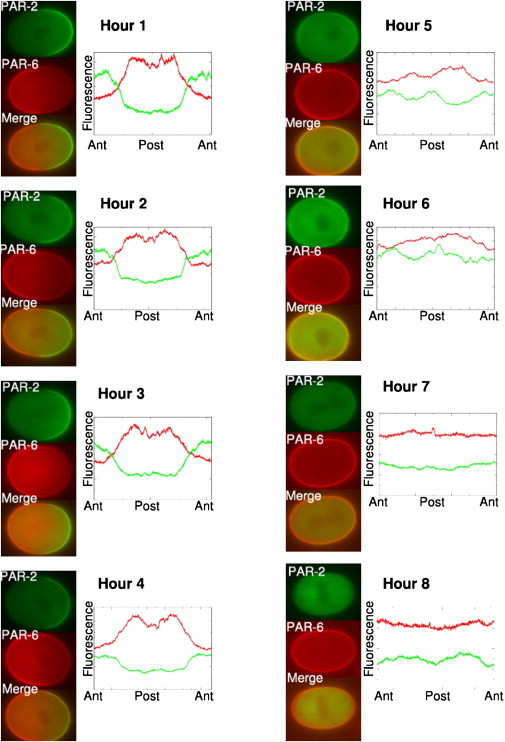
Time series showing embryos dissected from adult worms treated by par-6 RNA interference for progressively longer times to produce gradual depletion of PAR-6, an anterior Par protein. Midplane images of live embryos expressing PAR-2::GFP and PAR-6::mCHERRY were taken during the maintenance phase, after centration but before nuclear-envelope breakdown. The bright cortical halo seen in the red channel even after loss of polarity reflects autofluorescence of the eggshell which is weak relative to the PAR-6::mCHERRY signal in polarized embryos (see, for instance, 1 and 2 h timepoints). Each image has been rotated (so that the anterior pole is on the left and the posterior pole is on the right). Plots (right) show relative fluorescence intensities measured at the cortex as a function of position along the perimeter. Differences in intensity from one image to another reflect autoscaling and thus only the relative intensities are relevant. The abrupt loss of polarity seen ∼5 h after the start of RNAi treatment, in multiple experiments (see Fig. 8), is consistent with model predictions.
Figure 8.
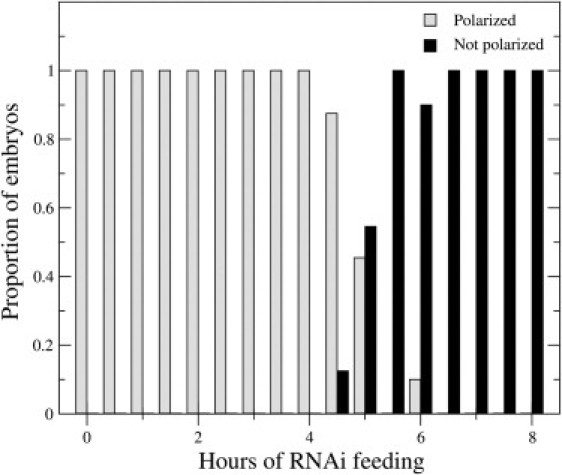
The loss of polarization in early C. elegans embryos resulting from PAR-6 depletion is consistently observed across repeated par-6 RNA interference experiments (n = 10–25 for each time point).
Our model shows that bistability can ensure quasistability of complementary domains in the presence of diffusion if interaction strengths and/or diffusivities are properly balanced. However, additional mechanisms may be involved in stabilizing the boundary against excess drift. Coupling our Par protein model to Tostevin and Howard's (39) actomyosin model effectively stabilized the boundary against drift without affecting bistability or its loss when ParA is depleted (see Fig. S2, Fig. S3, and Fig. S4). Another possibility is that depletion of a cytoplasmic pool of ParA and/or ParP could reduce or even eliminate boundary drift.
To test this idea, we assumed that the total pool of ParA and ParP are constant, and introduced a scaling factor α that relates the total effective volume of the cortex/PM compartment to the cytoplasmic volume as
| (4a) |
| (4b) |
where Ω denotes the total cross-sectional area of the cortex. We set initial values for ParAcyto and ParPcyto to 1, then we repeated the parameter space search for different values of α (0.02, 0.05, and 0.1), expecting an increased frequency of solutions if depletion buffers against drift (see Table S3). For α = 0.02, we observed minimal effects on solution frequency (1/254,600 vs. 1/239,000 for nonlimiting pool) and the boundary eventually drifted toward the anterior or posterior pole in all cases. However, for α = 0.1, we observed a 57-fold increase in the frequency of successful solutions. For 83% of these solutions, profiles of ParA and ParP (and thus the AP boundary) were stably held for a long simulation time. We conclude that cytoplasmic depletion can buffer a boundary against drift if the coupling is sufficiently strong.
Discussion
Polarity establishment in the C. elegans zygote involves dynamic segregation of Par proteins through active transport by actomyosin-based cortical flows (45,49). Once the Par domains are established, the zygote can maintain polarity in the absence of cortical actin for many minutes, with minimal drift in the boundary between the Par domains (1). Here we explored conditions under which cross-inhibition of anterior and posterior Par proteins could explain the actin-independent maintenance of preestablished polarities. We find that simple cross-promotion of dissociation via mass-action kinetics cannot do so for any choice of kinetic parameters. However, simple cross-inhibition plus PAR-3 dimerization can account for stable maintenance of complementary Par domains and for quasistability of AP boundary position in the face of diffusion.
A key feature of our model is that dimerization of PAR-3 (plus cross-inhibition) yields bistable dynamics, which in a spatially distributed system supports coexistence of complementary domains. Dimerization of PAR-3 replaces a monovalent linkage to the cortex with a divalent one, leading to cooperative binding kinetics via a nonlinear dependence of PAR-3 release on local density of PAR-3. Our model suggests that this nonlinearity plus cross-inhibition is sufficient to endow the Par system with intrinsic bistability. Although we limited consideration to PAR-3 dimers to reduce model complexity, PAR-3 may actually form higher-order structures such as filaments (21), which would yield even sharper nonlinearities in binding kinetics and thus increased potential for bistability. Studies in C. elegans, Drosophila, and mammalian cells have shown that inhibiting PAR-3 oligomerization abolishes or severely attenuates cortical localization of PAR-3 (19,21,23). These results are consistent with a loss of bistability, but could also be explained solely by a drop in PAR-3 affinity. Distinguishing these possibilities is an important goal for future experiments.
In the presence of diffusion, the boundary between Par protein domains in the model is unstable and tends to drift. This is not necessarily a failure of the model; Indeed, the AP boundary exhibits slow drift toward the posterior pole in embryos lacking functional myosin or treated with actin depolymerizing drugs (1,40). Such minor drift may be of little consequence because Par protein asymmetries need only be maintained until cleavage (i.e., for 5 min). Moreover, additional mechanisms operate during cell division to correct potential mismatch between the Par boundary and the cleavage plane (50). Our random sampling of parameter space shows that this drift can be readily minimized by tuning Par diffusivities and/or cross-interaction strengths and C. elegans embryos may rely in part on a similar tuning. However, additional mechanisms likely buffer the system against excess drift.
One potential mechanism to prevent drift in the Par protein boundary is that of wave-pinning—previously studied in the context of motile cells (51). In the wave-pinning model, rapid exchange between slow-diffusing active and fast-diffusing inactive proteins, whose total numbers are conserved in a one-dimensional geometry, results in stable boundary positioning. When we assume a weakly-limiting, well-mixed cytoplasmic pool (i.e., no turnover of Par protein; total cortical Par pool ∼10% of total cytoplasmic pool), we find a substantial increase in the number of randomly chosen parameters that yield acceptable drift rates, and many of these (83%) yield absolute boundary stability—suggesting that a variant of the wave-pinning mechanism could supply a significant buffer against boundary drift in this system.
Additional mechanisms may involve coupling of Par protein dynamics to polarized actomyosin. For example, Tostevin and Howard (39) proposed one such mechanism that involves enhanced recruitment of ParA to an anterior actomyosin-rich domain, which is stabilized by contracting against an elastic resistance. Incorporating that mechanism into our model can stabilize the boundary against drift (see Fig. S2). Another potential mechanism could involve active transport of Par proteins as occurs during polarity establishment. Indeed, there is a pronounced flow toward the AP boundary during maintenance in wild-type embryos (45), and we have recently identified a system of feedback interactions, involving Cdc42 and downstream effectors, that contributes to stabilizing the AP boundary through a balance of chemical flux, active contractility, and passive resistance to cortical deformation (C. Schoff, H. Clark, and E. Munro. unpublished).
A key prediction of our model is that there should be an abrupt loss of polarity as the levels of ParA (or ParP) are gradually decreased, and we confirmed this prediction experimentally using RNAi-based depletion of PAR-6. These results strongly support a role for bistable dynamics in polarity maintenance; whether Par-3 oligomerization is essential for bistability remains to be seen. Our model also reproduces spatial shifts in the Par boundary observed when anterior or posterior Par proteins are depleted and/or overexpressed (see Zonies et al. (46) and references therein); furthermore, it qualitatively accounts for the observation (46,52) that partial loss of function in anterior (or respectively, posterior) Par proteins can be rescued by reducing levels and/or activity of opposing posterior (or respectively, anterior) proteins. On the other hand, our model cannot account for certain mutant phenotypes, such as the retraction of the anterior Par domain when PAR-2 is depleted (45)—which is perhaps not surprising, as this retraction is accompanied, and likely caused by, ectopic actomyosin-based cortical flows (45).
Clearly a full account of polarization in C. elegans must integrate Par protein dynamics with actomyosin contractility. For example, the bistable switch mechanism we describe here likely synergizes with actomyosin-based cortical flows to break symmetry during polarity establishment (5): The anterior enrichment of ParA proteins by actomyosin-based flows could ensure that the switch remains in its initial high ParA/low ParP state at the anterior, while depletion of ParA at the posterior pole would throw the same switch toward a low ParA/high ParP state. An actin-independent, bistable switch mechanism may be sufficient to maintain quasistable Par protein domains, but as mentioned above, actomyosin dynamics and cortical flow almost certainly play a role in stabilizing the boundary.
Future work will incorporate spatial aspects of Par protein interactions, including possible mechanisms for initiation of cortical flow and mechanisms for spatial positioning of the Par protein boundary at the end of the establishment phase.
Acknowledgments
The authors thank members of the Center for Cell Dynamics, University of Washington, for helpful discussions and critical reading of the manuscript.
This work was supported by the National Institute of General Medical Sciences 5P50 GM66050-02 Center for Cell Dynamics, University of Washington (A.T.D. and E.M.M.), and a Natural Sciences and Engineering Research Council Discovery grant (A.T.D.).
Supporting Material
References
- 1.Goehring N.W., Hoege C., Hyman A.A. PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. J. Cell Biol. 2011;193:583–594. doi: 10.1083/jcb.201011094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda V., Nolan M.E., Muthuswamy S.K. PAR complex in cancer: a regulator of normal cell polarity joins the dark side. Oncogene. 2008;27:6878–6887. doi: 10.1038/onc.2008.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein B., Macara I.G. The PAR proteins: fundamental players in animal cell polarization. Dev. Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki A., Ohno S. The PAR-aPKC system: lessons in polarity. J. Cell Sci. 2006;119:979–987. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 5.Munro E., Bowerman B. Cellular symmetry breaking during Caenorhabditis elegans development. Cold Spring Harb. Perspect. Biol. 2009;1:a003400. doi: 10.1101/cshperspect.a003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuenca A.A., Schetter A., Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung T.J., Kemphues K.J. PAR-6 is a conserved PDZ domain-containing protein that colocalizes with PAR-3 in Caenorhabditis elegans embryos. Development. 1999;126:127–135. doi: 10.1242/dev.126.1.127. [DOI] [PubMed] [Google Scholar]
- 8.Beers M., Kemphues K. Depletion of the co-chaperone CDC-37 reveals two modes of PAR-6 cortical association in C. elegans embryos. Development. 2006;133:3745–3754. doi: 10.1242/dev.02544. [DOI] [PubMed] [Google Scholar]
- 9.Hird S.N., White J.G. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeks R.J., Canman J.C., Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 11.Petrásek Z., Hoege C., Schwille P. Characterization of protein dynamics in asymmetric cell division by scanning fluorescence correlation spectroscopy. Biophys. J. 2008;95:5476–5486. doi: 10.1529/biophysj.108.135152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joberty G., Petersen C., Macara I.G. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2000;2:531–539. doi: 10.1038/35019573. [DOI] [PubMed] [Google Scholar]
- 13.Lin D., Edwards A.S., Pawson T. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signaling and cell polarity. Nat. Cell Biol. 2000;2:540–547. doi: 10.1038/35019582. [DOI] [PubMed] [Google Scholar]
- 14.Nagai-Tamai Y., Mizuno K., Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7:1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 15.Izumi Y., Hirose T., Ohno S. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J. Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Etienne-Manneville S., Hall A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr. Opin. Cell Biol. 2003;15:67–72. doi: 10.1016/s0955-0674(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki A., Yamanaka T., Ohno S. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J. Cell Biol. 2001;152:1183–1196. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Kim H., Kemphues K.J. Binding to PKC-3, but not to PAR-3 or to a conventional PDZ domain ligand, is required for PAR-6 function in C. elegans. Dev. Biol. 2010;340:88–98. doi: 10.1016/j.ydbio.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li B., Kim H., Kemphues K. Different domains of C. elegans PAR-3 are required at different times in development. Dev. Biol. 2010;344:745–757. doi: 10.1016/j.ydbio.2010.05.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krahn M.P., Klopfenstein D.R., Wodarz A. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 2010;20:636–642. doi: 10.1016/j.cub.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 21.Feng W., Wu H., Zhang M. The Par-3 NTD adopts a PB1-like structure required for Par-3 oligomerization and membrane localization. EMBO J. 2007;26:2786–2796. doi: 10.1038/sj.emboj.7601702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benton R., St. Johnston D. A conserved oligomerization domain in Drosophila Bazooka/PAR-3 is important for apical localization and epithelial polarity. Curr. Biol. 2003;13:1330–1334. doi: 10.1016/s0960-9822(03)00508-6. [DOI] [PubMed] [Google Scholar]
- 23.Mizuno K., Suzuki A., Ohno S. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J. Biol. Chem. 2003;278:31240–31250. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- 24.Munro E.M. PAR proteins and the cytoskeleton: a marriage of equals. Curr. Opin. Cell Biol. 2006;18:86–94. doi: 10.1016/j.ceb.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 25.Hurov J.B., Watkins J.L., Piwnica-Worms H. Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Kusakabe M., Nishida E. The polarity-inducing kinase Par-1 controls Xenopus gastrulation in cooperation with 14-3-3 and aPKC. EMBO J. 2004;23:4190–4201. doi: 10.1038/sj.emboj.7600381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki A., Hirata M., Ohno S. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 28.Hao Y., Boyd L., Seydoux G. Stabilization of cell polarity by the C. elegans RING protein PAR-2. Dev. Cell. 2006;10:199–208. doi: 10.1016/j.devcel.2005.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plant P.J., Fawcett J.P., Pawson T. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian LGL. Nat. Cell Biol. 2003;5:301–308. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 30.Hoege C., Constantinescu A.T., Hyman A.A. LGL can partition the cortex of one-cell Caenorhabditis elegans embryos into two domains. Curr. Biol. 2010;20:1296–1303. doi: 10.1016/j.cub.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 31.Benton R., St. Johnston D. Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- 32.Hurd T.W., Fan S., Margolis B. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr. Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 33.Dawes A.T., Edelstein-Keshet L. Phosphoinositides and Rho proteins spatially regulate actin polymerization to initiate and maintain directed movement in a one-dimensional model of a motile cell. Biophys. J. 2007;92:744–768. doi: 10.1529/biophysj.106.090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onsum M., Rao C.V. A mathematical model for neutrophil gradient sensing and polarization. PLOS Comput. Biol. 2007;3:e36. doi: 10.1371/journal.pcbi.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong Y., Huang C.H., Devreotes P.N. Cells navigate with a local-excitation, global-inhibition-biased excitable network. Proc. Natl. Acad. Sci. USA. 2010;107:17079–17086. doi: 10.1073/pnas.1011271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narang A. Spontaneous polarization in eukaryotic gradient sensing: a mathematical model based on mutual inhibition of frontness and backness pathways. J. Theor. Biol. 2006;240:538–553. doi: 10.1016/j.jtbi.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 37.Meinhardt H. Complex pattern formation by a self-destabilization of established patterns: chemotactic orientation and phyllotaxis as examples. C. R. Biol. 2003;326:223–237. doi: 10.1016/s1631-0691(03)00018-0. [DOI] [PubMed] [Google Scholar]
- 38.Iglesias P.A., Devreotes P.N. Navigating through models of chemotaxis. Curr. Opin. Cell Biol. 2008;20:35–40. doi: 10.1016/j.ceb.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Tostevin F., Howard M. Modeling the establishment of PAR protein polarity in the one-cell C. elegans embryo. Biophys. J. 2008;95:4512–4522. doi: 10.1529/biophysj.108.136416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J., Maduzia L.L., Mello C.C. NMY-2 maintains cellular asymmetry and cell boundaries, and promotes a SRC-dependent asymmetric cell division. Dev. Biol. 2010;339:366–373. doi: 10.1016/j.ydbio.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill D.P., Strome S. An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev. Biol. 1988;125:75–84. doi: 10.1016/0012-1606(88)90060-7. [DOI] [PubMed] [Google Scholar]
- 42.Press W., Flannery B., Vetterling W. 2nd Ed. Cambridge University Press; New York: 2002. Numerical Recipes in C: The Art of Scientific Computing. [Google Scholar]
- 43.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timmons L., Court D.L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 45.Munro E., Nance J., Priess J.R. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Zonies S., Motegi F., Seydoux G. Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development. 2010;137:1669–1677. doi: 10.1242/dev.045823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montgomery M.K., Xu S., Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fire A., Xu S., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 49.Velarde N., Gunsalus K.C., Piano F. Diverse roles of actin in C. elegans early embryogenesis. BMC Dev. Biol. 2007;7:142. doi: 10.1186/1471-213X-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schenk C., Bringmann H., Cowan C.R. Cortical domain correction repositions the polarity boundary to match the cytokinesis furrow in C. elegans embryos. Development. 2010;137:1743–1753. doi: 10.1242/dev.040436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mori Y., Jilkine A., Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys. J. 2008;94:3684–3697. doi: 10.1529/biophysj.107.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kay A.J., Hunter C.P. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 2001;11:474–481. doi: 10.1016/s0960-9822(01)00141-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


