Abstract
Uridine at the wobble position of tRNA is usually modified, and modification is required for accurate and efficient protein translation. In eukaryotes, wobble uridines are modified into 5-methoxycarbonylmethyluridine (mcm5U), 5-carbamoylmethyluridine (ncm5U) or derivatives thereof. Here, we demonstrate, both by in vitro and in vivo studies, that the Arabidopsis thaliana methyltransferase AT1G31600, denoted by us AtTRM9, is responsible for the final step in mcm5U formation, thus representing a functional homologue of the Saccharomyces cerevisiae Trm9 protein. We also show that the enzymatic activity of AtTRM9 depends on either one of two closely related proteins, AtTRM112a and AtTRM112b. Moreover, we demonstrate that AT1G36310, denoted AtALKBH8, is required for hydroxylation of mcm5U to (S)-mchm5U in tRNAGlyUCC, and has a function similar to the mammalian dioxygenase ALKBH8. Interestingly, atalkbh8 mutant plants displayed strongly increased levels of mcm5U, and also of mcm5Um, its 2′-O-ribose methylated derivative. This suggests that accumulated mcm5U is prone to further ribose methylation by a non-specialized mechanism, and may challenge the notion that the existence of mcm5U- and mcm5Um-containing forms of the selenocysteine-specific tRNASec in mammals reflects an important regulatory process. The present study reveals a role in for several hitherto uncharacterized Arabidopsis proteins in the formation of modified wobble uridines.
INTRODUCTION
tRNA molecules typically contain several different post-transcriptional modifications, and such modifications are essential for tRNA function. Located at specific sites throughout the tRNA body they facilitate correct folding and contribute to stabilizing the tertiary structure (1,2). Moreover, modifications on the anticodon stem–loop (ASL) are important to ensure efficient and accurate decoding during translation, as well as to maintain a correct reading frame (3–5).
The nucleoside at the wobble position (position 34), which base pairs with the nucleoside in the third position of the codon, is frequently modified. In particular, a uridine at this position (U34) almost invariably carries a modification. Cytoplasmic tRNAs in eukaryotes usually harbour the U34 modifications 5-methoxycarbonylmethyluridine (mcm5U), 5-carbamoylmethyluridine (ncm5U) or derivatives thereof (6). The biogenesis of these modifications have been studied in most detail in Saccharomyces cerevisiae. Here, the Elongator complex, consisting of the six proteins Elp1–6, is required for the early steps in the synthesis of modified wobble uridines and, interestingly, the phenotypes of Elongator mutants are suppressed by over-expression of certain tRNAs that are hypomodified in these mutants (7,8). The formation of mcm5U from 5-carboxymethyluridine (cm5U) is catalysed by the methyltransferase Trm9 (9). Several mcm5U-containing tRNAs are further modified by thiolation, a modification that restricts wobbling, yielding 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) (10). Accordingly, these thiolated tRNAs decode in the split codon boxes, where purine- and pyrimidine ending codons encode different amino acids. In contrast, ncm5U-containing tRNAs generally decode in the ‘family’ codon boxes, where all four codons encode the same amino acid.
Several additional wobble uridine modifications not found in S. cerevisiae are present in mammals. The selenocysteine specific tRNA (tRNASec) exists as two differentially modified subpopulations; one containing mcm5U at the wobble position and another, which contains its 2′-O-ribose methylated derivative 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um) in this position (11). In addition, two diastereomers of hydroxylated mcm5U, (R)-mchm5U and (S)-mchm5U are found in mammalian  and
and 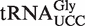 , respectively (12).
, respectively (12).
Mammals have nine homologues, ALKBH1-ALKBH9, of the Escherichia coli dioxygenase AlkB, a DNA repair protein that uses an oxidative mechanism to remove deleterious methyl groups from DNA (13–18). Mammalian ALKBH8 consists of three domains, i.e. an N-terminal RNA recognition motif (RRM), followed by the characteristic AlkB-like dioxygenase domain, and a C-terminal methyltransferase (MT) domain. The MT domain of ALKBH8 was recently shown to represent the mammalian homologue of S. cerevisiae Trm9, and to be required for mcm5U formation in mice (19,20). Similar to yeast Trm9, the MT domain of ALKBH8 forms a heterodimeric complex with TRM112, and this interaction is required for MT activity. In two recent studies, the RRM/AlkB moiety of mammalian ALKBH8 was demonstrated to catalyse the hydroxylation of mcm5U into (S)-mchm5U in  (12,21).
(12,21).
In plants, the knowledge about tRNA modifications and their formation is limited, and only a handful studies have focused on tRNA modifying enzymes. The yeast Mod5 protein is an isopentenyltransferase responsible for generating isopentenyladenosine (i6A) at position 37, and the corresponding Arabidopsis homologue has been identified and functionally characterized (22). Additionally, an Arabidopsis orthologue of the mammalian methyltransferases Dnmt2, which generates 5-methylcytosine in tRNA, has been identified (23). Recently, the modifications ncm5U and mcm5s2U were reported to be present in Arabidopsis total tRNA, and their formation was shown to depend on a functional Elongator complex (24). Finally, a reverse genetics approach recently identified five tRNA modification genes in Arabidopsis (25).
In the present study, we have identified and characterized two novel tRNA modification enzymes in Arabidopsis, denoted AtTRM9 and AtALKBH8. While AtTRM9 displays similarity to the yeast Trm9 protein, AtALKBH8 shows similarity to the RRM/AlkB part of mammalian ALKBH8. Using AtTRM9 and AtALKBH8 loss-of-function mutants, as well as in vitro enzymatic assays, we demonstrate that AtTRM9 is a methyltransferase required for mcm5U formation, while AtALKBH8 is the hydroxylase generating (S)-mchm5U from mcm5U. Finally, we show that the MT-activity of AtTRM9 is dependent on either of the two Arabidopsis TRM112 homologues, AtTRM112a and AtTRM112b.
MATERIALS AND METHODS
Protein sequence analysis
Multiple sequence alignments were generated using MAFFT algorithm (26) embedded in the JalView alignment editor (27).
Plant materials
All plant lines were obtained from the Nottingham Arabidopsis Stock Centre (NASC) unless otherwise stated. Wild-type (WT) Arabidopsis (Arabidopsis thaliana ecotype Col-0) were grown in perlite soil at 18°C under 16 h of light at 100 μE/m2/s. For axenic growth, seeds were surface sterilized in 7.5% hypochlorite solution containing 0.1% Tween-20, followed by treatment with 70% EtOH and washing in distilled water. Sterilized seeds were plated on solidified MS medium (28), supplemented with 2% sucrose (MS-2), kept in the dark at 4°C for 2 days and then transferred to 18°C.
For genotyping of T-DNA insertional mutants, genomic DNA was isolated from true leaves using the Ultraprep Plant DNA kit (AHN Biotechnologie). Homozygous atalkbh8-1 plants were screened by PCR using a T-DNA right border primer 1 (RB1) and genomic primer G1, as well as a T-DNA left border primer 1 (LB1) combined with genomic primer G2 (See Supplementary Table S1 for details on DNA primers used in the present work). For screening of atalkbh8-2 plants, the genomic primers G3 and G4 were used in combination with the LB primer. For attrm9 plants, the PCR was conducted using the primer LB2 in combination with genomic primers G5 and G6. PCR products were cloned into the TOPO Blunt vector (Invitrogen), and sequenced to verify the integration site.
To analyse the expression of AtALKBH8 and AtTRM9 in the T-DNA lines, RNA was first isolated from liquid N2 frozen tissue utilizing MagNA Lyser Green Beads (Roche Diagnostics), and subsequently purified using Spectrum Plant Total RNA Kit (Fluka/Sigma-Aldrich) followed by cDNA synthesis using Superscript II Reverse Transcriptase (Invitrogen) and a poly dT primer (Invitrogen). The expression of different regions upstream or downstream of the T-DNA insertion sites in the homozygous mutant lines was analysed as follows: for AtALKBH8, primers Up8-f/Up8-r were utilized to analyze expression upstream of the T-DNA integration site; primers Dwn8-f/Dwn8-r to examine expression downstream of the T-DNA insertion and primers Sp8-f/Sp8-r are spanning the T-DNA insertion. For AtTRM9, the following primers were utilized: primers Dwn9-f/Dwn9-r for investigating expression downstream of the T-DNA insertion and primers Sp9-r/Sp9-f that span the T-DNA insertion.
For scoring of seed germination in the different mutant plant lines, seeds were incubated in Petri dishes containing MS-2 medium supplemented with 1% (w/v) agar, for 48 h at 4°C, and then moved to a growth chamber at 22°C. All mutants were compared with WT from the same harvest. Testa rupture was scored every day using a LEICA EZ4 microscope (New York Microscope Company Inc, New York, USA). For assessing root growth, seeds were processed as above. Plates were placed vertically and the number of lateral roots were determined over a 8-day period using a LEICA EZ4 microscope.
For tRNA isolation, seeds from homozygous mutant and WT plants were surface sterilized and plated on MS2. Two weeks after germination, the seedlings were frozen in liquid N2 before grinding and isolation of tRNA.
Plasmid construction
The coding sequences of AtTRM9 and AtALKB8 were amplified by PCR from Arabidopsis cDNA templates using primer sets AtTRM9-f/AtTRM9-r and AtALKBH8-f/AtALKBH8-r, respectively, and subsequently cloned between the NdeI and BamHI restriction sites in the expression vector pET28a to generate pET28-AtTRM9 and pET28-AtALKB8. The coding sequence of AtTRM112a was PCR amplified from cDNA clone RAFL22-14-D01 [RIKEN Bio Research Centre (http://www.brc.riken.go.jp/)] using primers AtTRM112a-r and AtTRM112a-f, whereas AtTRM112b was amplified from cDNA clone U22673 {ABRC [Arabidopsis Biological Research Centre (www.abrc.osu.edu)]} using primers AtTRM112b-r and AtTRM112b-f. The PCR products were subsequently cloned between the NdeI and BglII sites of the second MCS in pET-Duet1 (Novagen), yielding pDuet-AtTRM112a and pDuet-AtTRM112b, respectively. To obtain pDuet-AtTRM9/AtTRM112a and pDuet-AtTRM9/AtTRM112b, a XbaI–BamHI fragment from pET28 to AtTRM9 was transferred to the first MCS of either pDuet-AtTRM112a or pDuet-AtTRM112b, respectively.
Protein purification
All recombinant proteins were expressed and purified from E. coli BL21-CodonPlus(DE3)-RIPL (Stratagene). Bacteria were grown at 16°C in LB medium containing 0.075 mM IPTG. Cell pellets were resuspended in ice-cold equilibration buffer [50 mM sodium phosphate pH 7.6, 150 mM NaCl, 10% glycerol, 0.5% NP-40, protease inhibitors (complete EDTA-free protease inhibitor tablets, Roche) and 0.5 mg/ml lysozyme (Fluka/Sigma-Aldrich], and lysates were generated utilizing a French press (Thermo IEC). Proteins were subsequently purified from pre-cleared lysates using TALON Metal Affinity Resin (Clontech) according to the manufacturer’s instructions.
Total tRNA and tRNA isoacceptor isolation from plants
Six-hundred milligram of seedlings were harvested and homogenized in liquid nitrogen. Lysates were prepared by adding 9 ml of ice-cold lysis buffer [4 M guanidine thiocyanate, 100 mM Tris–HCl, 25 mM MgCl2, 2% Triton X-100 (v/v), 25 mM EDTA, pH 7.5] to the homogenized tissue, followed by the addition of 9 ml of 3 M NaOAc. Total RNA was precipitated from the lysates with isopropanol, and the tRNA fraction was subsequently isolated using a RNA/DNA maxi kit (Qiagen). 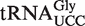 was isolated from Arabidopsis total tRNA by hybridization to an immobilized oligonucleotide (Supplementary Table S1) using a protocol described previously (20).
was isolated from Arabidopsis total tRNA by hybridization to an immobilized oligonucleotide (Supplementary Table S1) using a protocol described previously (20).
Liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) analysis
LC-MS/MS analysis of nucleosides was essentially done as described (20). In brief, tRNA was enzymatically digested to nucleosides, which were separated by reverse phase high performance liquid chromatography (HPLC) on a Zorbax SB-C18 column, using a mobile phase consisting of 0.1% formic acid and a gradient of 5–50% methanol. Online mass spectrometry detection was performed using an Applied Biosystems/MDS Sciex 5000 triple quadrupole mass spectrometer (Applied Biosystems Sciex) with TurboIonSpray probe operating in positive electrospray ionization mode. The nucleosides were monitored by multiple reaction monitoring (MRM) using the nucleoside to base ion mass transitions 317.2→185.1 (mcm5U), 333.2→201.1 (mchm5U), 303.2→171.1 (cm5U), 333.2→201.1 (mcm5s2U), 302.2→170.1 (ncm5U), 318.2→186.1 (ncm5s2U), 268.2→136.1 (A), 244.2→112.1 (C), 284.2→152.2 (G) and 245.2→113.1 (U). Quantitation was accomplished by comparison with pure nucleoside standards run intermediate the samples. In the case of mcm5Um, no nucleoside standard was available, but our previous studies on mammalian 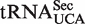 indicate that mcm5U and mcm5Um display similar responses in the mass spectrometer, and this was used to estimate the mcm5Um levels.
indicate that mcm5U and mcm5Um display similar responses in the mass spectrometer, and this was used to estimate the mcm5Um levels.
RNase T1 digestion and MALDI-TOF mass spectrometry
RNase T1 digestion of nucleosides and MALDI-TOF mass spectrometry were done as described previously (20).
Saponification of S. cerevisiae tRNA
To remove the methyl moiety from the methoxycarbonylmethyl group, the methyl ester linkage was broken by saponification of the tRNA substrate as described previously (20). Briefly, 10 μl of a tRNA solution was mixed with 1.25 μl of 1 M NaOH and the mixture was incubated at room temperature for 8 min. The solution was neutralized by the addition of 1.25 μl of 1 M acetic acid.
In vitro enzyme assays
For the methyltransferase assay, 5 and 10 µg total tRNA from plant and yeast (Roche), respectively, was incubated with AtTRM9 in the absence or presence of co-expressed AtTRM112a/b for 12 min at 37°C in 50 µl reaction buffer containing 1 µl S-adenosyl-l-[methyl-3H]methionine (GE Healthcare), 50 mM Tris–HCl pH 7.5, 25 mM KCl, 25 mM NH4OAc, 0.5 mM MgCl2, 0.5 mM EDTA, 1 U RNasin Plus RNase Inhibitor (Promega). The reaction was stopped by adding 10% ice-cold TCA followed by 30 min incubation on ice. The reaction mixture was carefully applied to glass microfiber filters (GE-Healthcare) mounted to a filter holder manifold (Millipore) under vacuum. Subsequently, the filters were washed with 10% TCA followed by 96% ethanol, dried and added to scintillation vials for measuring incorporation of radiolabelled methyl groups.
In the assay for hydroxylation of mcm5U in atalkbh8 tRNA, 5 μg tRNA was incubated with different amounts of the N-terminal part (amino acids 1–354) of human ALKBH8 for 30 min at 37°C in a 50 µl reaction mixture containing 50 mM Tris–HCl pH 7.5, 0.5 mM MgCl2, 2 mM ascorbic acid, 100 µM 2-oxoglutarate, 40 µM FeSO4, 10 U RNase inhibitor and then precipitated with 1 volume of isopropanol in the presence of 1 M NH4Ac and 20 μg of glycogen. Pellets were washed with 70% EtOH and dried, before they were further processed for LC–MS/MS analysis.
RESULTS
In silico identification of ALKBH8/Trm9-like proteins in plants
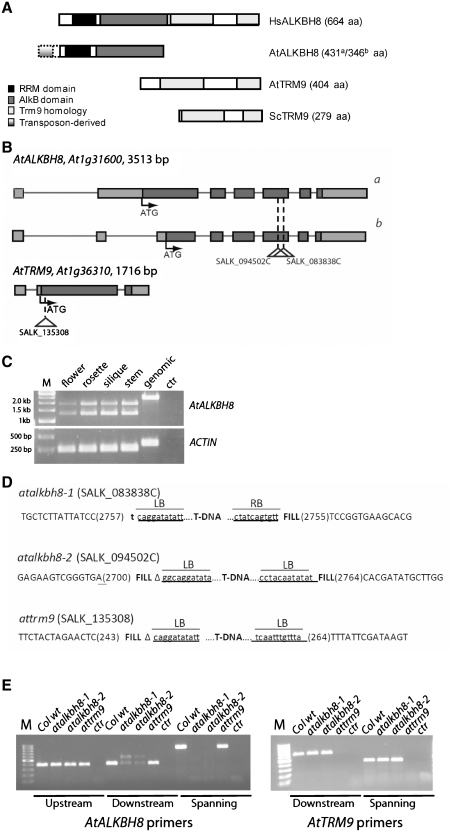
During our efforts to characterize the mammalian ALKBH8 protein, we became intrigued by the observation that the RRM/AlkB and MT functions of ALKBH8 appeared to be encoded by two separate genes in plants. A protein BLAST search using human ALKBH8 as query retrieved two major hits in Arabidopsis, encoded by the genes At1g31600 and At1g36310. The protein encoded by At1g31600 displays high similarity to the N-terminal part of ALKBH8, encompassing the RRM and AlkB domains, and will be referred to as AtALKBH8. The protein encoded by At1g36310 is highly similar to the C-terminal MT domain of human ALKBH8, as well as to the S. cerevisiae Trm9 protein, and was denoted AtTRM9. Putative orthologues of these two ALKBH8/Trm9-like proteins were found in other plants, such as rice (Oryza sativa) and grape (Vitis vinifera), showing that the separation of the dioxygenase and MT functions of ALKBH8 into two proteins, as observed in Arabidopsis, applies to both monocots and eudicots, and therefore might apply to land plants in general. Alignments of AtALKBH8 and AtTRM9 with putative orthologues from various organisms are shown in Figure 1A and B, respectively.
Figure 1.
Protein sequence alignment of AtALKB8 and AtTRM9 with putative orthologues from selected organisms. Functional domains are indicated by dotted (RRM motif), solid (AlkB-like) and dashed (Trm9-like) lines above the alignment. (A) Alignment of AtALKBH8 and putative orthologues. The upstream T-DNA insertion sites in the atalkbh8-1 and atalkbh8-2 lines are indicated by closed and open triangles, respectively. Gene identifier numbers: Hs, Homo sapiens, gi|195927059; Dm, Drosophila melanogaster, gi|24658267; At, Arabidopsis thaliana, gi|42571711; Vv, Vitis vinifera, gi|225427651; Os, Oryza sativa, gi|50928501 and Ce, Caenorhabditis elegans, gi|17552176. (B) Alignment of AtTRM9 and putative orthologues. The upstream T-DNA insertion site in the attrm9 line corresponds to Arg9 in the protein sequence (which is located in the non-conserved N-terminal part of the protein not included in the alignment). Gene identifier numbers: At, gi|18400083; Vv, gi|225459401; Os, gi|115448705; Hs, gi|195927059; Dm, gi|24658267; Ce, Caenorhabditis elegans, gi|17552176 and Sc, Saccharomyces cerevisiae, gi|6323627 (note that the gi numbers for AtALKBH8 and AtTRM9 homologues are identical in animals, where these two functions are carried out by a single protein).
Annotation data, as well as available ESTs and cDNAs, suggest that alternative splicing may give rise to two different forms of the AtALKBH8 protein (Figure 2A and B). A 7 exon gene model (At1g31600.1) encodes a 431 amino acids protein (AtALKBH8a), whereas the alternative 8 exon model (At1g31600.2) encodes a 344 amino acids protein (AtALKBH8b) (Figure 2B). AtALKBH8a represents an extension of AtALKBH8b by 83 amino acids, but this extension shares no detectable sequence homology with any other proteins in the NCBI protein sequence database. Putative ALKBH8 orthologues from other plants lack a corresponding extension, and they are generally very similar in size to the 344 amino acids AtALKBH8b protein, to which they align throughout their entire sequence. Interestingly, the 159 nt DNA sequence encoding the N-terminal part (53 amino acids) of the 83 amino acid extension is highly similar (94% sequence identity) to the Ac-type transposon Tag2. This transposon, and inactive remnants thereof, are found in numerous copies throughout the Arabidopsis genome, indicating that the alternative splice form may have arisen through a transposition event (29). RT–PCR experiments showed that mRNAs corresponding to both forms of AtALKBH8 were expressed in all tissues examined (stamen, rosette leaves, flowers and silique) (Figure 2C).
Figure 2.
Architecture of the AtALKBH8 and AtTRM9 genes and characterization of corresponding T-DNA insertion lines. (A) Schematic view of the domain architecture of ALKBH8 and TRM9 proteins from various organisms. The N-terminal extension present in the 431 amino acid splice variant of AtALKBH8 is indicated by a dotted box, and gradient shading indicates the part of this extension, which shows strong homology to the Tag2 transposon. The RNA recognition motif is shown in black, the AlkB oxygenase in dark grey, and the methyltransferase domain is shown in light grey. (B) Schematic view of AtALKBH8 and AtTRM9. Exons are indicated by boxes, where putatively untranslated regions are shown in light grey and translated regions are shown in dark grey. Annotated start codons (ATG) are indicated by arrows. The upstream T-DNA insertion sites are indicated by triangles. The two alternative gene models for the AtALKBH8 locus, which give rise to the two different forms of the AtALKBH8 protein, are indicated. (C) Expression of mRNA encoding the two forms of AtALKBH8 in various tissues. RT–PCR on mRNA from different tissues was performed, using primers amplifying fragments of 1882 and 1302 bp representing gene model AtALKBH8a and AtALKBH8b, respectively. Genomic DNA was used as positive control (note that the genomic fragment is larger due to intron sequences). The ACTIN2-7 gene, giving a fragment of 255 bp with primers spanning intron 2, amplified at comparable levels from all tissues, with no genomic contamination as indicated by the band of 340 bp fragment representing genomic DNA that is only seen in the genomic DNA control lane. M: Molecular weight marker. (D) Sequence determination of the insertion sites in the T-DNA lines. Precise location of the T-DNA in the AtALKBH8 and AtTRM9 genes are shown for the different lines. Exon sequences are shown in uppercase letters and left and right border (LB and RB) T-DNA sequences are underlined. Filler sequences are indicated as ‘fill’, while deletion of genomic DNA is marked by Δ. Numbers indicate the position of the insertion sites in the gene sequence. (E) Expression analysis of WT and mutant T-DNA lines by RT–PCR. Regions of the respective AtALKBH8 (left panel) and AtTRM9 (right panel) genes, together with relevant primer pairs (upstream, downstream or spanning the T-DNA integration site) are indicated below. For each primer set, expression analysis was conducted on mRNA from WT and different T-DNA lines as indicate above. Ctr refers to a negative control PCR sample run without cDNA, and M indicates marker lane.
Two different gene models for the At1g36310 gene have been predicted, consisting of two or three exons. The transcripts resulting from these gene models only differ in their 5′-UTR, and both encode the same 404 amino acid AtTRM9 protein. Available cDNAs and ESTs primarily support the three exon model, which is shown in Figure 2B. RT–PCR experiments showed that the mRNA for AtTRM9, were expressed in all tissues examined (stamen, rosette leaves, flowers and silique) (data not shown).
Genomic characterization of mutant plants with T-DNA insertions in the genes-encoding AtALKBH8 and AtTRM9
To investigate the role of the AtALKBH8 and AtTRM9 proteins in tRNA modification in planta, T-DNA insertion lines were obtained for AtALKBH8 (atalkbh8-1, SALK_083838C; atalkbh8-2, SALK_094502C) and AtTRM9 (attrm9, SALK_135308). The T-DNA in the atalkbh8 lines is inserted into the gene in such a way that it disrupts the AlkB domain, while the T-DNA in the attrm9 line is inserted shortly downstream of the translational start site in the AtTRM9 gene (Figures 1 and 2B).
The annotated T-DNA insertions were further verified by sequencing of PCR products spanning the individual integration sites. At the LB junction in SALK_083838C (atalkbh8-1), the insertion point was determined to be after position 2757 in the genomic sequence of the At1g31600 gene, corresponding to position 1634 in the mRNA sequence (NM_102896) (Figure 2D). A filler of 1 bp had been inserted between the genomic DNA and the T-DNA at this border. At the RB integration site in atalkbh8-1, the insertion point was determined to be at genomic position 2755 in the At1g31600 gene, corresponding to position 1632 in the mRNA sequence (NM_102896). At this junction, a filler consisting of 20 bp scrambled genomic DNA had been integrated. Also, part of the RB sequence of the T-DNA had been deleted. In SALK_094502C (atalkbh8-2), there are two LB sequences integrated (Figure 2D). The integration was verified to be at position 2700 in the genomic sequence of the At1g31600 gene, corresponding to position 1577 in the mRNA sequence (NM_102896). A filler of 30 bp was inserted between the genomic DNA and the T-DNA at this junction. The other LB junction was determined to be at genomic position 2764, corresponding to position 1642 in the mRNA sequence, showing a deletion of 64 bp at the integration site. A filler of 25 bp had been integrated at this site. Also in the SALK_135308 (attrm9) line, two LB sequences have been integrated (Figure 2D). The integration site of the upstream LB was determined to be at position 243 in the genomic sequence of the At1g36310 gene, corresponding to position 161 in the mRNA sequence (NM_103320). The downstream LB junction was determined to be at position 264 in the genomic sequence, corresponding to position 183 in the mRNA sequence, giving a deletion of 20 bp genomic DNA. In addition, a filler sequence of 27 bp of unknown origin was integrated at the junction.
We performed RT–PCR experiments in order to determine the expression level in the homozygous mutant lines. For the atalkbh8-1 and atalkbh8-2 alleles, primer pairs annealing upstream as well as downstream of the T-DNA integration site were utilized. Expression could be detected on both sides of the T-DNA insert, although considerably weaker expression was observed downstream as compared to upstream of the integration site (Figure 2E, left panel). Such downstream expression due to cryptic promoters inside the T-DNA has been shown for several other T-DNA insertion lines (30,31). However, since the T-DNA insertions in the atalkbh8-1 and atalkbh8-2 lines both disrupt the conserved AlkB domain of AtALKBH8, we consider it highly unlikely that a functional protein will be generated. In the attrm9 mutant, expression downstream of the T-DNA integration site was virtually absent (Figure 2E, right panel). Full-length mRNA spanning the insertion site could not be detected in any of the mutants.
Wobble uridine modification pattern of attrm9 and atalkbh8 plants
The characterization of the T-DNA insertion lines shown above clearly indicated that these lines represented knockout plants with respect to the AtTRM9 or AtALKBH8 functions. However, when grown in soil, none of the homozygous mutant lines displayed any obvious morphological or developmental phenotype when compared to WT. Furthermore, the germination efficiency of seeds from these plants, as measured by scoring of testa rupture, as well as root growth were similar to that of WT plants (data not shown).
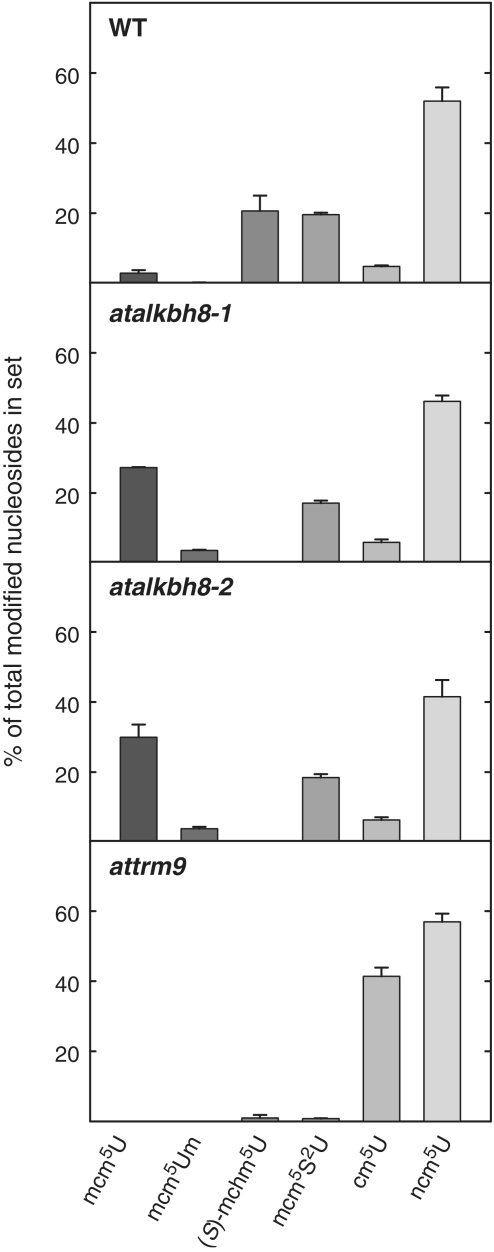
In order to investigate the potential role of these proteins in the modification of wobble uridines, we measured the levels of various modified uridines (Table 1) in total tRNA from WT and mutant plants. Total tRNA from Arabidopsis seedlings was digested into nucleosides and subsequently analysed by LC–MS/MS. The modifications ncm5U, mcm5s2U and (S)-mchm5U were abundant in total tRNA from WT Arabidopsis, and small amounts of cm5U, and mcm5U were also detected (Figure 3). In addition, small amounts of ncm5s2U appeared to be present, but we were not able to firmly establish the presence of this modification, due to an interfering peak with similar LC retention time, and identical MS fragmentation pattern (data not shown). In contrast to the modification pattern observed in WT plants, mcm5s2U, (S)-mchm5U and mcm5U were all virtually absent in tRNA from attrm9 mutant plants, while the level of cm5U, the unmethylated precursor of mcm5U, was strongly increased (Figure 3). The levels of ncm5U were similar in attrm9 and WT plants, which is different from results obtained in mice and yeast, where inactivation of the Trm9 (ALKBH8-MT) function lead to an increase in the levels of this modification, as well as of the 2-thiolated derivative ncm5s2U (4,20). These results strongly indicate that AtTRM9 is a functional Trm9 homologue, which is required for the generation of mcm5U in plants.
Table 1.
Overview of uridine modifications studied in the present work
 |
The –CH3 and –OH groups introduced by AtTRM9 and AtTRM8, respectively, are indicated in bold.
Figure 3.
Wobble uridine modification status in WT, atalkbh8 and attrm9 plants. Total tRNA isolated from WT or mutant plants was degraded to nucleosides, and the levels of the indicated modified nucleosides were determined by LC–MS/MS analysis. The relative level of each individual modification is expressed relative to the total amount of all the shown modifications. Error bars indicate the standard deviation between triplicate samples.
tRNA isolated from AtALKBH8-deficient plants completely lacked (S)-mchm5U, but showed a strong increase in the levels of its precursor, mcm5U (Figure 3), supporting the notion that AtALKBH8, like the mammalian orthologue, catalyses the hydroxylation of mcm5U into (S)-mchm5U. Both atalkbh8-1 and atalkbh8-2 mutant plants showed a virtually identical pattern of aberrant modification. Interestingly, mcm5Um, the 2′-O-ribose methylated form of mcm5U, which is present at very low levels in WT plants, was dramatically increased (∼20-fold) in atalkbh8 tRNA, indicating that a substantial portion of the accumulated mcm5U is subjected to ribose methylation (Figure 3).
Modification status of  in attrm9 and atalkbh8 plants
in attrm9 and atalkbh8 plants
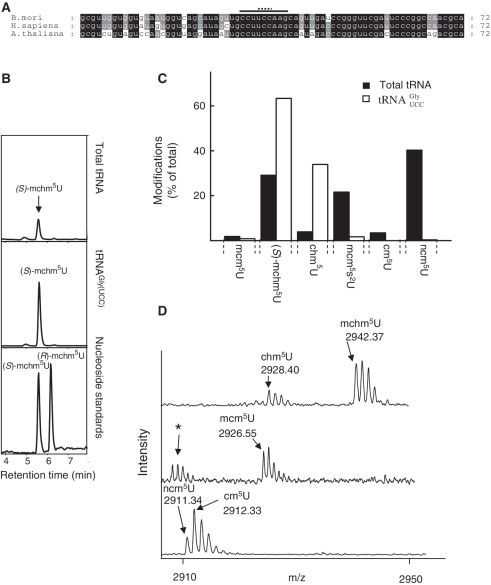
(S)-mchm5U has been detected in a variety of ALKBH8-containing eukaryotes, including Arabidopsis, and it has been demonstrated that 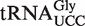 from mouse, calf and silkworm contains this modification in the wobble position (12,32). The primary sequence of Arabidopsis
from mouse, calf and silkworm contains this modification in the wobble position (12,32). The primary sequence of Arabidopsis 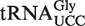 (retrieved from the Genomic tRNA database; http://lowelab.ucsc.edu/GtRNAdb/) is highly similar to that of silkworm and mammals, and the anticodon loop is identical (Figure 4A), suggesting that also this tRNA may carry an (S)-mchm5U modification. To investigate whether this is the case, an immobilized oligonucleotide of complementary sequence to
(retrieved from the Genomic tRNA database; http://lowelab.ucsc.edu/GtRNAdb/) is highly similar to that of silkworm and mammals, and the anticodon loop is identical (Figure 4A), suggesting that also this tRNA may carry an (S)-mchm5U modification. To investigate whether this is the case, an immobilized oligonucleotide of complementary sequence to 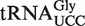 was used to specifically isolate this isoacceptor from total tRNA. Nucleoside analysis by LC–MS/MS clearly demonstrated a strong enrichment of (S)-mchm5U and the corresponding demethylated derivative, 5-[carboxy(hydroxy)methyl]uridine (chm5U), in the isolated
was used to specifically isolate this isoacceptor from total tRNA. Nucleoside analysis by LC–MS/MS clearly demonstrated a strong enrichment of (S)-mchm5U and the corresponding demethylated derivative, 5-[carboxy(hydroxy)methyl]uridine (chm5U), in the isolated 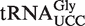 isoacceptor, relative to total tRNA (Figure 4B and C). Note that the LC–MS/MS method used was not able to distiguish between the R and S diastereomers of chm5U. The observed chm5U is probably the result of spontaneous hydrolysis of the ester bond in (S)-mchm5U, as previously reported (12). These results strongly indicated that the (S)-mchm5U modification is present in
isoacceptor, relative to total tRNA (Figure 4B and C). Note that the LC–MS/MS method used was not able to distiguish between the R and S diastereomers of chm5U. The observed chm5U is probably the result of spontaneous hydrolysis of the ester bond in (S)-mchm5U, as previously reported (12). These results strongly indicated that the (S)-mchm5U modification is present in 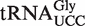 also in Arabidopsis.
also in Arabidopsis.
Figure 4.
Wobble uridine modifications on  in WT, atalkbh8 and attrm9 plants. (A) Alignment of
in WT, atalkbh8 and attrm9 plants. (A) Alignment of 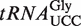 from selected organisms. A solid line indicates the sequence corresponding to the anticodon loop, while the anticodon is indicated by a dotted line. (B) Enrichment of (S)-mchm5U in
from selected organisms. A solid line indicates the sequence corresponding to the anticodon loop, while the anticodon is indicated by a dotted line. (B) Enrichment of (S)-mchm5U in 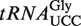 relative to WT total tRNA. LC–MS/MS analysis of (S)-mchm5U in Arabidopsis
relative to WT total tRNA. LC–MS/MS analysis of (S)-mchm5U in Arabidopsis 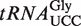 and in total tRNA. (C) Comparison of modification patterns in isolated
and in total tRNA. (C) Comparison of modification patterns in isolated 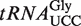 with that in total tRNA. Nucleosides were quantitated by LC–MS/MS analysis. (D) MALDI-TOF mass spectrometry of the anticodon fragment of
with that in total tRNA. Nucleosides were quantitated by LC–MS/MS analysis. (D) MALDI-TOF mass spectrometry of the anticodon fragment of 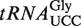 from WT, atalkbh8 and attrm9 plants. Digestion of
from WT, atalkbh8 and attrm9 plants. Digestion of 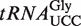 with RNase T1, which cleaves 3′ to G residues generated the anticodon fragment (indicated by a solid line in A) of primary sequence CCUUCCAAG (the wobble nucleotide is indicated). Peaks corresponding to mchm5U, chm5U, mcm5U cm5U and ncm5U in the wobble position, as well as the measured masses, are indicated. An asterisk indicates minor signals from the 2′-3′-cyclic phosphate version of the fragment (the 3′-phosphate version represents the major signal). Theoretical monoisotopic mass of an anticodon fragment containing the various modifications: chm5U, 2928.38; (S)-mchm5U, 2942.40; mcm5U, 2926.40; ncm5U, 2911.40; cm5U, 2912.39.
with RNase T1, which cleaves 3′ to G residues generated the anticodon fragment (indicated by a solid line in A) of primary sequence CCUUCCAAG (the wobble nucleotide is indicated). Peaks corresponding to mchm5U, chm5U, mcm5U cm5U and ncm5U in the wobble position, as well as the measured masses, are indicated. An asterisk indicates minor signals from the 2′-3′-cyclic phosphate version of the fragment (the 3′-phosphate version represents the major signal). Theoretical monoisotopic mass of an anticodon fragment containing the various modifications: chm5U, 2928.38; (S)-mchm5U, 2942.40; mcm5U, 2926.40; ncm5U, 2911.40; cm5U, 2912.39.
To study the roles of AtALKBH8 and AtTRM9 in wobble uridine modification specifically in 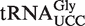 , this isoacceptor was isolated from attrm9 and atalkbh8-1 plants. The purified isoacceptor was then digested with RNase T1, which specifically cleaves 3′ to G residues, and the resulting fragments were analysed by matrix assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry. Based on genomic tRNA sequence, the anticodon triplet of
, this isoacceptor was isolated from attrm9 and atalkbh8-1 plants. The purified isoacceptor was then digested with RNase T1, which specifically cleaves 3′ to G residues, and the resulting fragments were analysed by matrix assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry. Based on genomic tRNA sequence, the anticodon triplet of 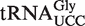 is expected to be present in a 9-mer fragment of sequence CCUUCCAAG (the putatively modified wobble uridine is underlined). Indeed, fragments corresponding to mchm5U and chm5U modifications were detected in
is expected to be present in a 9-mer fragment of sequence CCUUCCAAG (the putatively modified wobble uridine is underlined). Indeed, fragments corresponding to mchm5U and chm5U modifications were detected in 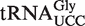 from WT plants (Figure 4D, upper panel); a modification pattern very similar to that observed for mammalian
from WT plants (Figure 4D, upper panel); a modification pattern very similar to that observed for mammalian 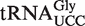 (12). It should be noted that, due to isotope distributions, each fragment gives rise to a cluster of peaks of ∼1 Da spacing, where the leftmost peak corresponds to the monoisotopic mass.
(12). It should be noted that, due to isotope distributions, each fragment gives rise to a cluster of peaks of ∼1 Da spacing, where the leftmost peak corresponds to the monoisotopic mass.
When 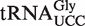 from atalkbh8-1 plants was analysed, the peak cluster corresponding to mchm5U was absent, but instead a cluster corresponding to mcm5U was observed (Figure 4D, middle panel), indicating that the (S)-mchm5U modification on
from atalkbh8-1 plants was analysed, the peak cluster corresponding to mchm5U was absent, but instead a cluster corresponding to mcm5U was observed (Figure 4D, middle panel), indicating that the (S)-mchm5U modification on 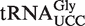 is generated by AtALKBH8-mediated hydroxylation of mcm5U.
is generated by AtALKBH8-mediated hydroxylation of mcm5U. 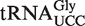 derived from attrm9 plants gave rise to a peak cluster that was shifted 31 Da relative to the mchm5U-containing cluster. Here, the first (leftmost) peak corresponds to the monoisotopic mass of ncm5U, whereas the second peak corresponds to the monoisotopic mass of cm5U (Figure 4D, lower panel). Since the first peak is substantially smaller than the second one, and thereby deviates from the expected isotope pattern of a single oligonucleotide, it is highly likely that the peak cluster represents both ncm5U and cm5U. This is also supported by our analysis of total tRNA from attrm9 plants, which showed an accumulation of cm5U (Figure 3), as well as by our previous study from mice lacking ALKBH8-MT (the functional equivalent of Trm9), which revealed the presence of both cm5U and ncm5U in
derived from attrm9 plants gave rise to a peak cluster that was shifted 31 Da relative to the mchm5U-containing cluster. Here, the first (leftmost) peak corresponds to the monoisotopic mass of ncm5U, whereas the second peak corresponds to the monoisotopic mass of cm5U (Figure 4D, lower panel). Since the first peak is substantially smaller than the second one, and thereby deviates from the expected isotope pattern of a single oligonucleotide, it is highly likely that the peak cluster represents both ncm5U and cm5U. This is also supported by our analysis of total tRNA from attrm9 plants, which showed an accumulation of cm5U (Figure 3), as well as by our previous study from mice lacking ALKBH8-MT (the functional equivalent of Trm9), which revealed the presence of both cm5U and ncm5U in  (12). The already high levels of ncm5U in total tRNA, due to the natural occurrence of this modification in many different tRNA isoacceptors, may explain why the partial modification of attrm9
(12). The already high levels of ncm5U in total tRNA, due to the natural occurrence of this modification in many different tRNA isoacceptors, may explain why the partial modification of attrm9 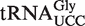 by ncm5U was not manifested as a significant increase in the ncm5U level in total attrm9 tRNA (Figure 3). Taken together, the data presented above show that
by ncm5U was not manifested as a significant increase in the ncm5U level in total attrm9 tRNA (Figure 3). Taken together, the data presented above show that 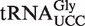 in Arabidopsis harbors an (S)-mchm5U modification in the wobble position, which depends on both AtALKBH8 and AtTRM9 for its biogenesis.
in Arabidopsis harbors an (S)-mchm5U modification in the wobble position, which depends on both AtALKBH8 and AtTRM9 for its biogenesis.
The activity of AtTRM9 depends on co-expressed AtTRM112a or AtTRM112b
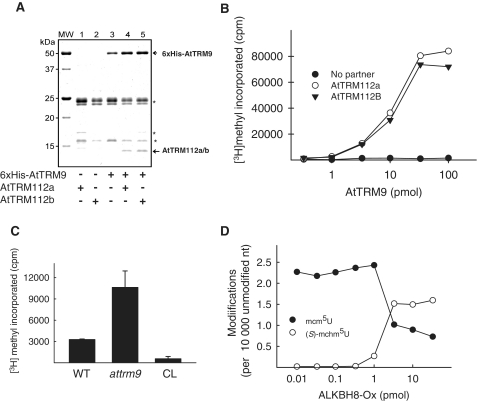
In mammals, the MT domain of ALKBH8 requires a small accessory protein, TRM112, to form a functional enzymatic complex (20), and it has also been shown that a similar complex is formed between yeast Trm112 and Trm9 (33). Recently, the gene At1g22270 was shown to encode a functional Arabidopsis homologue of TRM112, denoted SMALL ORGAN 2 (SMO2), since smo2 mutants show retarded organ growth due to inhibition of G2-M phase progression during organogenesis (34). In addition to SMO2, Arabidopsis has another similar protein, SMO2L (small organ 2-like), encoded by the gene At1g78190, and this protein has 78% sequence identity to SMO2. To better indicate their similarity to yeast Trm112, and to comply with the nomenclature from the NCBI protein database, we have chosen to denote these proteins AtTRM112a (SMO2) and AtTRM112b (SMO2L). Since both Trm9 and Trm112 are conserved between mammals, yeast and plants, we considered it likely that AtTRM9 may, similarly, form a functional complex with AtTRM112a or AtTRM112b. To test this, 6xHis-tagged AtTRM9 (6xHis-AtTRM9) was co-expressed in E. coli together with untagged AtTRM112a or AtTRM112b, and then affinity purified on TALON beads, to which 6xHis tagged proteins bind strongly. As shown in Figure 5A, a band of ∼14 kDa, corresponding in size to AtTRM112a or AtTRM112b, was present on the gel after co-expression of AtTRM112a or AtTRM112b with 6xHis-AtTRM9, but not after expression of either protein alone. These results suggested that untagged AtTRM112a/b co-purified with 6xHis-AtTRM9, and that these proteins form an enzymatic complex.
Figure 5.
Activity of recombinant enzymes on WT and mutant tRNA. (A) Co-expression of untagged AtTRM112a and AtTRM112b with 6xHis-AtTRM9. Recombinant proteins were expressed in E. coli, purified by affinity chromatography on TALON beads, and then analysed by SDS–PAGE. Protein bands corresponding to AtTRM112a or AtTRM112b and AtTRM9 are indicated by arrows. Non-specific bands representing endogenous E. coli proteins are indicated by asterisks. (B) Dependence of the MT activity of AtTRM9 on co-expressed AtTRM112a or AtTRM112b. The activity was assayed on 10 µg saponified yeast tRNA using different amounts of recombinant enzyme. (C) AtTRM9 methyltransferase activity on tRNA from attrm9 plants. 20 pmol AtTRM9/AtTRM112a complex was incubated with 5 µg of total tRNA from attrm9 or WT plants, or from calf liver (CL). (D) Cross-species activity of human ALKBH8-Ox on atalkbh8 tRNA. Different amounts of human ALKBH8-Ox was incubated with total tRNA from atalkbh8 plants. The tRNA was then digested to nucleosides, and the level of mcm5U (closed circles) and (S)-mchm5U (open circles) was measured by LC–MS/MS analysis.
The altered wobble uridine modification pattern in the AtTRM9-deficient plants indicated that this enzyme may have a function equivalent to that of yeast Trm9 and the MT moiety of mammalian ALKBH8. To investigate the putative enzymatic activity of AtTRM9 and to address the functional significance of the suggested interaction between AtTRM9 and AtTRM112a/b, purified 6xHis-AtTRM9, in the absence or presence of co-expressed AtTRM112a/AtTRM112b, was assayed for tRNA methyltransferase activity. We have previously observed that yeast tRNA, which is easily obtained in large quantities, is a good substrate for the MT activity of human ALKBH8 (20), and we therefore, considered it likely that this also may be the case for AtTRM9. To generate a substrate for the putative AtTrm9 MT activity, the yeast tRNA was subjected to mild alkaline hydrolysis referred to as saponification. This treatment causes hydrolysis of the methyl ester bond in mcm5U and mcm5s2U, resulting in their conversion to the putative AtTRM9 substrates cm5U and 5-carboxymethyl-2-thiouridine (cm5s2U), respectively. When saponified yeast tRNA was incubated with the radiolabelled methyl donor S-adenosyl-l-[methyl-3H]methionine and with 6xHis-AtTRM9 that had been co-expressed with AtTRM112a or AtTRM112b, incorporation of radioactivity into tRNA was observed, supporting the notion that AtTRM9 is a functional Trm9 homolog (Figure 5B). Importantly, no incorporation of radioactivity was observed when AtTRM9 had been expressed and purified in the absence of either AtTRM112a or AtTRM112b, suggesting that interaction with AtTRM112a or AtTRM112b is required for AtTRM9 to exert its MT activity (Figure 5B).
The accumulation of cm5U in tRNA from the attrm9 mutant plants suggested that this tRNA may function as a MT-substrate even without saponification. Indeed, non-saponified tRNA from attrm9 plants gave a substantially higher level of AtTRM9/AtTRM112a-mediated methylation, compared to tRNA from WT plants (Figure 5C), further supporting the notion that AtTRM9 catalyses the methylation of cm5U into mcm5U.
Human ALKBH8-Ox hydroxylates mcm5U to (S)-mchm5U in tRNA obtained from AtALKBH8 knock-out plants
To investigate the enzymatic activity of AtALKBH8 in vitro, we attempted to express and purify the recombinant protein from E. coli, but we were unable to recover soluble protein. Instead, we addressed whether the mcm5U-modified tRNA found in the atalkbh8 mutant plant can function as a substrate for the ALKBH8 oxygenase activity, by treating this tRNA with the dioxygenase domain (amino acids 1–354) of human ALKBH8 (ALKBH8-Ox) and subsequently analyzed the modification status by LC–MS/MS. With increasing concentrations of enzyme, we observed an increase of (S)-mchm5U levels accompanied by a simultaneous decrease in mcm5U (Figure 5D). However, a fraction of the mcm5U-modified tRNA appeared refractory to HsALKBH8-Ox-mediated hydroxylation, probably reflecting mcm5U-containing isoacceptors that are not subject to further hydroxylation. These data further support that mcm5U is the precursor of (S)-mchm5U also in plants, and that AtALKBH8 is the enzyme catalyzing the hydroxylation of mcm5U to (S)-mchm5U.
DISCUSSION
Wobble uridine modifications in eukaryotic cytoplasmic tRNAs have so far mainly been studied in yeast and mammals, where a number of such modifications have been identified, along with several enzymes involved in their biogenesis. In this study, we have investigated the wobble uridine modification pattern in WT Arabidopsis plants and in mutants defective in enzymes putatively involved in formation of modified wobble uridines. These experiments have been complemented by enzymatic assays with recombinant enzymes, and our results establish a role for four yet uncharacterized Arabidopsis proteins, AtALKBH8, AtTRM9, AtTRM112a and AtTRM112b in the formation of modified wobble uridines in tRNA.
In mammals, mcm5U and its 2′-O-ribose methylated derivative, mcm5Um, are found in the wobble position of the selenocysteine specific tRNA (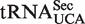 ), which is responsible for the incorporation of selenocysteine into selenoproteins. The abundance of the mcm5Um-modified isoform of
), which is responsible for the incorporation of selenocysteine into selenoproteins. The abundance of the mcm5Um-modified isoform of 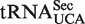 , relative to its mcm5U-modified counterpart, has been shown to increase in response to elevated selenium levels, and a concomitant increase in the expression of certain stress-linked selenoproteins was observed (35,36). These findings, together with studies of mice that lack the mcm5Um modification, suggest that mcm5Um promotes the expression of a subset of the selenoproteome and that the levels of this isoacceptor may be subject to active regulation (37,38). Thus, ribose methylation of mcm5U in
, relative to its mcm5U-modified counterpart, has been shown to increase in response to elevated selenium levels, and a concomitant increase in the expression of certain stress-linked selenoproteins was observed (35,36). These findings, together with studies of mice that lack the mcm5Um modification, suggest that mcm5Um promotes the expression of a subset of the selenoproteome and that the levels of this isoacceptor may be subject to active regulation (37,38). Thus, ribose methylation of mcm5U in 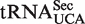 has been proposed to be a highly specialized event, and the selenocysteine incorporation factor SECp43 was launched as a candidate specialized methyltransferase, but this has not been confirmed experimentally (39). In WT Arabidopsis, we detected small amounts of the mcm5U modification, but the levels were elevated ∼10-fold in atalkbh8 plants. Moreover, in the atalkbh8 mutant, we also observed substantial amounts of of mcm5Um, which was virtually absent from WT plants, probably due to ribose methylation of accumulated mcm5U by a non-specialized enzyme not normally dedicated to this purpose. An obvious candidate enzyme is the Arabidopsis homologue of S. cerevisiae Trm7p, which is responsible for ribose methylation of several different tRNAs in Positions 32 and/or 34 (40). Based on these results, the possibility should be considered that the mcm5Um found in
has been proposed to be a highly specialized event, and the selenocysteine incorporation factor SECp43 was launched as a candidate specialized methyltransferase, but this has not been confirmed experimentally (39). In WT Arabidopsis, we detected small amounts of the mcm5U modification, but the levels were elevated ∼10-fold in atalkbh8 plants. Moreover, in the atalkbh8 mutant, we also observed substantial amounts of of mcm5Um, which was virtually absent from WT plants, probably due to ribose methylation of accumulated mcm5U by a non-specialized enzyme not normally dedicated to this purpose. An obvious candidate enzyme is the Arabidopsis homologue of S. cerevisiae Trm7p, which is responsible for ribose methylation of several different tRNAs in Positions 32 and/or 34 (40). Based on these results, the possibility should be considered that the mcm5Um found in 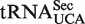 also is a result of ribose methylation by a non-specialized enzyme, e.g. a Trm7p-homologue, and that the biological significance of this ribose methylation could be smaller than previously thought.
also is a result of ribose methylation by a non-specialized enzyme, e.g. a Trm7p-homologue, and that the biological significance of this ribose methylation could be smaller than previously thought.
When the yeast Trm9 function is inactivated, tRNAs that normally contain mcm5U or mcm5s2U instead carry a ncm5U modification. When the corresponding ALKBH8 methyltransferase function is inactivated in mammals, a similar accumulation of ncm5U/ncm5s2U was observed, as well as an accumulation of the mcm5U precursor cm5U. In the attrm9 plants, the tRNAs that normally contain mcm5U or its derivatives primarily seem to accumulate cm5U, although some ncm5U also appeared to be present in  . Thus, the accumulation of cm5U and/or ncm5U in tRNAs that normally carry a mcm5U modification seems be a general consequence of Trm9 inactivation, but the relative amounts of the accumulated cm5U versus ncm5U appear to vary considerably between different organisms.
. Thus, the accumulation of cm5U and/or ncm5U in tRNAs that normally carry a mcm5U modification seems be a general consequence of Trm9 inactivation, but the relative amounts of the accumulated cm5U versus ncm5U appear to vary considerably between different organisms.
Upon inactivation of the AtALKBH8 and AtTRM9 genes, we could see an alteration in the modification pattern that corresponded well with previous observations in transgenic mice, where only the oxygenase-function [KI(Ox+)] or the MT-function [KI(MT+)] of murine ALKBH8 was expressed (12). Total tRNA from the KI(Ox+) mice showed a similar modification pattern as tRNA from attrm9 mutant plants, as it completely lacks the mcm5U modification while the levels of cm5U were strongly increased. The KI(MT+) tRNA showed a modification pattern similar to the atalkbh8 plants, where (S)-mchm5U was absent, and mcm5U was strongly increased. This clearly indicates that AtTRM9 and AtALKBH8 together perform the functions that in animals are carried out by the bifunctional ALKBH8 enzyme. The existence of bifunctional tRNA modification enzymes is not unprecedented, and one may speculate that the catalytic efficiency is improved when the product of the MT reaction can be directly channeled to the oxygenase.
Yeast Trm9 and the mammalian ALKBH8 methyltransferase both depend on the small accessory protein Trm112/TRM112 for activity (20,41). Here, we show that co-expression of AtTRM112a or AtTRM112b results in a protein of the expected size co-purifying with 6xHis-AtTRM9, and we show that the in vitro activity of 6x-His-AtTRM9 requires the co-expression with either AtTrm112a or AtTrm112b. Furthermore, tRNA isolated from attrm9 plants was a better substrate for AtTRM9, as compared to WT tRNA. Taken together, these results strongly indicate that AtTRM9 has a similar function as its orthologues from yeast and mammals. The attrm112a (smo2) mutant of Arabidopsis shows a severely reduced organ growth, but this phenotype could not be reversed by expression of AtTRM112b (SMO2-L) from the AtTRM112a promoter. This may suggest that the in vivo functions of TRM112a and TRM112b are different, and that the organ growth defects seen in the smo2 mutant are not related to the AtTRM9 MT activity.
Studies from yeast have suggested a role for Trm9 in the protection against DNA damage (42). Trm9-deficient cells are sensitive towards the methylating agent MMS; a phenotype that was explained by impaired translation of the DNA damage response genes RNR1 and RNR3 (42). These genes are particularly rich in the Arg codon AGA, which is decoded by a tRNA that depends on Trm9 for proper modification. We could, however, not observe any significant difference in the sensitivity towards MMS between the attrm9 mutant and the WT plant (data not shown). This may indicate that yeast Trm9 and AtTRM9 are important for expression of different subsets of genes, and that AtTRM9, as opposed to its yeast counterpart, is not a component in the DNA damage response machinery.
Two different forms of AtALKBH8 were identified on the mRNA level, corresponding to proteins of 344 and 431 amino acids, where the longer form contains an additional N-terminal extension. Interestingly, the major part of this extension is derived from the transposon Tag2, indicating that the long form, which is not present in other plants, has arisen through a transposition event. It has been frequently observed during genome evolution that transposon insertions generate new coding sequence or provide regulatory elements for gene expression (43). The addition of the transposon-derived extension to the AtALKBH8 coding sequence may have influenced the gene function in several conceivable ways, e.g. by adding a targeting signal for a non-cytosolic localization, by expanding the substrate specificity of the protein to allow hydroxylation of additional tRNAs, or simply by improving protein expression or increasing protein stability.
The present work represents, together with the recent characterization of mammalian ALKBH8, the only functional study of an ALKBH8 dioxygenase so far. Proteins with similarity to both the RRM and AlkB-domains of ALKBH8 appear to be present in all multicellular eukaryotes, as well as in some unicellular fungi, parasites and algae. The observation that the AtALKBH8 oxygenase, as its mammalian counterpart, is responsible for the final hydroxylation reaction leading to (S)-mchm5U formation in 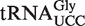 indicates that ALKBH8 orthologues catalyse this reaction in a wide range of organisms. Interestingly, AlkB homologues from some bacteria display a high degree of sequence similarity to the dioxygenase domain of eukaryotic ALKBH8, but they lack the RRM moiety (44,45). Several of these bacteria are plant pathogens, which may suggest that they have acquired the ALKBH8-like gene from the host during infection. Since the mcm5U modification appears to be absent from bacteria, these proteins probably have an enzymatic activity different from that of eukaryotic ALKBH8, and it will be a challenge to unravel their function in future studies.
indicates that ALKBH8 orthologues catalyse this reaction in a wide range of organisms. Interestingly, AlkB homologues from some bacteria display a high degree of sequence similarity to the dioxygenase domain of eukaryotic ALKBH8, but they lack the RRM moiety (44,45). Several of these bacteria are plant pathogens, which may suggest that they have acquired the ALKBH8-like gene from the host during infection. Since the mcm5U modification appears to be absent from bacteria, these proteins probably have an enzymatic activity different from that of eukaryotic ALKBH8, and it will be a challenge to unravel their function in future studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
FRIBIO and FUGE programs in the Research Council of Norway. Funding for open access charge: The Research Council of Norway.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We appreciate the contribution of Marius Ulleland to the initial phase of this project. We are grateful to Anders Bekkelund, Solveig H. Engebretsen, Roy Falleth, and Mari Kjos for excellent technical assistance. We thank Angela Ho for a critical reading of the manuscript. We are grateful to Grazyna Leszczynska Andrzej Malkiewicz for providing nucleoside standards for the LC-MS/MS analysis.
REFERENCES
- 1.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 3.Björk GR, Wikström PM, Byström AS. Prevention of Translational Frameshifting by the Modified Nucleoside 1-Methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 4.Johansson MJ, Esberg A, Huang B, Björk GR, Byström AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell. Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasvall SJ, Chen P, Björk GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esberg A, Huang B, Johansson MJ, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 8.Huang B, Johansson MJ, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amberg R, Urban C, Reuner B, Scharff P, Pomerantz SC, McCloskey JA, Gross HJ. Editing does not exist for mammalian selenocysteine tRNAs. Nucleic Acids Res. 1993;21:5583–5588. doi: 10.1093/nar/21.24.5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Born E, Vagbø CB, Songe-Møller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat. Commun. 2011;2:172. doi: 10.1038/ncomms1173. [DOI] [PubMed] [Google Scholar]
- 13.Aas PA, Otterlei M, Falnes PØ, Vagbø CB, Skorpen F, Akbari M, Sundheim O, Bjoras M, Slupphaug G, Seeberg E, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 14.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl Acad. Sci. USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 16.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 19.Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell. Biol. 2010;30:2449–2459. doi: 10.1128/MCB.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Songe-Møller L, van den Born E, Leihne V, Vagbø CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PØ, Klungland A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 2010;30:1814–1827. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew.Chem. Int. Ed. Engl. 2010;49:8885–8888. doi: 10.1002/anie.201001242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golovko A, Sitbon F, Tillberg E, Nicander B. Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana. Plant Mol. Biol. 2002;49:161–169. doi: 10.1023/a:1014958816241. [DOI] [PubMed] [Google Scholar]
- 23.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 24.Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jager G, Gong Z, Byström AS, Schaffrath R, Breunig KD. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010;76:1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P, Jager G, Zheng B. Transfer RNA modifications and genes for modifying enzymes in Arabidopsis thaliana. BMC Plant Biol. 2010;10:201. doi: 10.1186/1471-2229-10-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murashige T, Skoog F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- 29.Henk AD, Warren RF, Innes RW. A new Ac-like transposon of Arabidopsis is associated with a deletion of the RPS5 disease resistance gene. Genetics. 1999;151:1581–1589. doi: 10.1093/genetics/151.4.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen IP, Mannuss A, Orel N, Heitzeberg F, Puchta H. A homolog of ScRAD5 is involved in DNA repair and homologous recombination in Arabidopsis. Plant Physiol. 2008;146:1786–1796. doi: 10.1104/pp.108.116806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2007;104:18836–18841. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami M, Takemura S, Kondo T, Fukami T, Goto T. Chemical structure of a new modified nucleoside located in the anticodon of Bombyx mori glycine tRNA2. J. Biochem. 1988;104:108–111. doi: 10.1093/oxfordjournals.jbchem.a122403. [DOI] [PubMed] [Google Scholar]
- 33.Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z, Qin Z, Wang M, Xu C, Feng G, Liu J, Meng Z, Hu Y. The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J. 2010;61:600–610. doi: 10.1111/j.1365-313X.2009.04085.x. [DOI] [PubMed] [Google Scholar]
- 35.Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA, et al. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec) J. Biol. Chem. 1993;268:14215–14223. [PubMed] [Google Scholar]
- 36.Jameson RR, Diamond AM. A regulatory role for Sec tRNA[Ser]Sec in selenoprotein synthesis. RNA. 2004;10:1142–1152. doi: 10.1261/rna.7370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J. Biol. Chem. 2005;280:5542–5548. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 38.Moustafa ME, Carlson BA, El-Saadani MA, Kryukov GV, Sun QA, Harney JW, Hill KE, Combs GF, Feigenbaum L, Mansur DB, et al. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol. Cell. Biol. 2001;21:3840–3852. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatfield DL, Carlson BA, Xu XM, Mix H, Gladyshev VN. Selenocysteine incorporation machinery and the role of selenoproteins in development and health. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:97–142. doi: 10.1016/S0079-6603(06)81003-2. [DOI] [PubMed] [Google Scholar]
- 40.Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002;21:1811–1820. doi: 10.1093/emboj/21.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Studte P, Zink S, Jablonowski D, Bar C, von der HT, Tuite MF, Schaffrath R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- 42.Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falnes PØ, Rognes T. DNA repair by bacterial AlkB proteins. Res. Microbiol. 2003;154:531–538. doi: 10.1016/S0923-2508(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 45.van den Born E, Bekkelund A, Moen MN, Omelchenko MV, Klungland A, Falnes PØ. Bioinformatics and functional analysis define four distinct groups of AlkB DNA-dioxygenases in bacteria. Nucleic Acids Res. 2009;21:7124–7136. doi: 10.1093/nar/gkp774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.