Abstract
Reverse transcriptases (RTs) are RNA-dependent DNA polymerases that usually function in the replication of selfish DNAs such as retrotransposons and retroviruses. Here, we have biochemically characterized a RT-related protein, AbiK, which is required for abortive phage infection in the Gram-positive bacterium Lactococcus lactis. In vitro, AbiK does not exhibit the properties expected for an RT, but polymerizes long DNAs of ‘random’ sequence, analogous to a terminal transferase. Moreover, the polymerized DNAs appear to be covalently attached to the AbiK protein, presumably because an amino acid serves as a primer. Mutagenesis experiments indicate that the polymerase activity resides in the RT motifs and is essential for phage resistance in vivo. These results establish a novel biochemical property and a non-replicative biological role for a polymerase.
INTRODUCTION
Reverse transcriptases (RTs) play many roles in biology, including in retroviruses, retrotransposons, hepadnaviruses, telomerase and organellar retroplasmids (1–5). Most eukaryotic RTs are components of selfish DNAs, and their polymerization activities allow the elements to replicate and spread without necessarily contributing benefits to their host organism. In spite of this general pattern of selfishness, a few eukaryotic RTs have evolved useful functions, such as telomerase and HetA-TART retroelements, both of which protect chromosomal ends from sequence loss during DNA replication (4,6).
RTs in bacteria are less prevalent and less well studied than those in eukaryotes. In fact, we have an incomplete knowledge of the types of bacterial RTs that exist in nature. Two recent compilations have identified a surprising number of uncharacterized RTs and RT-like proteins in bacterial genomes, including at least 20 phylogenetically based groupings with 11 domain architectures (7,8). Of these, only three types have been substantially characterized: group II introns, diversity-generating retroelements (DGRs) and retrons.
Group II introns are the most abundant RTs in bacteria by far (8), and have the classical properties of retrotransposons, being capable of efficient retromobility (9,10). Their mobility mechanism is known as target-primed reverse transcription (TPRT), and uses the cooperative activities of a catalytic, self-splicing intron and an intron-encoded RT. Group II introns insert at high efficiencies into homing sites of defined sequences, and at lower frequencies into ectopic sites.
DGRs in contrast are not selfish, but contribute a useful function to their hosts through the generation of sequence diversity in target genes. The Bordetella phage DGR, for example, uses a template-dependent, RT-mediated process to introduce nucleotide substitutions into the variable region of the phage gene mtd. The mtd gene encodes a tail protein responsible for host recognition, and the variability allows the phage to adapt its tropism to the dynamic Bordetella cell surface, which changes between in vivo and ex vivo phases (11). Other DGRs are not phage-associated, and may diversify cellular genes (12). The third characterized bacterial retroelement, the retron, carries out a specialized reverse transcription reaction to produce branched RNA–DNA molecules called multicopy single-stranded DNAs (msDNAs) (13). Retrons are not independently mobile, nor has a biological function been identified to date (14).
Among the remaining putative RTs in bacteria, there is little indication for active retromobility, as would be suggested by multiple RT copies in a genome. Instead, it has been predicted that the RTs contribute beneficial functions, analogous to telomerase and DGRs (7,8). Supporting this hypothesis, two uncharacterized RT classes are associated with bacterial CRISPR/Cas defense systems, and presumably help protect against invading phages and plasmids (7,8,15,16). Three other uncharacterized RTs (AbiK, AbiA, Abi-P2) have been experimentally associated with abortive phage infection (17,18) or phage exclusion mechanisms (19), but none of these has been studied extensively.
Abortive infection (Abi) is a type of bacterial defense system in which a virulent phage DNA is injected into bacteria, but phage maturation is blocked and most infected cells die (20,21). Abi systems are prevalent across bacteria, and can act specifically against a single phage group, or more generally against multiple groups of phages. Abi systems are typically mediated by a single host gene, which is often plasmid-encoded. In many cases the exact modes of action are incompletely understood.
The AbiK system of Lactococcus lactis is encoded by the native plasmid pSRQ800 of a lactococcal strain isolated from raw milk, an ecological niche known to contain many virulent phages (17). AbiK provides broad immunity against the most predominant lactococcal phage groups, typically reducing infectivity by 106. The system is encoded by a single gene, abiK, which is constitutively transcribed. It has been noted for some time that the N-terminal sequence of AbiK is related to RTs (22).
In the present study, we investigate the biochemical properties of the AbiK protein and show that it is an active polymerase; however, it does not possess the activities expected for an RT. Rather AbiK polymerizes ‘random’ DNA sequence analogous to a terminal transferase. Furthermore, the DNA appears to be attached covalently to AbiK, putatively through an amino acid that serves as primer for DNA polymerization. Our results expand the mechanistic repertoire of RT-related enzymes, and suggest that the AbiK system is a biochemically novel mechanism of phage resistance that is mediated by an RT-related polymerase.
MATERIALS AND METHODS
Plasmid constructs
The wild-type AbiK expression construct was made by PCR-amplifying AbiK sequence from the plasmid pSRQ818 (17) with the primers AbiK-S-B and AbiK-AS-K (Supplementary Table S1), and cloning into the BamHI and KpnI sites of pQE30 (Qiagen, Valencia, CA, USA) to generate pQE30-AbiK. The BamHI-SmaI fragment of pQE30-AbiK was subcloned into pGEX4T1 (GE Healthcare, Piscataway, NJ, USA) to generate the plasmid pGEX4T1-AbiK. Mutations were introduced into AbiK sequence by recombinant PCR of pGEX4T1-AbiK using the primer pairs AbiK-m-S and AbiK(DI-DVII)AS, and AbiK-m-AS and AbiK(DI-DVII)S (Supplementary Table S1).
Constructs to test abortive infection in vivo were made by subcloning mutant sequences into the BglII and XhoI sites of the lactococcal plasmid pSRQ823. pSRQ823 is a pNZ123-based vector containing the EcoRI-XbaI abiK-fragment from pSRQ818 which originated from the natural L. lactis plasmid pSRQ800 (17). For the gst-abiK fusion construct in pNZ123, the gst-abiK fragments from pGEX4T1-AbiK wt were PCR-amplified with Pwo polymerase and primers rbs-gst-abiKFor and 3′_pGEX sequencing primer (Supplementary Table S1), thereby introducing a ribosome binding site. The PCR fragment was digested with XhoI and cloned into the XhoI site and Klenow-filled-in BglII site of pSRQ823. All plasmid constructs in this study were verified by sequencing.
Preparation of recombinant AbiK-GST
An overnight culture of Escherichia coli BL21 harboring the plasmid pGEX4T1-AbiK was grown in LB (23) at 37°C with ampicillin (100 µg/ml), and then diluted 100-fold into one litre of 2× YT medium (23), and grown to an OD600 of 0.5. Cells were induced with 0.1 mM IPTG for two hours at room temperature, harvested, and stored at −70°C. Protein purification was carried out according to the manufacturer’s manual for Glutathione Sepharose 4B beads, using the column binding protocol (GE Healthcare), with the following minor modifications and an additional step to remove Hsp70 (24). Frozen pellets were resuspended in 35 ml lysate buffer (16 mM Na2HPO4, 4 mM NaH2PO4, pH 7.3, 150 mM NaCl, 1% Triton X-100, 5 mM DTT, 1 mM PMSF) and broken with a French press. Then, 3.5 ml of 10× ATP stock (16 mM Na2HPO4, 4 mM NaH2PO4, pH 7.3, 150 mM NaCl, 1% Triton X-100, 5 mM DTT, 100 mM ATP, 200 mM MgCl2, 500 mM KCl) were added to the clarified lysate and incubated at 37°C for 10 min, followed by addition of 200 µl (7.6 mg) of denatured E. coli cellular proteins (24), and an additional incubation at 37°C for 20 min. The lysate was clarified by centrifugation, and applied to a 300 µl bed of Glutathione Sepharose 4B beads (GE Healthcare). Two washes were performed with 10 and 5 ml of lysate buffer containing either added 10 mM ATP, 20 mM MgCl2, and 50 mM KCl (wash buffer 1) or 5 mM ATP, 20 mM MgCl2 and 50 mM KCl (wash buffer 2). Purified protein was stored at −20°C in 25 mM Tris–HCl, pH 8.0, 50 mM NaCl, 5 mM reduced glutathione, 5 mM DTT and 50% glycerol. Mutant AbiK preparations were made in the same way, except that only 3.6 mg of denatured protein were used to remove Hsp70.
For thrombin treatment of AbiK-GST, 1 µg of the AbiK-GST preparation was incubated with 1 U of thrombin (GE Healthcare) at room temperature, either for 1.5 h in 1× RT buffer (50 mM Tris–HCl, pH 8.5, 100 mM KCl, 2 mM MgCl2, 5 mM DTT) (Figure 3A, Table 1), or for 1.5 or 17.5 h in 20 mM Tris–HCl, pH 8.4, 150 mM NaCl and 2.5 mM CaCl2 (Supplementary Figure S2B).
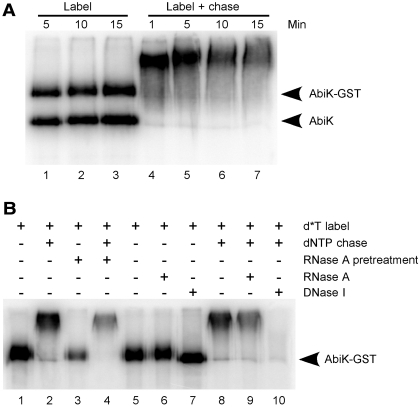
Figure 3.
Resolution of products by SDS–PAGE. (A) An AbiK preparation was cleaved (partially) with thrombin and then incubated with [α-32P]dTTP for 5–15 min (label), followed by a dNTP chase for 1–15 min (label + chase). Products were resolved by SDS–PAGE and visualized by phosphorimaging. The positions of AbiK-GST and AbiK were determined by Coomassie staining of the gel. (B) Nuclease susceptibility. AbiK-GST was incubated with [α-32P]dTTP (d*T) for 10 min (lane 1) followed by a dNTP chase for 10 min (lane 2). Pretreatment of AbiK-GST with RNase A for 10 min prior to the reactions had little effect (lanes 3 and 4). Products of the labeling reaction were either untreated or subjected to RNase A or DNase I (lanes 5–7). Products of the label-chase reaction were similarly untreated or incubated with RNase A or DNase I (lanes 8–10).
Table 1.
In vitro polymerization assays with purified AbiK-GST proteina
| Assay | 32P incorporation (cpm)b |
|---|---|
| AbiK-GST + [α-32P]dTTP + poly rA/oligo(dT)18 | 141 000 ± 1700 |
| AbiK-GST + [α-32P]dTTP | 141 000 ± 4000 |
| AbiK-GST + [α-32P]dTTP + RNase A | 157 000 ± 7800 |
| AbiK-GST + [α-32P]dTTP + RNase A pretreatment (10 min) | 109 000 ± 2300 |
| AbiK-GST + [α-32P]dTTP + 0.2 mM dATP, dCTP, dGTP | 6500 ± 700 |
| AbiK-GST + [α-32P]dTTP + 0.05 mM dATP, dCTP, dGTP | 22 000 ± 1500 |
| AbiK-GST + [α-32P]dTTP + 0.2 mM dATP, dCTP | 5400 ± 300 |
| AbiK-GST + [α-32P]dTTP + 0.2 mM dATP, dGTP | 5600 ± 200 |
| AbiK-GST + [α-32P]dTTP + 0.2 mM dCTP, dGTP | 4400 ± 800 |
| AbiK-GST + thrombin + [α-32P]TTP + poly rA/oligo(dT)18 | 215 000 ± 21 000 |
| AbiK-GST + thrombin + [α-32P]TTP | 230 000 ± 23 000 |
aSee Supplementary Data for experimental description.
bMean ± standard deviation, n = 3.
AbiK polymerization and protein labeling assays
Unless noted otherwise, 1 µg AbiK-GST preparation was incubated with 10 µCi [α-32P]dTTP in 1× RT buffer at 37°C for 10 min. This basic reaction was followed by different treatments. For chased reactions, the labeling reactions were followed by addition of either 0.2 mM each dNTP or 0.2 mM dTTP, and a further 10 min incubation. For some reactions, 2 µg proteinase K was added and incubated for 10 min at 37°C. For nuclease analyses, reactions were incubated at 37°C for 10 min with either 2 µg RNase A or with 10 U DNase I (Invitrogen, Carlsbad, CA, USA). For resolution by SDS–PAGE, samples were mixed with SDS loading dye (23), boiled and resolved on a 10% gel, and either dried and subjected to phosphorimaging or stained with Coomassie blue (23) or silver (25). For resolution on PAGE–urea gels, samples were extracted with phenol/chloroform/isoamyl alcohol (25:24:1) with 1% acrylamide carrier and precipitated with ethanol in the presence of 0.3 M NaOAc (pH 5.2), followed by heating to 80°C in 50% formamide. Samples were resolved on an 8% PAGE–urea gel, dried and phosphorimaged, or on a 20% PAGE–urea gel followed by phosphorimaging. Assays with poly rA/oligo (dT)18 were carried out as previously described (26).
Cloning and sequencing of AbiK-synthesized DNAs
The cloning strategy was based on the observation that AbiK can use an oligonucleotide as a primer if provided in excess. Five micrograms of AbiK preparation were incubated with 0.5 µg oligonucleotide AD96 or PQEREV and 0.2 mM each dNTP (or 0.2 mM dATP, dCTP, dGTP, or 0.2 mM dATP, dGTP, dTTP) in 25 µl of 1× RT buffer at 37°C for 15 min. The reaction was stopped by heating to 95°C for 5 min, and digesting with 2 µg of proteinase K at 37°C for 1 h. After purification using a DNA Clean kit (Zymo Research, Orange, CA, USA), the eluted DNA was tailed with 0.5 mM dGTP and 20 U terminal transferase (New England Biolabs, Beverly, MA, USA) according to the manufacturer’s protocol. DNAs were amplified by PCR with primer pairs AD96 and AD98, or PQEREV and AD98, and the PCR products were gel-purified, cloned and sequenced.
In vivo assays for abortive infection
Clones containing AbiK mutants in pSRQ823 were transformed into L. lactis MG1363 by electroporation (27). The plasmid content of each transformant was confirmed by plasmid isolation (28) and sequencing. Some lactococcal plasmids were also extracted using QIAquick Spin Miniprep columns but with a slightly modified protocol. Cells were first treated by adding sucrose (25%) and lysozyme (30 mg/ml) into the P1 buffer, and incubated at 37°C for 15 min. L. lactis MG1363 strains were grown at 30°C in M17 medium (OXOID, Nepean, Ontario, Canada) supplemented with 0.5% glucose (GM17) and 5 µg/ml chloramphenicol.
To measure the phage resistance phenotype, the efficacy of plaquing (EOP) of lactococcal phage p2 (936 group, double-stranded DNA genome, Siphoviridae family, Caudovirales order) was estimated as described previously (22). Briefly, phage p2 was propagated on the wild-type phage-sensitive laboratory strain L. lactis MG1363 in GM17 medium supplemented with 10 mM CaCl2 (29). The EOP was calculated by dividing the phage titer of an AbiK construct in MG1363 by the titer of the sensitive control (pNZ123 in MG1363).
RESULTS
The family of AbiK-related proteins contains RT domains
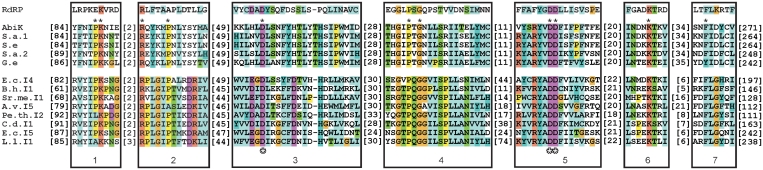
To gain further insight into the domain structure of the AbiK protein, its 599 amino acid sequence was aligned with its four closest relatives in GenBank, as well as representative group II intron RT sequences (Figure 1, Supplementary Figure S1). Only four relatives were chosen for the alignment because more distant sequences do not align across the entire length of AbiK sequence. The seven domains characteristic of RTs are identifiable in the AbiK-related sequences, although RT2 and RT7 align ambiguously. The identified RT motifs correspond to the finger and palm domains of the polymerase protein structure (by analogy with the HIV RT crystal structure), with the putative active site residues located in RT3 and RT5 (outlined asterisks in Figure 1, Supplementary Figure S1) (30). Thumb domains of RTs are typically located downstream of RT7, but are not highly conserved among RT groups. For AbiK, the region expected to be the thumb domain does not align with the X/thumb domain of group II introns. In fact, the ∼200 amino acid C-terminal sequence of AbiK and its relatives does not have similarity to any other proteins in GenBank. The region is however inferred to have an Abi-specific role, in addition to being the polymerase thumb domain, because a 46 amino acid C-terminal deletion was previously shown to eliminate the phage resistance phenotype (22).
Figure 1.
Alignment of AbiK sequence with group II intron RTs and RNA-dependent RNA polymerases. An amino acid alignment is shown for the AbiK protein, four closely related proteins, and representative group II intron RTs. Above AbiK is the consensus sequence for the seven domains of RNA-dependent RNA polymerases (RdRPs), derived from the Pfam database (31). Only the seven domains conserved among RT proteins are shown; full alignments are available in Supplementary Figure S1. Asterisks directly above AbiK sequence indicate residues mutated in this study, while the three outlined asterisks below the group II intron sequences mark the putative active site aspartate residues. Bracketed numbers are amino acids not shown in the alignment. Sequences are: AbiK (Lactococcus lactis, gi|32455447), S.a.1 (Staphylococcus aureus, gi|17227201), S.a.2 (Staphylococcus aureus, gi|24636606), S.e. (Staphylococcus epidermidis, gi|281296383), G.e. (Granulicatella elegans, gi|260157297), E.c.I.4 (Escherichia coli, class A; gi|6009435), B.h.I1 (Bacillus halodurans, class C; gi|10172711), Sr.me.I1 (Sinorhizobium meliloti, class D; gi|4586216), A.v.I5 (Azotobacter vinelandii, class E; gi|226719487), Pe.th.I2 (Pelotomaculum thermopropionicum, class F; gi|146274844), C.d.I1 (Clostridium difficile, class B; gi|1418413), E.c.I5 (Escherichia coli, class CL; gi|188574179), L.l.I1 (Lactococcus lactis, class ML; gi|125624188).
In light of the unusual biochemical properties found for AbiK in this study (below), it is of interest to reexamine the relationship between AbiK and other polymerase families. BLAST searches consistently identify AbiK as containing RT motifs, based on the Conserved Domain Database (CDD) of NCBI (not shown); however, AbiK sequence is only distantly related to characterized RTs [see ref. (8) for alignments among bacterial RTs]. Because RTs belong to the superfamily of RNA-dependent RNA polymerases (RdRPs) (31), AbiK was aligned to the RdRP1 family. Alignment is quite limited, with only domains 3, 4 and 5 aligning unambiguously (Figure 1, Supplementary Figure S1 and data not shown). Thus, sequence alignments do not suggest that AbiK is more closely related to RdRPs than to group II intron RTs. It can be concluded that AbiK belongs to the sequence family of RTs, regardless of its biochemical properties.
Expression and purification of AbiK in E. coli
The AbiK protein was expressed in E. coli as a fusion with a GST tag. In the initial affinity purification method, three bands were produced (Supplementary Figure S2A), the largest of which was the size expected for AbiK-GST, while the smallest band comigrated with GST, indicating that the majority of AbiK-GST is degraded, leaving the 26 kDa GST domain. The third band was determined by mass spectrometry to be Hsp70. Consequently, a modified purification protocol was used to eliminate the Hsp70 contamination along with substantial background biochemical activities (see ‘Materials and Methods’ section). In the resulting AbiK preparation, the GST tag could be cleaved from AbiK-GST with thrombin (Supplementary Figure S2B), while in the initial preparation it could not.
Purified AbiK has DNA polymerase activity
Purified AbiK was assayed for RT activity using conventional RT assays with poly rA/oligo(dT)18 substrate. High levels of [α-32P]dTTP incorporation were observed; however, incorporation occurred even without the poly rA/oligo(dT)18 substrate (Table 1). To test for a copurifying RNA in the AbiK preparation that might be a template, the preparation was treated with RNase A, either before or during the assay. RNase A does not degrade poly rA/oligo(dT)18 because it cleaves at pyrimidine residues. The fact that polymerization occurred regardless of the RNase treatment argues against the presence of an RNA template in the AbiK preparation. Interestingly, addition of non-radiolabeled dATP, dCTP and dGTP drastically reduced [α-32P]dTTP incorporation, which would not be expected for templated polymerization, but would occur for non-templated polymerization such as for a terminal deoxytransferase. Treatment of the AbiK-GST preparation with thrombin had little effect on [α-32P]dTTP incorporation, indicating that the GST tag minimally perturbs AbiK’s activity in vitro. Subsequently, the GST tag was found to also have a minimal effect on AbiK function in vivo (see below and Table 2).
Table 2.
In vivo abortive infection assays with AbiK mutants
| AbiK allele | Domain mutated | Efficacy of plaquinga | 32P incorporation (cpm)b | Mutation |
|---|---|---|---|---|
| Mut1 | 1 | 8.9 ± 0.7 × 10−1 | − | P89A, K90A |
| Mut2a | 2 | N.D.c | ± | R96A |
| Mut2b | 2 | 2.6 ± 0.7 × 10−8 | + | P101A |
| Mut3 | 3 | N.D.c | − | D163A |
| Mut4 | 4 | 8.8 ± 0.9 × 10−1 | − | P214A, G216A |
| Mut5 | 5 | 8.7 ± 1.1 × 10−1 | − | D247A, D248A |
| Mut6 | 6 | 9.0 ± 2.4 × 10−1 | − | K281A |
| Mut7 | 7 | 5.3 ± 2.2 × 10−8 | ++ | F321A |
| AbiK-GST | – | 3.7 ± 1.0 × 10−7 | +++ | − |
| pSRQ823 | WT | 2.6 ± 1.0 × 10−8 | N.D.d | − |
aMean ± standard deviation, n = 3.
bRT activities are summarized from Figure 5, with weak activities determined by overexposure.
cNo data. Construct could not be cloned in L. lactis.
dNo data. Wild-type protein was not purified without the GST tag.
The polymerized DNA appears to be attached to AbiK through a covalent bond
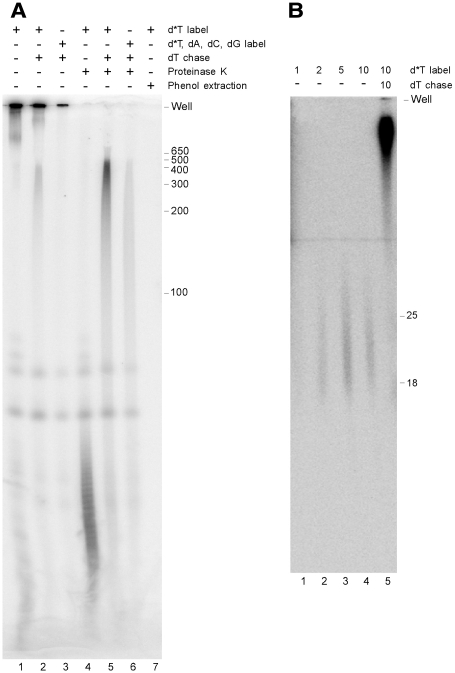
When AbiK reaction products were resolved on a polyacrylamide–urea gel, most products remained in the wells (Figure 2A, lanes 1 and 2). Treatment with protease allowed the resolution of heterogeneous products of <100 nt (Figure 2A, lane 4), which were extended to several hundred nucleotides after a chase with dNTPs (Figure 2A, lanes 5 and 6). Similar to the RT assay data (Table 1), the inclusion of dATP, dCTP and dGTP reduced the incorporation of the radiolabeled dTTP (Figure 2A, lanes 3 and 6). Phenol extraction of the products resulted in complete loss of signal in the gel (Figure 2A, lane 7), suggesting a covalent attachment between the radiolabeled DNA and AbiK. Together, the data indicate that the AbiK reaction product is a short radiolabeled DNA attached to the AbiK protein, which becomes elongated upon chasing with dTTPs.
Figure 2.
Resolution of polymerization products on a polyacrylamide–urea gel. (A) AbiK-GST protein was incubated with 10 µCi [α-32P]dTTP (d*T) without (lane 1) or with (lane 2) a chase with dTTP. In lane 3, 0.2 mM of dATP, dCTP and dGTP were included in the labeling incubation. Reactions equivalent to lanes 1–3 were subsequently treated with proteinase K at 37°C for 30 min (lanes 4–6). The reaction in lane 1 was phenol extracted and precipitated (lane 7). All reactions were heated to 80°C in 80% formamide before resolving on an 8% polyacrylamide/8 M urea gel. The sizes of a DNA ladder are to the right of the gel (nt). (B) AbiK-GST protein was incubated with 10 µCi [α-32P]dTTP (d*T) for 1, 2, 5 and 10 min (lanes 1–4), with the 10 min reaction being further chased with dTTP for 10 min (lane 5). Reactions were purified on a G-50 Sephadex column to remove unincorporated radiolabel, treated with proteinase K, and resolved on a 20% acrylamide/8 M urea gel. Positions of oligonucleotide DNA markers are to the right (nt).
To determine the shortest DNA linked to AbiK, AbiK-GST was incubated with [α-32P]dTTP for different time points, with some sample chased with dNTPs. Products were purified on a G-50 Sephadex column to remove unincorporated nucleotides, digested with proteinase K, and resolved on a 20% polyacrylamide–urea gel. The shortest detectable products migrated as heterogeneous bands of 18–25 nt (Figure 2B). Because protease digestion will not remove all amino acids from the DNA, and because it is not certain how the attached amino acids will affect gel migration, the shortest DNAs may be smaller than 18–25 nt. Other experiments supported the hypothesis of a short DNA produced in the labeling reaction. In initial cloning attempts, AbiK-generated DNAs were produced by incubating AbiK with a low concentration of dTTP, chasing with dNTPs, and then tailing with G’s. The resulting DNAs failed to PCR amplify with (dT)18 and (dC)18 primers, suggesting that the labeling reaction polymerizes too few T’s to anneal with the (dT)18 primer.
To test more directly for a covalent linkage between AbiK and DNA, a thrombin-treated preparation containing both AbiK-GST and AbiK was incubated with [α-32P]dTTP, boiled in SDS loading buffer, and resolved by SDS–PAGE (Figure 3). Radiolabeled bands were observed that comigrated with each form of AbiK, indicating that each polypeptide undergoes the same radiolabeling reaction, and confirming that the attached DNA is short and does not affect protein migration on the SDS–PAGE gel (Figure 3A, lanes 1–3). Chasing of the reaction with dNTPs resulted in greatly retarded migration for both protein forms (Figure 3A, lanes 4–7).
The reaction was further characterized by nuclease digestions. Consistent with previous data (Table 1), preincubation of AbiK with RNase A for 10 min did not abolish either labeled or chased reaction products (Figure 3B, lanes 1–4). In addition, the products of the AbiK reaction were resistant to RNase treatment (Figure 3B, lane 9). However, the reaction products had a differential susceptibility to DNase I. The chased products were sensitive to DNase I as expected (Figure 3B, lane 10), while the unchased products were resistant (Figure 3B, lane 7). One possible explanation for the difference is a conformational change of the AbiK protein, which exposes the attached DNA during the chasing reaction (see Discussion section).
The sequence of AbiK-polymerized DNAs indicates non-templated polymerization
Additional experiments failed to identify a candidate RNA or DNA template in the AbiK preparation. The preparation was treated with protease and/or phenol extraction followed by SDS–PAGE and silver staining. No nucleic acids were detectable (not shown). In another approach, the radiolabeled DNAs produced by AbiK were used as a probe in Southern and northern blot experiments, using blots of either DNA or RNA isolated from E. coli harboring the AbiK expression plasmid. No hybridization to E. coli cellular DNA or RNA was detectable (not shown), indicating that the AbiK-synthesized DNA sequence does not correspond to a DNA or RNA in the E. coli strain in which AbiK is expressed.
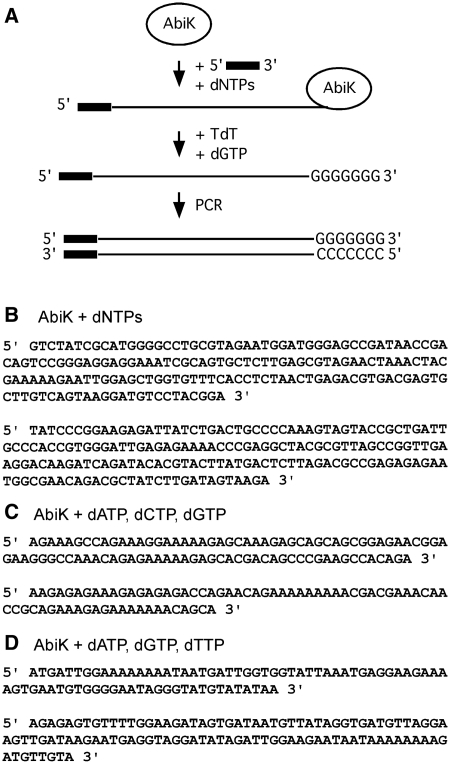
Consequently, the AbiK-synthesized DNAs were cloned and sequenced, taking advantage of the fact that AbiK can use an oligonucleotide as a primer to a small extent when the oligonucleotide is provided in excess (Figure 4A, Supplementary Figure S3). The sequences from 10 independent clones did not match either E. coli sequence or other sequence in GenBank, and were unique with respect to each other (Figure 4B, Supplementary Figure S4A), suggesting the absence of a template. To further test our hypothesis of non-templated, ‘random’ polymerization, AbiK was incubated with either dATP+dCTP+dGTP, or dATP+dGTP+dTTP. As hypothesized, the 10 cloned sequences from each reaction were composed of the corresponding nucleotides, and had no matches in GenBank (Figures 3D and 4 C, Supplementary Figure S4B and S4C). We conclude that AbiK polymerizes DNA without a template, and incorporates bases without sequence specificity, analogous to a terminal transferase. While there may be a modest bias for adenosines in the sequences (Supplementary Figure S4), nevertheless, all four bases are readily incorporated.
Figure 4.
Sequence of DNAs polymerized by AbiK. (A) Strategy for cloning of AbiK-generated DNAs. AbiK was incubated with an oligonucleotide and dNTPs, tailed with dGTP and terminal deoxytransferase, and then PCR amplified and cloned (see ‘Materials and Methods’ section for details). For panels B–D, purified AbiK was incubated with either all dNTPs (B), or combinations of 3 nt triphosphates (C and D), and the resulting polymerized DNAs were cloned and sequenced. None of the sequenced DNAs have significant similarity to sequences in GenBank, or to each other. Representative sequences are shown, with full data available in Supplementary Figure S3.
Mutations in the RT motifs abolish polymerization in vitro
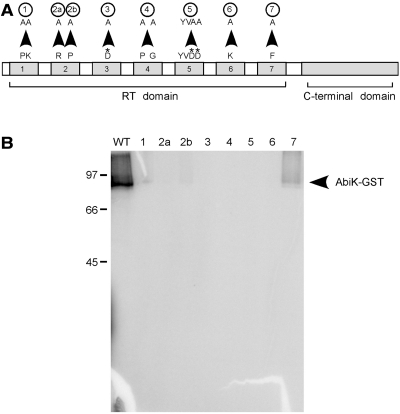
To test whether the RT motifs of AbiK are responsible for the polymerization observed in vitro, a series of mutations was made across the seven RT domains (Figures 1 and 5, Supplementary Figure S1). Mutant proteins were expressed in E. coli, affinity purified, and tested for polymerization activity using the protein-labeling assay. Protein amounts were calibrated on a silver-stained gel to account for differing amounts of GST in the preparations (Supplementary Figure S5). All RT domain mutants were found to have significantly lower activity than wild-type AbiK (Figure 5B, Table 2), including Mut3 and Mut5, which lack the presumed catalytic residues. Of the RT mutants, only Mut2b and Mut7 had faint signals in long exposures; these mutants correspond to the least conserved of AbiK’s RT domain sequences (Figure 1, Supplementary Figure S1). Overall the loss of activity supports the hypothesis that polymerase activity of AbiK lies in its RT motifs. We cannot exclude the possibility that the AbiK mutations affected protein folding rather than polymerase activity per se; however, we note that eight different mutations showed a similar effect in greatly reducing or abolishing polymerase activity in vitro.
Figure 5.
Activities of AbiK mutants. (A) Schematic of AbiK mutations (not to scale). Arrowheads indicate the amino acid mutations, which are further specified in Supplementary Figure S1 and Table 2. Asterisks denote the putative catalytic residues. (B) AbiK wild-type and mutant proteins were incubated with [α-32P]dTTP, resolved on a 10% SDS–PAGE gel, dried and phosphorimaged. Lane labeling corresponds to the mutations in (A). Positions of molecular weight markers (kDa) and AbiK-GST are indicated.
Mutations in the RT domains block abortive infection in vivo
AbiK mutants were also tested for the abortive phage infection phenotype in vivo by introducing their sequences into a high-copy plasmid (pNZ123) in L. lactis and challenging the cells with phages. Consistent with previous work (22), the wild-type AbiK sequence provided an efficacy of plaquing (EOP) of 2.6 × 10−8 (Table 2) relative to a sensitive strain, while the AbiK-GST construct remained substantially active with an EOP of 3.7 × 10−7. In contrast, mutations in the RT domains had levels of abortive infection that were reduced or undetectable, with the exception of Mut2b and Mut7 (Table 2). The less drastic impairment for these two mutants in vivo compared to in vitro (Table 2) might reflect the instability of AbiK activity in vitro or the availability of accessory proteins in vivo. Nonetheless, the assays provide good correlation between in vitro polymerization and in vivo abortive infection, and indicate that an activity of the RT domains (DNA polymerization) is critical to the abortive infection phenotype.
DISCUSSION
AbiK is a DNA polymerase with novel biochemical properties
This study establishes a novel type of DNA polymerase that synthesizes long DNAs of non-specific sequence (>500 nt) that are linked to the polymerase protein, likely through a covalent bond. These properties are not typical of RTs; however, they reside in the RT sequence motifs, because mutations in the motifs abolish the self-labeling and DNA polymerization activities. To our knowledge, this is the first example of an enzyme possessing activities of both self-priming and random sequence synthesis.
Several lines of evidence support the unusual finding of non-templated polymerization by AbiK: (i) no nucleic acids were detectable in the AbiK preparation by silver staining or hybridization experiments; (ii) incorporation of [α-32P]dTTP competes with incorporation of dATP, dCTP and dGTP; (iii) long DNAs are produced by either incubation with dNTPs, or with dTTP alone; and (iv) the cloned sequences generated by AbiK do not correspond to E. coli sequence or any other sequence in GenBank. The most critical experiment, however, was the incubation of AbiK with only three nucleotides (either dATP+dCTP+dTTP or dATP+dGTP+dTTP), with the resulting cloned sequences consisting of multiple ‘random’ sequences of only those three nucleotides. Thus, AbiK appears to incorporate any available nucleotides into long polymers, similar to a terminal transferase.
Although non-templated incorporation was unanticipated, some eukaryotic RTs have been reported to carry out non-templated incorporation, albeit on a smaller scale and usually under artificial conditions. The RT of the non-long terminal repeat (non-LTR) element R2 of Bombyx mori, for some mutant RNA templates, is capable of adding more than 50 non-templated nucleotides to its cleaved DNA target (i.e. its primer) before engaging its RNA template. These additions may be pseudotemplated rather than truly random (32,33). Another example is the RT of the mitochondrial retroplasmid Varkud, which can polymerize several non-templated nucleotides, either before engaging an RNA template in the absence of a primer, or when switching from one RNA template to another (34–36). Finally, yeast and human telomerase RT in the presence of manganese can add up to 20 non-templated nucleotides to the 3′-ends of DNA primers, which was noted to be analogous to terminal transferase activity (37). These different eukaryotic RT examples help to rationalize the ability of the bacterial AbiK to carry out non-templated polymerization, but they also suggest that like other RTs, AbiK may perform multiple reactions in vitro, and be capable of reverse transcription under conditions not tested, or in vivo. Experiments so far have failed to detect RNA-templated polymerization by AbiK in vitro (Table 1; data not shown), and so it appears that under our in vitro conditions, AbiK favors non-templated polymerization.
The self-priming property of lactococcal AbiK was likewise unexpected, but has been seen for the reverse transcriptase of the hepatitis B virus (HBRT) (3). Self-priming appears probable as well for the RTs of the fungal mitochondrial pFOXC retroplasmids (38). For HBRT, DNA polymerization is primed by a tyrosine residue located in the protein’s ∼175 amino acid N-terminal domain. Initiation begins at a specific RNA structure and after reverse transcribing this priming motif, the RT relocates to the 3′-end of the viral RNA intermediate and reverse transcribes the entire genome. The synthesized DNA remains covalently linked to the RT throughout this reaction. For AbiK, we hypothesize that the priming domain is located in its C-terminal region, because this region does not yet have an identified function in abortive phage infection, yet is essential (17). The C-terminal sequence of AbiK contains multiple Y, S and T residues that might serve as priming residues (Figure 1, Supplementary Figure S1).
For the viral HBRT enzyme, there is a conformational change during the transition from priming to elongation phases of reverse transcription, which removes the N-terminal protein priming domain from the RT’s active site, and replaces it with the 3′-end of the RNA genome (39). An analogous rearrangement for AbiK may help explain the nuclease susceptibility data and unusual kinetics of polymerization. Low concentrations of [α-32P]dTTP produce only a short DNA that is resistant to DNase I treatment (Figure 3B). This may correspond to a ‘closed’ priming conformation in which the DNA is protected in the active site by the priming domain. (As noted above, the initially polymerized DNA may be significantly shorter than 18–25 nt.) Upon chasing with high concentrations of dNTPs, the DNA rapidly extends to several hundred nucleotides (Figure 2) and is sensitive to DNase I; this may correspond to an ‘open’ conformation in which the DNA is not protected. Other explanations for the nuclease susceptibility data may involve differential steric effects caused by long and short DNAs.
A novel biological function for a polymerase
As a rule, polymerases function in genome replication; however, here we uncover a non-replicative role for a polymerase, in which an RT-related protein has evolved a function to provide resistance to phage infections. Polymerase activity of AbiK appears to be required for abortive infection, because mutations in the RT domains block both polymerization activities in vitro and abortive infection in vivo. Previously, it was shown that abiK is the only gene on the plasmid pSRQ800 that is required for abortive infection in Lactococcus lactis (22).
Additional clues to the AbiK mechanism come from mutant phages that gain resistance to AbiK. For lactococcal phage ul36, suppressive mutations are mapped to the phage sak gene (sensitivity to AbiK) (40), while for other lactococcal phages, equivalent mutations are localized to sak2 (phage P335), sak3 (phage p2) and sak4 (phage ϕ31) (40). In fact, sak, sak2 and sak4 share sequence similarity with RAD52, Erf and RAD51 families of recombinases, respectively (41). Biochemical experiments have shown that purified Sak forms rings of 11 subunits around ssDNA and has single-strand annealing activity (42). Recently, Sak3 was also experimentally shown to have single-strand annealing activity (43). The role of Sak in phage replication remains unknown, but in coliphages, single-strand annealing proteins are involved in steps of DNA circularization, repair of DNA concatemers and DNA replication (40).
It can therefore be concluded that abortive infection occurs by the synergistic interaction of AbiK and a phage-encoded recombinase. Nonetheless, a detailed mechanism for abortive infection remains unclear, and multiple potential mechanisms are possible. Given available data, it appears that the mechanism of AbiK-mediated abortive infection is distinct from characterized phage resistance systems, none of which to our knowledge involve a polymerase.
Two key remaining questions are whether non-templated polymerization occurs in vivo, and whether such a polymerization product is involved in the abortive infection pathway. Studies are underway to address these issues.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was funded by a strategic grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada to S.M. and S.Z [03601-CG083160]. Salary support for S.Z. was from the Alberta Heritage Foundation for Medical Research. Funding for open access charge: Natural Sciences and Engineering Research Council (Canada).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelman A. Reverse transcription and integration. In: Kurth R, Bannert N, editors. Retroviruses: Molecular Biology, Genomics and Pathogenesis. Berlin: Caister Academic Press; 2010. pp. 114–142. [Google Scholar]
- 3.Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn EH. Telomerase RNA. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3 rd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2006. pp. 419–436. [Google Scholar]
- 5.Galligan JT, Kennell JC. Retroplasmids: linear and circular plasmids that replicate via reverse transcription. In: Meinhardt F, Klassen R, editors. Microbial Linear Plasmids. Vol. 7. Heidelberg: Springer; 2007. pp. 163–186. [Google Scholar]
- 6.Pardue ML, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Annu. Rev. Genet. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. [DOI] [PubMed] [Google Scholar]
- 7.Kojima KK, Kanehisa M. Systematic survey for novel types of prokaryotic retroelements based on gene neighborhood and protein architecture. Mol. Biol. Evol. 2008;25:1395–1404. doi: 10.1093/molbev/msn081. [DOI] [PubMed] [Google Scholar]
- 8.Simon DM, Zimmerly S. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 2008;36:7219–7229. doi: 10.1093/nar/gkn867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. In: Gestland RF, Cech TR, Atkins JF, editors. RNA Worlds: From Life’s Origins to Diversity in Gene Regulation. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2011. pp. 103–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, Deora R, Doulatov SR, Gingery M, Eiserling FA, Preston A, Maskell DJ, Simons RW, Cotter PA, Parkhill J, et al. Reverse transcriptase-mediated tropism switching in Bordetella bacteriophage. Science. 2002;295:2091–2094. doi: 10.1126/science.1067467. [DOI] [PubMed] [Google Scholar]
- 12.Medhekar B, Miller JF. Diversity-generating retroelements. Curr. Opin. Microbiol. 2007;10:388–395. doi: 10.1016/j.mib.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lampson BC, Viswanathan M, Inouye M, Inouye S. Reverse transcriptase from Escherichia coli exists as a complex with msDNA and is able to synthesize double-stranded DNA. J. Biol. Chem. 1990;265:8490–8496. [PubMed] [Google Scholar]
- 14.Lampson BC, Inouye M, Inouye S. Retrons, msDNA, and the bacterial genome. Cytogenet. Genome. Res. 2005;110:491–499. doi: 10.1159/000084982. [DOI] [PubMed] [Google Scholar]
- 15.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 16.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emond E, Holler BJ, Boucher I, Vandenbergh PA, Vedamuthu ER, Kondo JK, Moineau S. Phenotypic and genetic characterization of the bacteriophage abortive infection mechanism AbiK from Lactococcus lactis. Appl. Environ. Microbiol. 1997;63:1274–1283. doi: 10.1128/aem.63.4.1274-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinsmore PK, Klaenhammer TR. Phenotypic consequences of altering the copy number of abiA, a gene responsible for aborting bacteriophage infections in Lactococcus lactis. Appl. Environ. Microbiol. 1994;60:1129–1136. doi: 10.1128/aem.60.4.1129-1136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odegrip R, Nilsson AS, Haggård-Ljungquist E. Identification of a gene encoding a functional reverse transcriptase within a highly variable locus in the P2-like coliphages. J. Bacteriol. 2006;188:1643–1647. doi: 10.1128/JB.188.4.1643-1647.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 22.Fortier LC, Bouchard JD, Moineau S. Expression and site-directed mutagenesis of the lactococcal abortive phage infection protein AbiK. J. Bacteriol. 2005;187:3721–3730. doi: 10.1128/JB.187.11.3721-3730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3 rd edn. NY: Cold Spring Harbor Laboratory, Cold Spring Harbor; 2001. [Google Scholar]
- 24.Rohman M, Harrison-Lavoie KJ. Separation of copurifying GroEL from glutathione-S-transferase fusion proteins. Protein Expr. Purif. 2000;20:45–47. doi: 10.1006/prep.2000.1271. [DOI] [PubMed] [Google Scholar]
- 25.Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 26.Zimmerly S, Moran JV, Perlman PS, Lambowitz AM. Group II intron reverse transcriptase in yeast mitochondria. Stabilization and regulation of reverse transcriptase activity by the intron RNA. J. Mol. Biol. 1999;289:473–490. doi: 10.1006/jmbi.1999.2778. [DOI] [PubMed] [Google Scholar]
- 27.Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Sullivan DJ, Klaenhammer TR. Rapid mini-prep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarvis AW. Serological studies of a host range mutant of a lactic streptococcal bacteriophage. Appl. Environ. Microbiol. 1978;36:785–789. doi: 10.1128/aem.36.6.785-789.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 31.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luan DD, Eickbush TH. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 1995;15:3882–3891. doi: 10.1128/mcb.15.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luan DD, Eickbush TH. Downstream 28S gene sequences on the RNA template affect the choice of primer and the accuracy of initiation by the R2 reverse transcriptase. Mol. Cell. Biol. 1996;16:4726–4734. doi: 10.1128/mcb.16.9.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kennell JC, Wang H, Lambowitz AM. The Mauriceville plasmid of Neurospora spp. uses novel mechanisms for initiating reverse transcription in vivo. Mol. Cell. Biol. 1994;14:3094–3107. doi: 10.1128/mcb.14.5.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang CC, Kennell JC, Wanner LA, Lambowitz AM. A mitochondrial retroplasmid integrates into mitochondrial DNA by a novel mechanism involving the synthesis of a hybrid cDNA and homologous recombination. Mol. Cell. Biol. 1994;14:6419–6432. doi: 10.1128/mcb.14.10.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen B, Lambowitz AM. De novo and DNA primer-mediated initiation of cDNA synthesis by the Mauriceville retroplasmid reverse transcriptase involve recognition of a 3′ CCA sequence. J. Mol. Biol. 1997;271:311–332. doi: 10.1006/jmbi.1997.1185. [DOI] [PubMed] [Google Scholar]
- 37.Lue NF, Bosoy D, Moriarty TJ, Autexier C, Altman B, Leng S. Telomerase can act as a template- and RNA-independent terminal transferase. Proc. Natl Acad. Sci. USA. 2005;102:9778–9783. doi: 10.1073/pnas.0502252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galligan JT, Marchetti SE, Kennell JC. Reverse transcription of the pFOXC mitochondrial retroplasmids of Fusarium oxysporum is protein primed. Mobile DNA. 2011;2:1. doi: 10.1186/1759-8753-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin L, Wan F, Hu J. Functional and structural dynamics of hepadnavirus reverse transcriptase during protein-primed initiation of reverse transcription: effects of metal ions. J. Virol. 2008;82:5703–5714. doi: 10.1128/JVI.02760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouchard JD, Moineau S. Lactococcal phage genes involved in sensitivity to AbiK and their relation to single-strand annealing proteins. J. Bacteriol. 2004;186:3649–3652. doi: 10.1128/JB.186.11.3649-3652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 2010;38:3952–3962. doi: 10.1093/nar/gkq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ploquin M, Bransi A, Paquet ER, Stasiak AZ, Stasiak A, Yu X, Cieslinska AM, Egelman EH, Moineau S, Masson JY. Functional and structural basis for a bacteriophage homolog of human RAD52. Curr. Biol. 2008;18:1142–1146. doi: 10.1016/j.cub.2008.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scaltriti E, Launay H, Genois M-M, Bron P, Rivetti C, Grolli S, Ploquin M, Campanacci V, Tegoni M, Cambillau C, et al. Lactococcal phage p2 ORF35-Sak3 is an ATPase involved in DNA recombination and AbiK mechanism. Mol. Microbiol. 2011;80:102–116. doi: 10.1111/j.1365-2958.2011.07561.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.