Abstract
The mTOR protein kinase is a master growth promoter that nucleates two complexes, mTORC1 and mTORC2. Despite the diverse processes controlled by mTOR, few substrates are known. We defined the mTOR-regulated phosphoproteome by quantitative mass spectrometry and characterized the primary sequence motif specificity of mTOR using positional scanning peptide libraries. We found that the phosphorylation response to insulin is largely mTOR-dependent and that mTOR exhibits a unique preference for proline, hydrophobic, and aromatic residues at the +1 position. The adaptor protein Grb10 was identified as an mTORC1 substrate that mediates the inhibition of PI3K typical of cells lacking TSC2, a tumor suppressor and negative regulator of mTORC1. Our work clarifies how mTORC1 inhibits growth factor signaling and opens new areas of investigation in mTOR biology.
The serine-threonine kinase mechanistic target of rapamycin (mTOR) is a major controller of growth that is deregulated in cancer and diabetes (1, 2). mTOR is the catalytic subunit of two multi-protein complexes, mTORC1 and mTORC2. mTORC1 is activated by growth factors and nutrients through a pathway that involves the tuberous sclerosis complex (TSC1-TSC2) tumor suppressors as well as the Rag and Rheb guanosine triphosphatases (GTPases). mTORC1 phosphorylates the translational regulators S6 Kinase 1 (S6K1) and the eIF-4E binding proteins (4E-BP1 and 4E-BP2) while mTORC2 activates Akt and serum/glucocorticoid regulated kinase 1 (SGK1) and is part of the growth factor-stimulated phosphoinositide-3-kinase (PI3K) pathway. Collectively, mTORC1 and mTORC2 regulate processes that control cell growth and proliferation, including protein synthesis, autophagy, and metabolism. mTOR inhibitors derived from rapamycin, an allosteric mTORC1 inhibitor, have been in trials for anti-cancer uses, but the feedback activation of the PI3K-Akt pathway that occurs with mTORC1 inhibition may lessen their clinical efficacy (3).
The few mTOR substrates with defined phosphorylation sites likely cannot explain all processes under the control of mTOR (1, 2, Table S1). In order to discover additional substrates, we conducted a systematic investigation of the mTOR-regulated phosphoproteome using mass spectrometry and isobaric tags that permit 4-way multiplexed relative quantification of phosphopeptide abundances (iTRAQ) (4). With duplicate analyses for each, we analyzed phosphopeptides from two sets of cells in which the pathway was hyperactivated and then inhibited with Torin1, a recently developed ATP-competitive mTOR kinase domain inhibitor that blocks all known phosphorylations downstream of mTORC1 and mTORC2 (5). Human embryonic kidney (HEK)-293E cells were deprived of serum and then stimulated with insulin in the presence or absence of rapamycin or Torin1 (Fig. 1A). Wild-type (TSC2+/+) and TSC2-null (TSC2−/−) mouse embryonic fibroblasts (MEFs), which have increased mTORC1 signaling, were also treated with or without Torin1 (Fig. 1A). Under these conditions, phosphorylation events known to be downstream of mTORC1 (e.g. rapamycin-sensitive T389 S6K1 and rapamycin-insensitive T37 and T46 4E-BP1) and mTORC2 (e.g. S473 Akt, T246 PRAS40/AKT1S1, T346 NDRG1) behaved as expected (Fig. S1).
Fig. 1. Identification of the mTOR-regulated phosphoproteome.
(A) Phosphopeptide abundances were determined from two sets of samples: HEK-293E cells serum starved for 4 hrs, treated with 100 nM rapamycin, 250 nM Torin1, or vehicle control for 1 hr, and then stimulated with 150 nM insulin for 20 min and TSC2+/+ and TSC2−/− MEFs treated with 100 nM Torin1 or vehicle control for 1 hr. (B and C) Distributions of robust z-scores (median absolute deviations (MADs) away from the median (B) log2(Torin1/Insulin) for HEK-293Es or (C) log2(TSC2−/− Torin1/TSC2−/− vehicle) for MEFs). p-values associated with enrichment for known mTOR-modulated sites among the −2.5 MAD Torin1-sensitive phosphopeptides were determined by Fisher’s exact test. Phosphopeptides detected in both replicates had to meet the −2.5 MAD threshold both times to be considered mTOR-regulated. (D, E, and F) Correspondence between (D) Torin1 treatment and serum deprivation in HEK-293Es, (E) Torin1 and rapamycin treatment in HEK-293Es, and (F) Torin1 treatment and upregulation in TSC2−/− MEFs. The relevant robust z-scores for both replicates, phosphopeptides corresponding to known mTOR-modulated sites, Spearman’s rank correlation coefficient (ρ), and associated p-values are indicated. Outliers were excluded to aid in visualization.
From the HEK-293E cells, we identified 4256 unique phosphopeptides corresponding to 47 phosphotyrosine and 4204 phosphoserine-threonine sites on 1661 distinct proteins (FDR ~1%, Table S2). Using a cutoff of 2.5 median absolute deviations (MADs) below the median log2(Torin1/Insulin ratio) (robust z-score < −2.5), 127 phosphopeptides from 93 proteins were identified as sensitive to Torin1 and designated as mTOR-regulated (Fig. 1B). From the MEFs, 7299 unique phosphopeptides corresponding to 110 phosphotyrosine and 7145 phosphoserine-threonine sites on 2406 distinct proteins were identified (FDR~1%, Table S2), of which 231 phosphopeptides from 174 proteins were regulated by mTOR (∡2.5 MAD, log2(TSC2−/− Torin1/TSC2−/− vehicle) (Fig. 1C). By this −2.5 MAD cutoff for both the HEK-293E and MEF datasets, the mTOR-regulated sites were highly enriched in canonical mTOR pathway phosphorylations (Fisher’s exact test p-value = 5.2 × 10−24 and 6.5 × 10−23, respectively; Fig. 1B, 1C, Table S1), an indication of the predictive potential of the data to identify mTOR pathway components. Additionally, we identified sites on known mTOR substrates with less well-characterized sites (CAP-GLY domain containing linker protein 1 (CLIP1) S1158 (6), Unc-51 like kinase 1 (ULK1) S638 (7–9), and insulin receptor substrate 2 (IRS2) S616 (10)).
Global comparisons of the datasets revealed several interesting features. In the HEK-293E cells, phosphorylation changes resulting from Torin1 treatment were strikingly similar to those observed under serum deprivation (Spearman’s ρ = 0.66, p-value ~ 0, Fig. 1D), revealing that insulin-regulated phosphorylations (both down- and up-) are largely mTOR-dependent. The effects of rapamycin and Torin1 treatment were similar (Spearman’s ρ = 0.48, p-value ~ 0, Fig. 1E), but a subset of Torin1-sensitive sites were not rapamycin-sensitive (upper left quadrant, Fig. 1E), including T37 and T46 of 4E-BP1 and 4E-BP2 (5, 11, 12) and the mTORC2-mediated S472 Akt3 and S330 NDRG1. Analysis of the MEF dataset revealed that phosphoryations that increase with TSC2 loss are more likely to be inhibited by Torin1 (Spearman’s ρ = −0.25, p-value = 1.4 × 10−130) (Fig. 1F). Heirarchical clustering of the conditions and sorting of the phosphopeptide abundances in the HEK-293E cells also verified the similarity between serum starvation and Torin1 treatment (Fig. S2) and our ability to discriminate between known rapamycin-sensitive (top, Fig. S2) and -insensitive (bottom, Fig. S2) sites, and showed that phosphorylations that are rapamycin-sensitive tend to be inhibited to a greater extent by Torin1 treatment than those that are not (Fig. S2).
Pathway analysis of the candidate mTOR-regulated proteins revealed enrichment (FDR < 10%) in processes known to be downstream of mTOR, such as translation (GO:0006417) and regulation of cell size (GO:0008361), as well as some not generally considered to be under mTOR control (Table S3). These include RNA splicing (GO:0008380), DNA replication (GO:0006260), vesicle-mediated transport (GO:0016192), and regulation of mRNA processing bodies (GO:0000932), signifying a broader role for mTOR signaling than presently appreciated.
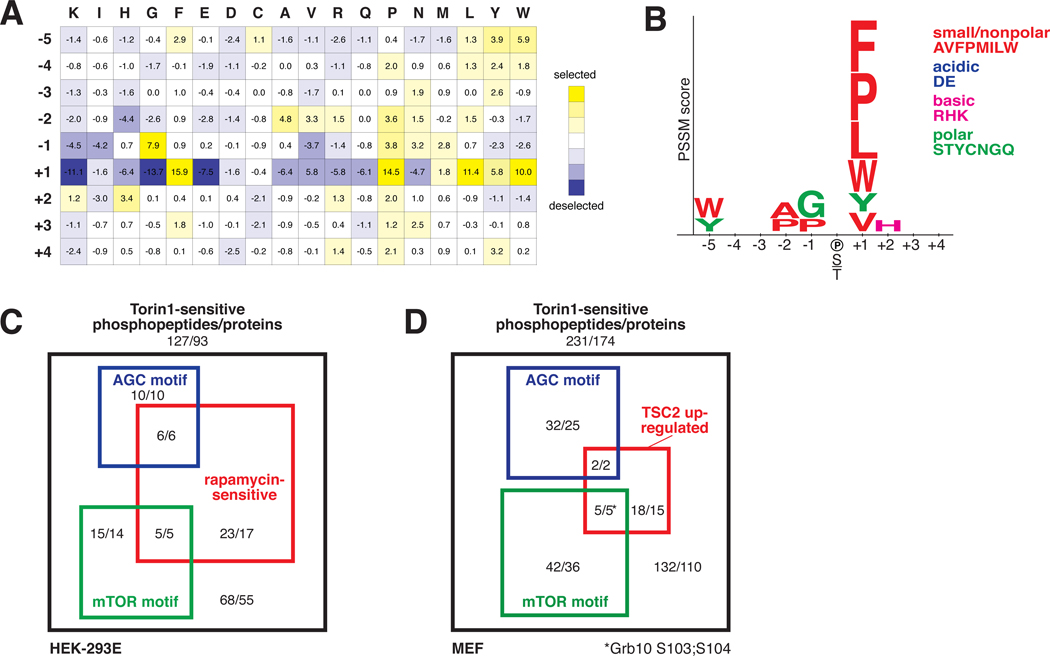
As the mTOR-regulated sites may be phosphorylated by mTOR or by downstream kinases we sought to distinguish direct substrates from indirect effectors by determining a consensus phospho-acceptor motif for mTOR. An example of such a motif is the (R/K)X(R/K)XX(S*/T*) sequence (X = any amino acid, * = phospho-acceptor) recognized by the mTOR substrates Akt, S6K1, and SGK1, all members of the AGC kinase family (13). Because mTOR phosphorylates hydrophobic motifs (HMs) of the AGC kinases as well as the quite distinct proline-directed sites of proteins such as 4E-BP1 and 4E-BP2 (Fig. S3), it is unknown if the kinase exhibits any motif specificity or if the choice of sites is entirely determined by factors beyond the primary substrate sequence. We found that when combined with its activator, GTP-bound Rheb, highly pure and intact mTORC1 (14) robustly phosphorylated an arrayed positional scanning peptide library (15) (Fig. S4, 2A). Although mTORC1 and mTORC2 phosphorylate distinct sets of substrates, they likely have similar motif preferences as they share the same catalytic domain. This unbiased assay revealed that mTOR possesses selectivity towards peptide substrates concordant with known mTOR sites (Fig. S3, S4, 2A, 2B), primarily at the +1 position at which mTOR prefers proline, hydrophobic (L, V), and aromatic residues (F, W, Y). This pattern of specificity at the +1 position is unique amongst all kinases previously profiled (16). mTOR also exhibits minor selectivity at other positions (Fig. S4, 2A, 2B). These data suggest that within the HM of the AGC kinases (Fig. 2A) the -4 and -1 hydrophobic residues are dispensable for mTOR recognition.
Fig. 2. Characterization of a consensus mTOR phosphorylation motif.
(A) The position-specific scoring matrix (PSSM) resulting from quantification of the in vitro phosphorylation of a position scoring peptide library (PSPL) by mTORC1. (B) The visualized mTOR consensus motif. Letter height is proportional to the PSSM score. Only those selected residues with scores greater than a standard deviation from the average PSSM score within a row are shown. (C and D) Classification of the mTOR-regulated phosphopeptides in (C) HEK-293E and (D) MEFs organized by rapamycin sensitivity (−2.5 MAD (log2 (Rapamycin/Insulin)) or TSC2 upregulation (+2.5 MAD log2(TSC2−/− vehicle/TSC2+/+ vehicle)), consistency with the mTOR motif (5th percentile by Scansite), or presence of an AGC motif ((R/K)X(R/K)XX(S*/T*)). The numbers represent the number of unique phosphopeptides or proteins. Refer to Figs. S5, S6 and Table S4 for more details.
Combining our two approaches, we classified the mTOR-regulated phosphorylation sites, first by rapamycin sensitivity (HEK-293E -2.5 MAD log2(Rapamycin/Insulin) or by increased phosphorylation in cells lacking TSC2 (MEFs, +2.5 MAD log2(TSC2−/− vehicle/TSC2+/+ vehicle) (Fig. 2C, 2D, S5, S6, Table S4). Rapamycin-sensitive sites or those upregulated in TSC2−/− cells are likely mTORC1-regulated while the remaining could be downstream of either complex. Second, we scored the sites by motif into the following categories: (1) candidate direct mTOR sites, (2) candidate AGC kinase substrates, or (3) mTOR-regulated but by an undetermined mechanism (Fig. 2C, 2D, S5, S6, Table S4).
Several candidate substrates implicate mTOR in new aspects of cell growth regulation. WD repeat domain, phosphoinositide interacting 2 (WIPI2) (Fig. S6), a sparsely characterized orthologue of the yeast Atg18p, is a potential substrate implicated in autophagosome formation (17). In addition, the candidate substrates protein associated with topoisomerase II homolog 1 (PATL1) (Fig. S5, S6) and La ribonucleoprotein domain family member 1 (LARP1) (Fig. S5, S6) bind RNA, localize to P-bodies, and control mRNA stability (18, 19). Pat1p phosphorylation is rapamycin-sensitive in yeast (20), and Pat1p-deficient yeast do not repress mRNA translation upon amino acid withdrawal (21), suggesting that the regulation of mRNA degradation may be important for growth control. Other potential substrates point to nascent areas of mTOR biology. mTOR putatively regulates the neural stem cell marker Nestin, the pleiotropic AP-1 transcription factor c-Jun, and the myogenic stem cell transcription factor forkhead box K1 (FoxK1) (Fig. S6).
One candidate of special interest was the adaptor protein growth-receptor bound protein 10 (Grb10) (Fig. 2D, S6). The abundance of a Grb10 phosphopeptide with putative mTOR motif sites was increased in the absence of TSC2 and decreased after Torin1 treatment in both TSC2+/+ and TSC2−/− MEFs (Table S2, S4, Fig. 2D, S6), patterns consistent with being in the mTORC1 pathway. Conserved among vertebrates, Grb10 negatively regulates growth factor signaling (22). It binds the insulin and insulin-like growth factor 1 (IGF-1) receptors, and mice without Grb10 are larger and exhibit enhanced insulin sensitivity (23–25). Although the ubiquitin ligase neural precursor cell expressed, developmentally down-regulated 4 (Nedd4) does not directly ubiquitinate Grb10, Nedd4-null mice have more Grb10 protein and are insulin- and IGF-resistant, a signaling phenotype reminiscent of cells lacking TSC1 or TSC2 (26). Therefore, we speculated that Grb10 might function downstream of mTORC1 to inhibit PI3K-Akt signaling.
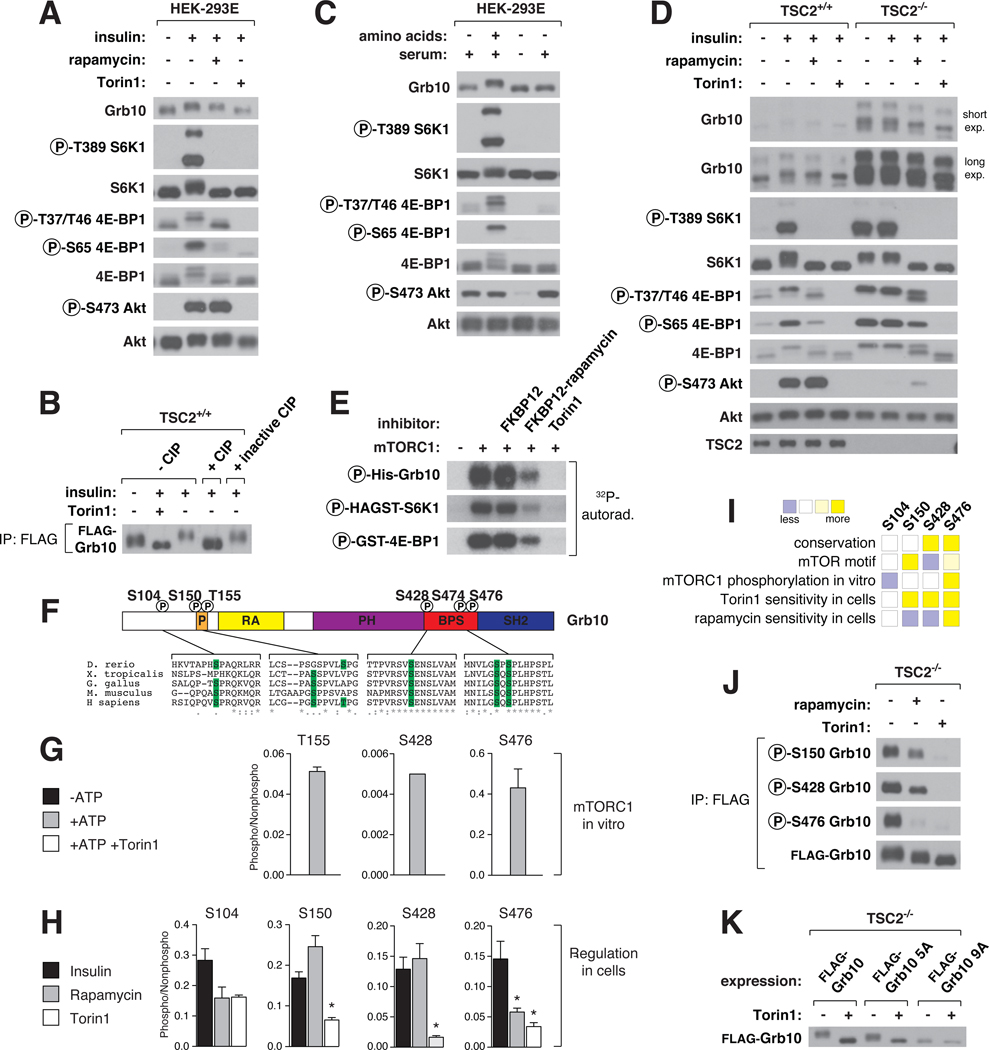
In SDS-PAGE analyses, Grb10 exhibited an insulin-stimulated mobility shift that is partially sensitive to rapamycin (Fig. 3A). In vitro phosphatase treatment eliminated the shift, as did Torin1, indicating that the shift results from phosphorylation and is dependent on mTOR activity (Fig. 3A, 3B). Amino acids stimulated Grb10 phosphorylation and were required for its serum-dependent phosphorylation (Fig. 3C), and in TSC2−/− MEFs, Grb10 phosphorylation was retained in the absence of serum but lost upon acute rapamycin and Torin1 treatment (Fig. 3D). These data point to mTORC1, but not mTORC2, as the main regulator of Grb10. Consistent with this conclusion, the loss of rictor, a core component of mTORC2, did not affect Grb10 phosphorylation (Fig. S7A, S7B).
Fig. 3. Grb10 as an mTORC1 substrate with rapamycin-sensitive and -insensitive sites.
(A) HEK-293E cells were deprived of serum for 4 hrs, treated with 100 nM rapamycin or 250 nM Torin1 for 1 hr, and then stimulated with 150 nM insulin for 15 min. Cell lysates were analyzed by immunoblotting. (B) TSC2+/+ MEFs stably expressing FLAG-Grb10 were serum deprived for 4 hours, treated with 250 nM Torin1 for 1 hr, and then stimulated with 150 nM insulin for 15 min. All FLAG-tagged Grb10 constructs correspond to isoform c of human Grb10. FLAG-immunoprecipitates were incubated in buffer, CIP, or heat-inactivated CIP and analyzed by immunoblotting. (C) HEK-293E cells were deprived of amino acids or both amino acids and serum for 50 min, and then stimulated with either amino acids or serum for 10 min and analyzed by immunoblotting. (D) TSC2+/+ and TSC2−/− MEFs were treated and analyzed as in (A). (E) mTORC1 in vitro kinase assays with substrates in the presence of the indicated inhibitors and radiolabeled ATP were analyzed by autoradiography. (F) Schematic representation of Grb10 protein structure with the phosphorylation sites from vertebrate orthologs aligned below. Numbering is according to human isoform a. (G) The phosphorylation state of Grb10 from kinase assays performed similarly to (E) were analyzed by targeted mass spectrometry (MS) and phosphorylation ratios determined from chromatographic peak intensities. (H) FLAG-immunoprecipitates from HEK-293E cells stably expressing FLAG-Grb10 treated as in (A) were analyzed as in (G). Data are means ± s.e.m (n=2–6). *Mann-Whitney t-test p-values < 0.05 for differences between stimulated and treated conditions. (I) A summary of (F), (G), and (H) for each Grb10 phosphorylation site. (J) FLAG-immunoprecipitates from TSC2−/− MEFs stably expressing FLAG-Grb10 treated with 100 nM rapamycin or 250 nM Torin1 for 1 hr were analyzed by immunoblotting with Grb10 phospho-specific antibodies. (K) TSC2−/− MEFs stably expressing FLAG-Grb10, 5A (S150A T155A S158A S474A S476A), or 9A (5A + S104A S426A S428A S431A) mutants treated with 250 nM Torin1 for 1 hr were analyzed by immunoblotting.
In cells lacking S6K1 and S6K2, Grb10 was still regulated in an mTOR-dependent manner (Fig. S7C), suggesting that it might be a direct substrate. Indeed, Grb10 was phosphorylated in vitro by mTORC1 to an extent comparable with known substrates (Fig. 3E). The sites regulated by mTOR in vitro (Fig. 3G) and in cells (Fig. 3H) were mapped to S104, S150, T155, S428, and S476, which are located in or near the proline-rich region or between the PH and SH2 domains (BPS) of Grb10 (Fig. 3F). In cells, all sites were Torin1-sensitive, while S476 was also rapamycin-sensitive (Fig. 3H). Grb10 is therefore similar to 4E-BP1, an mTORC1 substrate with both rapamycin-sensitive and -insensitive sites (Fig. 3I). We verified our characterization of these sites with phospho-specific antibodies against S150, S428, and S476 (Fig. 3J, S8A, S8B). Mutation of the identified sites along with a few neighboring residues eliminated the mobility shift (Fig. 3K), indicating that most if not all mTOR-regulated sites were localized.
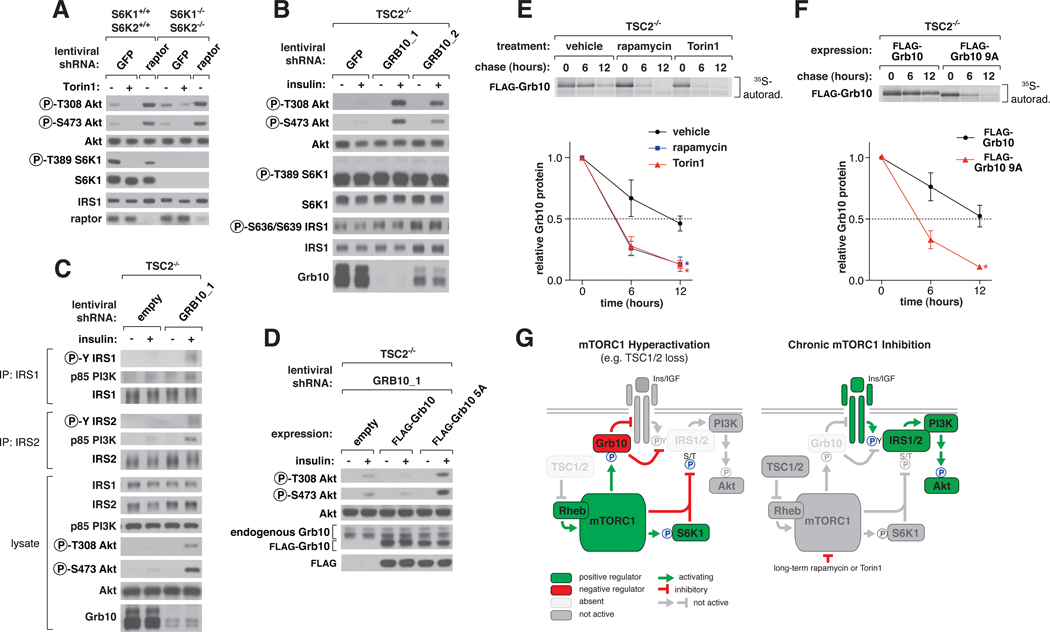
mTORC1 inhibits PI3K-Akt signaling, but the molecular connections involved are poorly understood. One mechanism is the destabilization of insulin receptor substrate 1 (IRS1) by S6K1 phosphorylation (10, 27). However, other mechanisms likely exist because loss of raptor, an essential mTORC1 component, in S6K1−/−S6K2−/− cells still activated Akt phosphorylation without affecting IRS1 abundance (Fig. 4A). Therefore, we tested whether mTORC1 might also inhibit the PI3K pathway through Grb10. Consistent with this possibility, the shRNA-mediated knockdown of Grb10 in HEK-293E and HeLa cells boosted Akt phosphorylation (Fig. S9A, S9B). This boost was increased with rapamycin treatment and, to a lesser extent, with S6K inhibition, suggesting that Grb10 is important for feedback but that other mTOR-dependent mechanisms are also at play (Fig. S9A, S9B) (28). Loss of Grb10 in TSC2−/− MEFs also restored insulin sensitivity to Akt phosphorylation without affecting total IRS1 levels or the phosphorylation of S636 and S639 on IRS1 (Fig. 4B, S9C). While in TSC2−/− cells Grb10 suppression or acute rapamycin treatment each did not rescue insulin signaling to the same level as in wild-type cells, the two in combination approximated the wild-type level of Akt activation (Fig. S9D). This restoration in growth factor sensitivity also applied to increased autophosphorylation of the insulin and IGF receptors, Erk1/2 activation, and IGF-1, but not EGF and PDGF, stimulation (Fig. S10A, S10B). Suppression of Grb10 also increased tyrosine phosphorylation of IRS1 and IRS2 and p85 PI3K recruitment by IRS, again independently of IRS protein levels (Fig. 4C). Compared to cells expressing wild-type Grb10, cells expressing an equivalent amount of non-phosphorylatable Grb10 had increased Akt phosphorylation, confirming that mTORC1 phosphorylation is necessary for its inhibitory function (Fig. 4D, S10C).
Fig. 4. mTORC1 inhibits PI3K-Akt signaling by regulating Grb10 function and stability.
(A) S6K1−/− S6K2−/− or control cells expressing short hairpin RNA (shRNA) constructs against GFP or raptor were treated with 250 nM Torin1 for 1 hr, and lysates were analyzed by immunoblotting. (B) TSC2−/− MEFs expressing shRNAs against GFP or Grb10 were deprived of serum for 4 hrs and then stimulated with 100 nM insulin for 15 min as indicated and analyzed by immunoblotting. (C) TSC2−/− MEFs expressing a control shRNA or shRNA against Grb10 were treated as in (B). IRS1 and IRS2 immunoprecipitates and cell lysates were analyzed by immunoblotting. (D) TSC2−/− MEFs coexpressing an shRNA against the mouse Grb10 3’UTR and an empty vector, FLAG-Grb10, or 5A cDNA expression construct were treated and analyzed as in (B). (E) TSC2−/− MEFs stably expressing FLAG-Grb10 were labeled for 2 hours with [35S]cysteine and methionine and then chased for the indicated times in the presence of vehicle control, 100 nM rapamycin, or 100 nM Torin1. FLAG-immunoprecipitates were analyzed by autoradiography. Data are means ± s.e.m (n=3). *Two-way ANOVA p-values < 0.05 for differences between vehicle and inhibitor treatment. (F) TSC2−/− MEFs stably expressing FLAG-Grb10 or 9A mutant were treated and analyzed as in (E) but without inhibitor treatment. (G) mTORC1 orchestrates feedback inhbition of PI3K-Akt signaling by activating and stabilizing Grb10 while inhibiting and destabilizing IRS proteins.
We suspected that mTORC1-mediated phosphorylation of Grb10 might affect its stability because the more sites we mutated to alanine, the more lentiviral expression construct was required to achieve expression levels equivalent to the wild-type protein. Grb10 is also highly abundant in the TSC2−/− cells with hyperactive mTORC1 signaling (Fig. 3D, S11A), and chronic mTOR inhibition decreased Grb10 protein abundance (Fig. S11A) without significantly affecting mRNA levels (Fig. S11B). Indeed, determination of Grb10 half-life by pulse-chase experiments revealed at least a two-fold decrease (~12 hrs. to ~5 hrs.) in stability with either mTOR inhibitor treatment (Fig. 4E) or mutation of the mTOR sites to alanines (Fig. 4F). Proteasome inhibition (Fig. S11C), suppression of Nedd4 (Fig. S11D), or phosphomimetic mutation of the mTOR sites (Fig. S11E) rescued the decrease in Grb10 protein caused by mTOR inhibition. Therefore, mTORC1 inhibits and destabilizes IRS1 and simultaneously activates and stabilizes Grb10 (Fig.S12).
These results confirm the importance of the mTORC1 pathway in regulating growth factor signaling and clarify the nature of the feedback loop to PI3K-Akt. While acute mTORC1 inhibition leads to dephosphorylation of IRS1 and Grb10, chronic mTORC1 inhibition leads to changes in the levels of IRS and Grb10 proteins which are likely to be the most important effects of mTOR inhibitors to consider in their clinical use (Fig. 4G). Our findings also support the idea (29, 30) that concomitant IGF-1 receptor inhibition may improve the anti-cancer efficacy of mTOR inhibitors. Finally, the discovery of Grb10 as an mTORC1 substrate validates our approach and suggests that the other potential downstream effectors we identified may also serve as starting points for new areas of investigation in mTOR biology.
Supplementary Material
Acknowledgments
We thank members of the Sabatini Lab for helpful discussion and especially thank B Joughin, G. Bell, H. Keys, K. Birsoy, N. Kory, C. Thoreen, J. Claessen, and D. Wagner for assistance with technical or conceptual aspects of this project. This work was supported by the National Institutes of Health (CA103866 and AI47389 to D.M.S.; ES015339, GM68762, and CA112967 to M.B.Y.), Department of Defense (W81XWH-07-0448 to D.M.S.), the W.M. Keck Foundation (D.M.S.), LAM Foundation (D.M.S.), Dana Farber Cancer Institute (N.S.G, J.M), the International Fulbright Science and Technology Award (J. R.), and American Cancer Society (S.A.K.). D.M.S. is an investigator of the Howard Hughes Medical Institute.
References and Notes
- 1.Laplante M, Sabatini DM. J Cell Sci. 2009;122:3589. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoncu R, Efeyan A, Sabatini DM. Nat Rev Mol Cell Biol. 2011;12:21. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowling RJ, Topisirovic I, Fonseca BD, Sonenberg N. Biochim Biophys Acta. 2010;1804:433. doi: 10.1016/j.bbapap.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Ross PL, et al. Mol Cell Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Thoreen CC, et al. J Biol Chem. 2009;284:8023. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JH, et al. EMBO Rep. 2002;3:988. doi: 10.1093/embo-reports/kvf197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jung CH, et al. Mol Biol Cell. 2009;20:1992. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganley IG, et al. J Biol Chem. 2009;284:12297. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hosokawa N, et al. Mol Biol Cell. 2009;20:1981. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah OJ, Wang Z, Hunter T. Curr Biol. 2004;14:1650. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Proc Natl Acad Sci U S A. 2008;105:17414. doi: 10.1073/pnas.0809136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman ME, et al. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pearce LR, Komander D, Alessi DR. Nat Rev Mol Cell Biol. 2010;11:9. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 14.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Mol Cell. 2010;38:768. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutti JE, et al. Nat Methods. 2004;1:27. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 16.Mok J, et al. Sci Signal. 2010;3:ra12. doi: 10.1126/scisignal.2000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polson HE, et al. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 18.Parker R, Sheth U. Mol Cell. 2007;25:635. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 19.Nykamp K, Lee MH, Kimble J. RNA. 2008;14:1378. doi: 10.1261/rna.1066008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber A, et al. Genes Dev. 2009;23:1929. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coller J, Parker R. Cell. 2005;122:875. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt LJ, Siddle K. Biochem J. 2005;388:393. doi: 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charalambous M, et al. Proc Natl Acad Sci U S A. 2003;100:8292. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith FM, et al. Mol Cell Biol. 2007;27:5871. doi: 10.1128/MCB.02087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, et al. Mol Cell Biol. 2007;27:6497. doi: 10.1128/MCB.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao XR, et al. Sci Signal. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrington LS, et al. J Cell Biol. 2004;166:213. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce LR, et al. Biochem J. 2010;431:245. doi: 10.1042/BJ20101024. [DOI] [PubMed] [Google Scholar]
- 29.O'Reilly KE, et al. Cancer Res. 2006;66:1500. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan X, Harkavy B, Shen N, Grohar P, Helman LJ. Oncogene. 2007;26:1932. doi: 10.1038/sj.onc.1209990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.