Abstract
Purpose
The generation of non-viable homozygous null mouse embryos from heterozygote null/+ breedings can be highly resource consuming, with only 25% of the embryos in the litter being null mutants. We hypothesized that 1) we could double the number of homozygous null mouse embryos in a litter without reducing litter size using Hypoxanthine-guanine phosphoribosyltransferase-Cre (Hprt-Cre) (which is active in the female germ line at the time of fertilization) and 2) these homozygous null mutants would be identical to mutants generated through traditional null/+ breedings.
Methods
To test this hypothesis we used a conditional allele Fgfr2IIIbflox. This allele when recombined is identical to the Fgfr2IIIbnull allele. An F1 generation of Fgfr2IIIbrec/+; HprtCre/+ females was created by mating Fgfr2IIIb+/+; Hprtcre′/cre females to a Fgfr2IIIbflox/flox male. The F1 females were then mated to a Fgfr2IIIbflox/flox male. F2 embryos were genotyped and the morphology and histology of the lungs, intestine, limbs and brain was analyzed.
Results
The Hprt-Cre mating strategy results in 51% of pups being genotypic homozygous null embryos (85/166) versus 23% for the standard null/+ approach (38/167). These embryos did not express the Fgfr2IIIb transcript and were phenotypically identical to null embryos generated through standard null/+ breedings.
Conclusions
The Hprt-Cre mating strategy increases the number of homozygous mutant embryos in a litter without decreasing litter size. Embryos generated through this approach are phenotypically identical to those from standard heterozygous breedings. We recommend this approach to investigators using an model system that relies on the generation of homozygous null embryos.
Keywords: Hprt-Cre, cre loxp system, breeding strategy, homozygous null mutants, efficiency, generation of mutants, knockout
Use of murine homozygous null mutant embryos has been critical for modeling a number congenital defects in humans. With this approach animal models for imperforate anus and cloaca [1], intestinal atresia [2, 3], diaphragmatic hernia [4], abdominal wall defects [5–7], limb defects [8], and cleft palate [9, 10] have been described. Generation of homozygous null embryos through null/+ (n/+) heterozygous breedings can be highly resource consuming. When employing the n/+ breedings for one gene, a single homozygous null (n/n) embryo is generated for every 4 embryos. Thus, a litter of 10 will yield 2 or 3 homozygous null mutant embryos. Generating compound homozygous null mutants for 2 or 3 genes with this process becomes even less efficient as the frequency of homozygous null mutants decreases to 1 in 16 embryos and 1 in 64 embryos, respectively. Researchers also face increasing pressure to minimize the number of animals used while maintaining, or even increasing, the quality of basic science research. This pressure is driven in part by both the animal rights movement [11, 12] and increasing animal care and housing costs at research universities. Strategies that can reduce the number of mice required to generate homozygous null embryos have the potential to significantly cut research costs for scientists in many fields.
We hypothesized that Hprt Cre [13] could be used to circumvent standard heterozygote breeding strategy inefficiencies. In these mice, a Cre gene under a cytomegalovirus enhancer/chicken β-globin promoter has been inserted into the X-linked locus for hypoxanthine-guanine phosphoribosyltransferase yielding Cre protein in the female germ line prior to the generation of oocytes. Thus, in a female heterozygous for Hprt-Cre, all ova receive the active Cre protein regardless of inheritance of the Hprt Cre gene. Once the oocyte with the Cre protein is fertilized with sperm, the flox allele derived from the male genome should be recombined by the Cre protein at the 1 or 2 cell stage. Hypothetically if a male homozygous for a conditional allele is mated to a female who is carrying a single copy of the Hprt-Cre gene and a copy of the recombined allele of interest then half of the progeny should be homozygous for the recombined allele. If the recombined allele is identical to the null allele for the same gene, this method should increase the number of null mutants in a litter as the active ova-derived Cre protein recombines a sperm-derived floxed allele at fertilization.
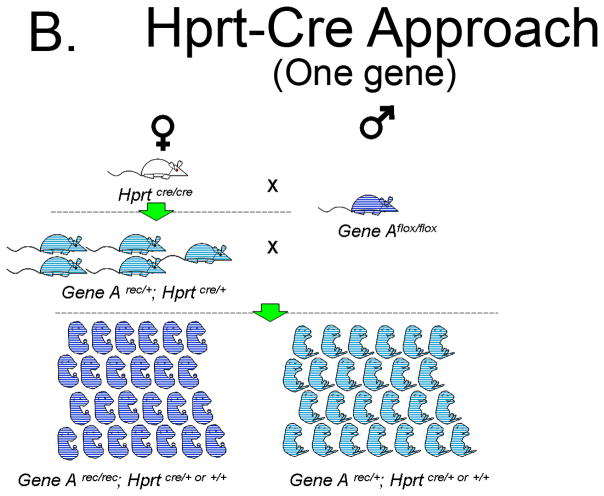
The novel Hprt-Cre approach depends on having a conditional floxed allele of the gene of interest (a hypothetical Gene Aflox). Ideally, following Cre dependent recombination of Gene Aflox, the Gene Arec should be genetically identical to a Gene Anull allele. Thereafter, the strategy is simple: First, generate Gene Arec/+; HprtCre/+ female progeny (F1) from a homozygous HprtCre/Cre to a homozygous Gene Aflox/flox mating. Next, breed the F1 Gene Arec/+; HprtCre/+ females to a Gene Aflox/flox male to generate homozygous mutants (F2). Half of the ova from the F1 females will carry the Gene Arec allele and all the ova will have an active Cre protein. Sperm-derived Gene Aflox alleles will be recombined upon fertilizing these ova. Presumably, all of the Gene Arec/rec embryos will be phenotypically equivalent to Gene Anull/null embryos. Thus, half of the F2 embryos generated with this strategy should be Gene Arec/rec homozygotes and half should be Gene Arec/+ heterozygotes.
To test the efficiency of the Hprt-Cre strategy, we determined the number of phenotypically null embryos in a litter as compared to the standard heterozygous null/+ breeding strategy. We utilized the floxed Fgfr2IIIb allele [14, 15] which, when recombined, is genetically identical to the Fgfr2IIIb null allele [2]. The Fgfr2IIIbnull/null embryos have been well characterized with defects in a number of organ systems including, but not limited to, the limbs, lungs, intestines, pituitary gland and eyes [2, 14, 16]. Here we report that Fgfr2IIIbrec/rec embryos generated with Hprt-Cre methodology are identical to previously characterized Fgfr2IIIbnull/null embryos and that the Hprt-Cre strategy increases the frequency of null embryos in a given litter from 1-in-4 to 1-in-2 without reduction in litter size.
1. Methods
1.1 Animals
IACUC approval for these studies was obtained from the University of Wisconsin School of Medicine and Public health (P.F.N. protocol # M02258). All animals were maintained in a clean facility with ad libitum access to fresh food and water, and kept on a 12 hour alternating light/dark cycle. For animals generated with Hprt-Cre, the recombined Fgfr2IIIb floxed allele is genetically identical to that of the Fgfr2IIIb null allele [14, 15]. Five Fgfr2IIIb+/+; HprtCre/Cre females were bred to Fgfr2IIIbflox/flox males generating a total of 40 pups. Male pups were sacrificed and female pups were genotyped by toe clipping before P10 for Hprt-Cre, Fgfr2IIIb wild-type, null and floxed alleles [14, 15]. The genotype of all female pups was Fgfr2IIIbrec/+; HprtCre/+. Fgfr2IIIbrec/rec embryos were generated by breeding mature Fgfr2IIIbrec/+; HprtCre/+ females to Fgfr2IIIbflox/flox males. Fgfr2IIIbnull/null embryos were generated through traditional heterozygous crossing of Fgfr2IIIbnull/+ females to Fgfr2IIIbnull/+ males. Genotyping of embryos for Fgfr2IIIb alleles was performed as described previously [14, 15]. Ratios of Fgfr2IIIbrec/rec and Fgfr2IIIbnull/null embryos were determined.
1.2 RT-PCR
Total RNA was prepared from fetal tissues on Embryonic day (E)18.5 using the GeneJet RNA extraction kit (Fermentas Inc., Glen Burnie, MD), according to the manufacturer’s protocol. 0.5 μg of RNA was used with the RevertAid cDNA Synthesis Kit (Fermentas Inc., Glen Burnie, MD), according to the manufacturer’s protocol. The following cDNA-specific primers were used; 5′ Fgfr2IIIb, ttc aat gtg acg gag atg; 3′ Fgfr2IIIb, ctg ttt ggg cag gac agt g; 5′ Gapdh, caa ggt cat cca tga caa ctt tg; 3′ Gapdh, gtc cac cac cct gtt gct gta g. Thermocycler conditions for Fgfr2IIIb were: 94°C/30 sec, 58°C/30 sec, 72°C/45 sec, for 35 cycles then 72°C/7 minutes for one cycle. Thermocycler conditions for Gapdh were: 94°C/30 sec, 58°C/30 sec, 72°C/45 sec, for 35 cycles then 72°C/5 minutes. PCR products were run on a 1.5% agarose gel and visualized by ethidium bromide staining.
1.3 Morphologic studies
Embryos generated using the Hprt-Cre strategy were harvested at either (E) 10.5, E14.5 or E18.5. Fgfr2IIIbnull/null embryos generated from Fgfr2IIIbnull/+ heterozygous crossings were harvested at E18.5. Whole mount photographs were taken of E18.5 embryos under a stereoscopic dissecting microscope. The thorax was opened and the embryos were fixed overnight in 4% paraformaldehyde at 4°C. The intestines were dissected out and photographs of the duodenal, cecal and colonic regions were obtained. Incidence of duodenal, colonic and cecal defects for all mutants was determined.
1.4 Histological studies
E10.5 and E14.5 embryos were dissected free of the yolk sac and embryos were fixed overnight at 4°C in 4% paraformaldehyde. They were then dehydrated through a series of escalating Methanol/PBST washes, isopropyl alcohol and xylene. They were washed 3 times in paraffin at 60 °C in a vacuum oven and then embedded head up in paraffin. Sections were taken a 5μm thickness. Sections were stained for standard H&E or TUNEL staining and then photographed under either a light microscope or a fluorescent microscope.
1.5 Three-dimensional reconstructions
Photomicrographs of transverse sections of the head region of E10.5 embryos were uploaded sequentially into Amira™ (Visage Imaging, Inc., San Diego, CA) and aligned using the auto align tool. They were labeled using the segmentation editor. Following this, the three dimensional structures of Rathke’s pouch and the Buccal cavity along with the Diencephalon were generated using the surface generator tool. A final image was generated as a JPEG file at 600 pixels per square inch and imported into Adobe Photoshop (San Jose, CA) for labeling.
Apoptosis studies
Embryos heterozygous and homozygous for the Fgfr2IIIbrec allele were harvested at E10.5, fixed and embedded in paraffin as described above. Transverse sections were generated at 5 μm and processed for detection of apoptosis. The DeadEnd Fluorometric TUNEL System (Promega, Madison, WI) was used per supplied instructions with appropriate positive and negative controls. Slides were cover slipped and processed sections were then photographed with a fluorescent microscope.
2. Results
Comparative efficiencies of Hprt-Cre and null/+ breedings in generating homozygous mutant embryos
We tested whether the Hprt-Cre strategy could double the number of homozygous mutant embryos compared to the standard heterozygous null/+ breeding strategy using the Fgfr2IIIbflox allele. Eighteen litters were generated by mating Fgfr2IIIbrec/+; HprtCre/+ females to Fgfr2IIIbflox/flox males and 18 litters were generated through standard Fgfr2IIIbnull/+ heterozygous matings. The 18 Hprt-Cre litters yielded 166 embryos (average of 9.22/litter), where fifty-one percent were homozygous mutants. The 18 standard heterozygous null/+ strategy litters yielded a total 167 embryos (average of 9.28/litter) of which 23% were homozygous mutants (Table 1). The results indicated that the Hprt-Cre doubles the number of homozygous mutant embryos within a litter without a reduction in total litter size.
Table 1.
Relative efficiencies of generating mutant embryos with Hprt-Cre vs. Traditional heterozygous null/+ breedings
| Breeding | Normal (n) | Mutant (n) | % Mutants | Litters | Embryos/litter(n) |
|---|---|---|---|---|---|
| Fgfr2IIIbrec/+; HprtCre/+ X Fgfr2IIIb flox/flox | 81 | 85 | 51.20% | 18 | 9.22 |
| Fgfr2IIIbnull/+ X Fgfr2IIIbnull/+ | 129 | 38 | 22.75% | 18 | 9.28 |
2.2 Expression of the Fgfr2IIIb exon in Fgfr2IIIbrec/rec embryos
To confirm Hprt-Cre removes all detectable expression of the conditional allele of interest, expression of the Fgfr2IIIb exon in Fgfr2IIIbrec/rec embryos was assessed by RT-PCR. By E18.5, there were no detectable levels of Fgfr2IIIb in the Fgfr2IIIbrec/rec embryos (Figure 1A). In contrast, levels of Gapdh (a standard housekeeping gene) in the wild-type embryos was identical to that of Fgfr2IIIbrec/rec embryos (Figure 1B). This indicated that Fgfr2IIIbrec/rec embryos generated with Hprt-Cre were genotypically and transcriptionally equivalent to Fgfr2IIIbnull/null embryos.
Figure 1. Fgfr2IIIbrec/rec mutants do not express the Fgfr2IIIb exon.
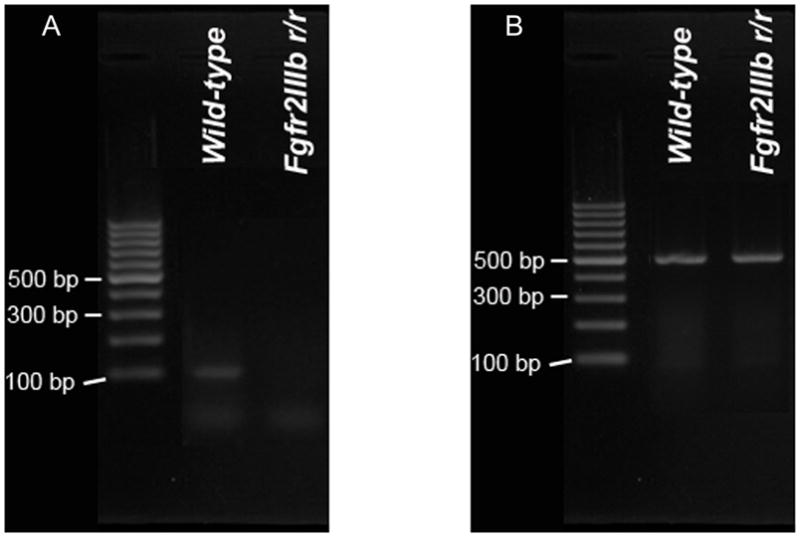
RT-PCR was used to assess expression of the Fgfr2IIIb exon in wild-type embryos and embryos homozygous for the Fgfr2IIIbrec allele. RNA was isolated from whole embryos at E18.5 and converted to cDNA. RT-PCR was performed with primers that hybridize within exon IIIb of Fgfr2 (A) or with primers for Gapdh (B). The product size for the Fgfr2IIIb exon is 102 bp and 496 bp for Gapdh. Note the lack of expression of the Fgfr2IIIb exon in the embryos homozygous for the Fgfr2IIIbrec allele.
2.3 External appearance, limb and pituitary development
To determine whether Fgfr2IIIbrec/rec embryos are phenotypically equivalent to Fgfr2IIIbnull/null embryos, we conducted morphological and histological studies of Fgfr2IIIbrec/rec embryos in several organ systems and compared these results to the published phenotype for Fgfr2IIIbnull/null embryos.
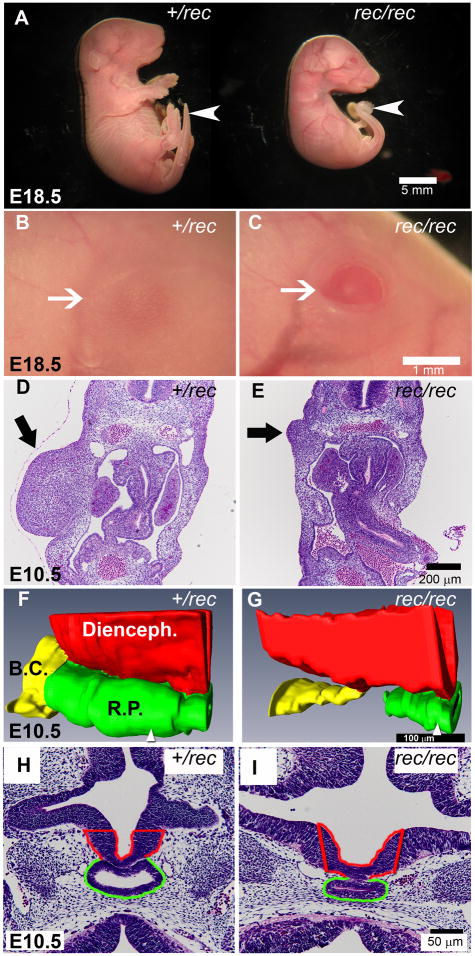
Fgfr2IIIbrec/rec embryos appeared externally identical to Fgfr2IIIbnull/null embryos [14]. At E18.5, all Fgfr2IIIbrec/rec mutants exhibited a curved tail and an eyelid defect (Figure 2A–C) with absent or significantly attenuated limbs (Figure 2A). Consistent with this, at E10.5 Fgfr2IIIbrec/rec embryos had diminished limb bud mesenchyme in comparison to heterozygous littermates (Figure 2D and E). Fgfr2IIIbrec/rec embryos were also smaller at E18.5 than heterozygous littermates (Figure 2A). Not surprisingly, these embryos also exhibited early defects in pituitary development at E10.5. Rathke’s pouch was narrowed along its dorsal-ventral axis and shortened along the rostral-caudal axis such that it was not in continuity with the buccal cavity (Figure 2F–I). These findings are equivalent to Fgfr2IIIbnull/null embryos at this stage [14].
Figure 2. Morphology of Fgfr2IIIbrec/rec mutants.
(A) Low magnification views of Fgfr2IIIbrec/+ and Fgfr2IIIbrec/rec embryos. At E18.5, the Fgfr2IIIbrec/rec mutant lacks limbs, is smaller than its heterozygous littermate, and also has a curved tail (white arrow). (B and C) Higher magnification of the eye (white arrow) from embryos shown in (A). The Fgfr2IIIbrec/rec embryo has an eyelid defect. (D and E) Transverse sections through the limb region at E10.5. The Fgfr2IIIbrec/rec mutant (E, black arrow) exhibits defects in the limb mesenchyme. (F–I) Pituitary development at E 10.5. (F and G) 3-dimensional reconstructions of Rathke’s pouch (RP), the Buccal cavity (BC), and the diencephalon (Di) generated from transverse sections. (H and I) Representative sections from the specimens reconstructed in F and G. Position of the sections (H and I) within each reconstruction are indicated by the white arrowhead at the bottom of panels F and G. In the Fgfr2IIIbrec/rec mutant, Rathke’s pouch is narrower in the dorsal-ventral axis and no longer continuous with the Buccal cavity.
2.4 Lungs
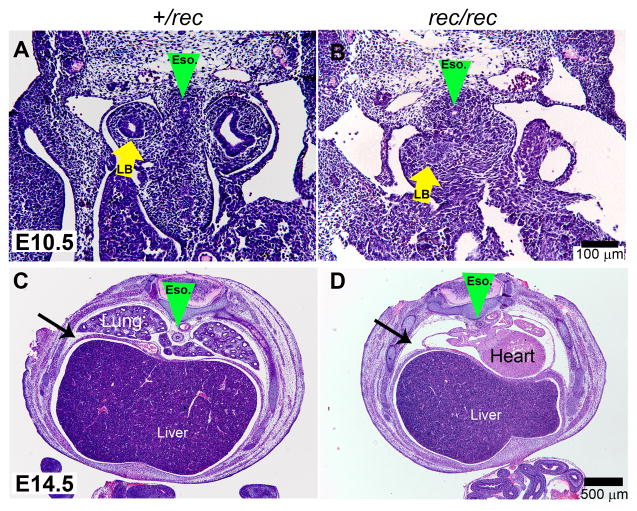
Fgfr2IIIbrec/rec embryos also had a lung defect that was identical to that described in Fgfr2IIIbnull/null embryos [14]. At day 10.5 a tracheal bud formed, but only minimal secondary branching occurred with bronchial buds terminating blindly in the early lung mesenchyme (Figure 3A and B, yellow arrow). Examination of Fgfr2IIIbrec/rec embryos at day 14.5 demonstrated the absence of lungs in the pleural cavity (Figure 3C and D).
Figure 3. Lung development in Fgfr2IIIbrec/rec mutants.
(A and B) Transverse sections through the thoracic region at E10.5. Green arrowhead indicates position of the esophagus. Yellow arrows indicate lung buds. (B) Note that Fgfr2IIIbrec/rec mutant lung bud has terminated into the mesenchyme. (C and D) Low magnification views of transverse sections through the thoracic cavity at E 14.5.. In the Fgfr2IIIbrec/rec mutant, lungs are absent from the thoracic cavity which is indicated by the black arrows.
2.5 Intestine
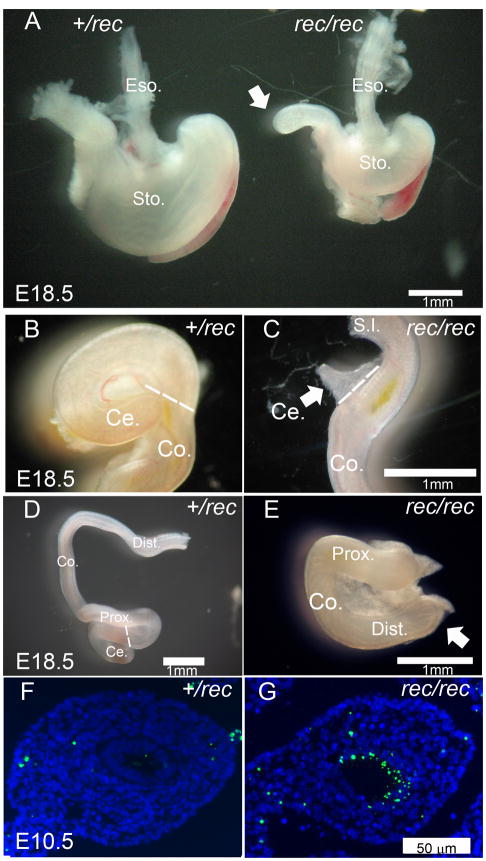
The morphology of the gastrointestinal tract of Fgfr2IIIbrec/rec embryos at E18.5 was identical to that reported for Fgfr2IIIbnull/null embryos. The Fgfr2IIIbrec/rec embryos had a hypomorphic stomach in comparison to heterozygous litter mates (Figure 4A). Forty-two percent of Fgfr2IIIbrec/rec embryos developed a type III duodenal atresia (Figure 4A, white arrow). The penetrance of this defect was equivalent to that reported for Fgfr2IIIbnull/null embryos (Table 2) [2]. Additionally, all Fgfr2IIIbrec/rec embryos had a cecal defect (Figure 4B and C, white arrow) and a distal colonic atresia (Figure 4E, white arrow), both morphologically identical to that reported in Fgfr2IIIbnull/null embryos [16, 17]. The incidence of the colonic and cecal defects (Table 2) was also identical to that reported for Fgfr2IIIbnull/null embryos [16, 17]. Finally, the colonic endoderm of Fgfr2IIIbrec/rec embryos demonstrated significant apoptosis at E10.5 (Figure 4G). Apoptosis in the colonic endoderm occurs in Fgfr2IIIbnull/null embryos and is the leading event in the formation of colonic atresia [3]. These morphological and histological findings indicated that Fgfr2IIIbrec/rec embryos derived from the Hprt-Cre system are phenotypically identical to Fgfr2IIIbnull/null embryos for all structures examined.
Figure 4. Alimentary track development in Fgfr2IIIbrec/rec mutants.
(A) Stomach and duodenum of Fgfr2IIIbrec/+ and Fgfr2IIIbrec/rec embryos at E18.5 (B and C) the cecum, (D and E) and the colon. White arrows in A and E indicate atresias. White arrow in C indicates malformed cecum. Broken white line in B, C, and D indicates boundary between cecum and colon. TUNEL staining of transverse sections of colon in (F) Fgfr2IIIbrec/+ and (G) Fgfr2IIIbrec/rec embryos at E10.5.
Table 2.
incidence of duodenal atresia in Fgfr2IIIbrec/rec emrbyos generated with Hprt-Cre
| Mutants (n) | Colonic Atresia | Cecal Atresia | Duod. Atresia |
|---|---|---|---|
| 45 | 45 (100%) | 45 (100%) | 19 (42%) |
3. Discussion
We hypothesized that Hprt-Cre could double the number of homozygous knockout embryos in a litter without decreasing the litter size. To test this we chose Fgfr2IIIb mutants because they have defects in many organs. Embryos homozygous for the recombined allele had defects of the limbs, eyes, lungs and gut that are identical in both penetrance and appearance to that published for Fgfr2IIIbnull/null embryos [2, 14, 16, 17]. This strongly suggests that deletion of flox allele occurred in all cells and no mosaicism with non-recombined cells. We also demonstrated that Hprt-Cre in the maternal germ line effectively removes Fgfr2IIIb expression in Fgfr2IIIbrec/rec embryos. Thus rendering the Fgfr2IIIbrec/rec embryos genetically and phenotypically equivalent to Fgfr2IIIbnull/null embryos.
Use of Hprt-Cre to generate homozygous mutant embryos has been reported previously [18]. However, a detailed comparison between homozygous mutant embryos generated from heterozygous null/+ breedings to those generated with Hprt-Cre has not been published. Furthermore, this is the first comprehensive, quantitative demonstration that this method doubles the number of null embryos in a litter. The use of Hprt-Cre to generate null embryos should double the efficiency by which investigators can generate null embryos for a single gene, while simultaneously reducing the cost.
We would caution the investigator to characterize rec/rec embryos of their gene of interest to confirm they are phenotypically equivalent to a homozygous null embryos generated by a standard heterozygous breeding strategy. It is also possible that the recombination of the gene of interest by the Cre-recombinase can vary based on the genomic structure thus investigators should confirm that there is no mosacism as well. There is also an additional drawback that no wild type embryos can be obtained from this cross. Nevertheless this method is far more efficient than standard heterozygous breedings.
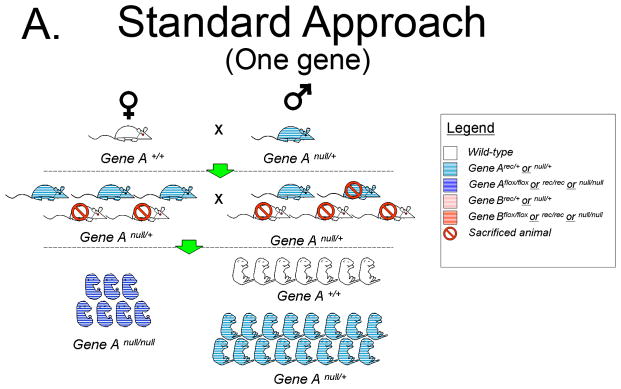
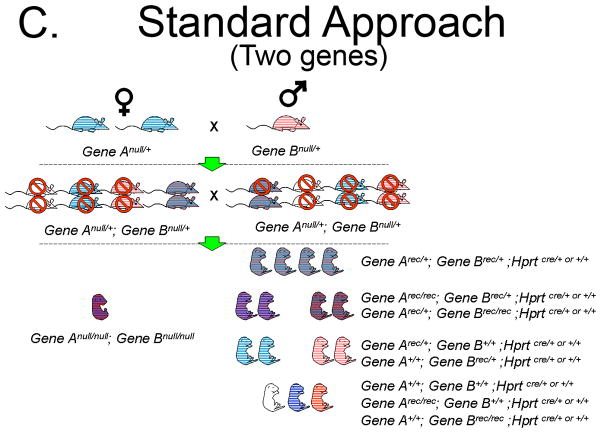
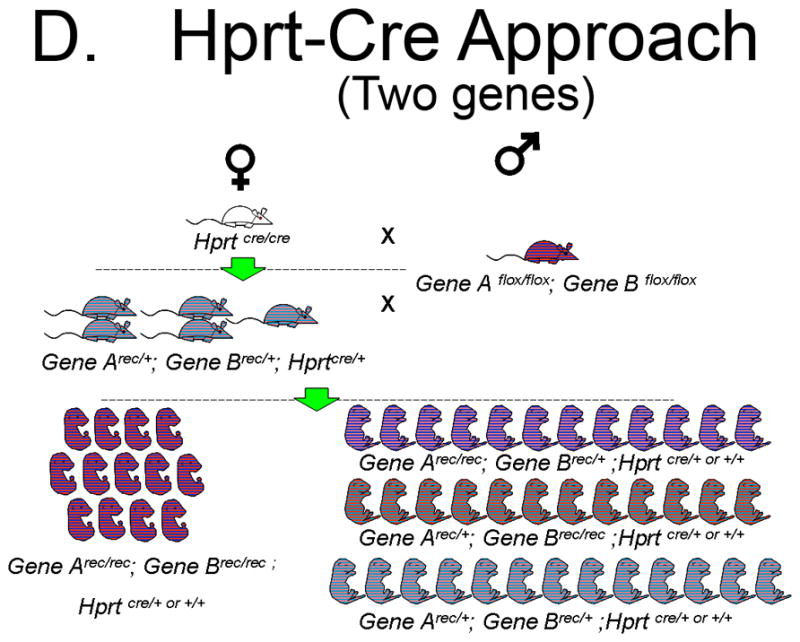
To illustrate the increased efficiency of Hprt-Cre in generating functional null embryos we conducted a comparison to the Standard Approach of a heterozygous mating strategy using a hypothetical Gene A. The Standard Approach One begins with one heterozygote for Gene Anull/+ male and one Gene A+/+ female (Figure 5A). The Hprt-Cre Approach begins with one HprtCre/Cre female and one Gene Aflox/flox male (Figure 5B). For purpose of comparison, the litter size of the F1 generation in each Approach will be 10 pups. The Standard Approach F1 progeny must be genotyped to identify those heterozygous for Gene Anull/+, of which there will be 5. For the sake of argument, three heterozygous pups will be females and two will be males, the remaining wild-type pups are sacrificed. By contrast, all of the F1 pups in the Hprt-Cre Approach will have the genotype Gene Arec/+; HprtCre/+. These pups are sexed, the females retained and the males are sacrificed (Figure 5B). When the F1 females of both approaches are greater than mature, they are bred to males. In the case of the Hprt-Cre Approach, the Gene Arec/+; HprtCre/+ females are bred to the original Gene Aflox/flox males. In the Standard Approach, the Gene Anull/+ females are bred to a Gene Anull/+ male. Each female becomes pregnant (5 F1 females in the Hprt-Cre Approach and 3 in the Standard Approach). Assuming a litter size of 10 embryos, the 5 Gene Arec/+; HprtCre/+ females will generate 50 embryos half of which (25) are Gene Arec/rec; and half (25) are Gene Arec/+. In the Standard Approach, the 3 Gene Anull/+ females will generate 30 pups of which 25% (7 or 8) will be Gene Anull/null embryos. Keeping the number of cages equal, the cost of generating a mutant with the Hprt-Cre Approach will be about 1/3 that of the Standard Approach. This calculation excludes the time and money required for genotyping the Standard Approach F1 generation, a step not required in the Hprt-Cre Approach.
Figure 5. Comparison of the efficiency of a standard heterozygous null/+ breeding strategy to the Hprt-Cre strategy.
A. Generation of homozygous null embryos for a single gene with the Standard Approach, a heterozygous null/+ breeding strategy. With this strategy the 3 F1 heterozygous females and single F1 heterozygous male will yield 7 to 8 null embryos. Thus, 25% of embryos in the F2 generation will be homozygous null embryos.
B. Generation of homozygous null embryos for a single gene with the Hprt-Cre Approach. With this strategy 50% of embryos in the F2 generation will be homozygous for the recombined allele.
C. Generation of homozygous null embryos for two genes with the Standard Approach, a heterozygous null/+ breeding strategy. With this strategy multiple females carrying the null allele are needed and 2 F1 double heterozygous females will be required to generate a single embryo. Only 6.25% (1/16) of embryos in the F2 generation will be homozygous null embryos for both genes.
D. Generation of homozygous null embryos for two genes with the Hprt-Cre Approach. With this strategy 5 F1 double heterozygous females will yield a total of 50 embryos, of which 12 or 13 will be homozygous null embryos for both genes. Therefore, 25% of embryos in the F2 generation will be homozygous for the recombined allele.
Differences in efficiency between the heterozygous mating strategy and the Hprt-Cre Approach increase dramatically when dealing with compound mutants. For example, one breeding pair of compound heterozygotes (Anull/+; Bnull/+ X Anull/+; Bnull/+) will generate a single double homozygous embryo for every 16 embryos (Figure 5C). In contrast, an Arec/+; Brec/+; HprtCre/+ X Aflox/flox; Bflox/flox breeding will generate double homozygous embryos at a frequency of 1 in 4 (Figure 5D). The real advantage, however, comes in generating the F1 progeny from an A+/+; B+/+; HprtCre/Cre X Aflox/flox; Bflox/flox mating which will yield 5 Arec/+; Brec/+; Hprt Cre/+; females from a single breeding pair. In contrast, a single mating pair of Anull/+;B +/+ X A+/+; Bnull/+ will generate both a single female and male Anull/+; Bnull/+ in the F1 generation only under ideal circumstances. Due to a 1 in 16 chance of generating an Anull/null; Bnull/null double homozygote from an Anull/+; Bnull/+X Anull/+; Bnull/+ breeding, a second Anull/+; Bnull/+ female is required. In the end, a single Anull/null; Bnull/null mutant will be generated from these Anull/+; Bnull/+ breedings. That is in stark contrast to the 12 or 13 Arec/rec; Brec/rec embryos that 5 F1 Arec/+; Brec/+; Hprt Cre/+; females should generate. The Hprt-Cre Approach yields a 12-fold increase in efficiency. This approach could be very useful when generating embryos that require homozygous mutations in multiple genes, such in the Msx1/Msx2 double knock-out model of omphalocele.
Existing Hprt-Cre technology represents an efficiency improvement for generating mutant murine animal models of human conditions. We believe this strategy will be useful to many investigators in helping to lower animal housing costs, reducing the time spent performing genotyping, and increasing the efficiency in generating null embryos.
Acknowledgments
Funding
PFN: American College of Surgeons Faculty Research Fellowship 2006–2008; Society for Surgery of the Alimentary Tract Career Development Award 2010–2012; NIH 1K08DK087854
KMZ: Hilldale Undergraduate/Faculty Research Fellowships 2011
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Peter F. Nichol, Email: Nichol@surgery.wisc.edu, Section of Pediatric Surgery Department of Surgery University of Wisconsin SMPH, Madison, WI, (O) (608) 263-9419, (F) (608) 261-1876.
Robert Botham, Specialist Section of Pediatric Surgery Department of Surgery University of Wisconsin SMPH Madison, WI.
Yukio Saijoh, Department of Neurobiology and Anatomy University of Utah Salt Lake City, UT.
Amy L. Reeder, Section of Pediatric Surgery, Department of Surgery University of Wisconsin SMPH, Madison, WI.
Krzyztoff M. Zaremba, Section of Pediatric Surgery, Department of Surgery, University of Wisconsin SMPH, Madison, WI.
References
- 1.Mo R, Kim JH, Zhang J, et al. Anorectal malformations caused by defects in sonic hedgehog signaling. Am J Pathol. 2001;159:765–774. doi: 10.1016/S0002-9440(10)61747-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fairbanks TJ, Kanard R, Del Moral PM, et al. Fibroblast growth factor receptor 2 IIIb invalidation--a potential cause of familial duodenal atresia. J Pediatr Surg. 2004;39:872–874. doi: 10.1016/j.jpedsurg.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 3.Fairbanks TJ, Sala FG, Kanard R, et al. The fibroblast growth factor pathway serves a regulatory role in proliferation and apoptosis in the pathogenesis of intestinal atresia. J Pediatr Surg. 2006;41:132–136. doi: 10.1016/j.jpedsurg.2005.10.054. discussion 132–136. [DOI] [PubMed] [Google Scholar]
- 4.Ackerman KG, Herron BJ, Vargas SO, et al. Fog2 is required for normal diaphragm and lung development in mice and humans. PLoS Genet. 2005;1:58–65. doi: 10.1371/journal.pgen.0010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manley NR, Barrow JR, Zhang T, et al. Hoxb2 and hoxb4 act together to specify ventral body wall formation. Dev Biol. 2001;237:130–144. doi: 10.1006/dbio.2001.0365. [DOI] [PubMed] [Google Scholar]
- 6.Ogi H, Suzuki K, Ogino Y, et al. Ventral abdominal wall dysmorphogenesis of Msx1/Msx2 double-mutant mice. Anat Rec A Discov Mol Cell Evol Biol. 2005;284:424–430. doi: 10.1002/ar.a.20180. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu Y, Thumkeo D, Keel J, et al. ROCK-I regulates closure of the eyelids and ventral body wall by inducing assembly of actomyosin bundles. J Cell Biol. 2005;168:941–953. doi: 10.1083/jcb.200411179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qu S, Niswender KD, Ji Q, et al. Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development. 1997;124:3999–4008. doi: 10.1242/dev.124.20.3999. [DOI] [PubMed] [Google Scholar]
- 9.Rice R, Spencer-Dene B, Connor EC, et al. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riley BM, Mansilla MA, Ma J, et al. Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci U S A. 2007;104:4512–4517. doi: 10.1073/pnas.0607956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayvel AC, Cross N. Animal welfare: a complex domestic and international public-policy issue--who are the key players? J Vet Med Educ. 2010;37:3–12. doi: 10.3138/jvme.37.1.3. [DOI] [PubMed] [Google Scholar]
- 12.Westly E. Animal rights activists try a more creative legal tactic. Nat Med. 2010;16:501. doi: 10.1038/nm0510-501. [DOI] [PubMed] [Google Scholar]
- 13.Tang SH, Silva FJ, Tsark WM, et al. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 14.De Moerlooze L, Spencer-Dene B, Revest JM, et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 15.Grose R, Fantl V, Werner S, et al. The role of fibroblast growth factor receptor 2b in skin homeostasis and cancer development. EMBO J. 2007;26:1268–1278. doi: 10.1038/sj.emboj.7601583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns RC, Fairbanks TJ, Sala F, et al. Requirement for fibroblast growth factor 10 or fibroblast growth factor receptor 2-IIIb signaling for cecal development in mouse. Dev Biol. 2004;265:61–74. doi: 10.1016/j.ydbio.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Fairbanks TJ, Kanard RC, Del Moral PM, et al. Colonic atresia without mesenteric vascular occlusion. The role of the fibroblast growth factor 10 signaling pathway. J Pediatr Surg. 2005;40:390–396. doi: 10.1016/j.jpedsurg.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 18.Urness LD, Paxton CN, Wang X, et al. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol. 2010;340:595–604. doi: 10.1016/j.ydbio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]