Abstract
Transcriptional control is dependent on a vast network of epigenetic modifications. One epigenetic mark of particular interest is tri-methylation of lysine 27 on histone H3 (H3K27me3), which is catalysed and maintained by Polycomb Repressive Complex 2 (PRC2). Although this histone mark is studied widely, the precise relationship between its local pattern of enrichment and regulation of gene expression is currently unclear. We have used ChIP-seq to generate genome-wide maps of H3K27me3 enrichment, and have identified three enrichment profiles with distinct regulatory consequences. First, a broad domain of H3K27me3 enrichment across the body of genes corresponds to the canonical view of H3K27me3 as inhibitory to transcription. Second, a peak of enrichment around the transcription start site (TSS) is commonly associated with ‘bivalent’ genes, where H3K4me3 also marks the TSS. Finally and most surprisingly, we identified an enrichment profile with a peak in the promoter of genes that is associated with active transcription. Genes with each of these three profiles were found in different proportions in each of the cell types studied. The data analysis techniques developed here will be useful for the identification of common enrichment profiles for other histone modifications that have important consequences for transcriptional regulation.
INTRODUCTION
Transcription is an intricate process that is regulated by both genetic and epigenetic factors. Epigenetic marks, such as DNA methylation and the modification of histone tails, play an important role in regulating transcription. These marks are inherently plastic and are redistributed during development to preserve cell fate decisions (1). Because of their widespread influence on gene expression, it is not surprising that epigenetic marks are disrupted in disease. Understanding the role and influence of epigenetic marks is at the heart of understanding transcriptional regulation across cell types and in disease states.
Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) is a powerful technique that can be used to map transcription complexes and epigenetic modifications throughout the genome (2,3). Numerous epigenetic modifications have been mapped in a variety of cell types and in different developmental contexts [e.g. see (4,5)]. Collectively, these studies have shown that the chromatin landscape is highly complex and that the precise regulation of gene expression is dependent on the coordinated interaction of many different modifications (4). Looking at the distribution of many epigenetic modifications has revealed common epigenetic profiles that are repeated throughout the genome, which have distinct functional consequences for gene expression (6,7). As a great assortment of chromatin marks have been identified, there are an enormous number of possible combinations of marks and marking patterns at any given locus (8).
A common approach in analyzing ChIP-seq data is to classify genes as ‘marked’ based on the detection of a peak of enrichment at any position within a gene. This approach disregards variation in the pattern of marking across a gene, i.e. the enrichment profile. Some recent studies have investigated the relationship between enrichment profile and regulatory function. One of the most striking examples is the finding that H3K36me3 is enriched specifically on the exons of actively transcribed genes, revealing cross talk between chromatin and the splicing machinery (9). An analysis of H3K4me2 in CD4+ T cells showed that marking in the body of genes is associated with higher expression of tissue-specific genes, compared with marking at the transcription start site (TSS) (10). Therefore, it is not only important to consider the presence of different combinations of epigenetic marks, but also that modifications occur with distinct profiles on genes, and these can have important consequences for transcriptional regulation (7,8,10,11).
Polycomb Repressive Complex 2 (PRC2) is a histone methyl-transferase that catalyses tri-methylation of Histone 3 at Lysine 27 (H3K27me3). H3K27me3 is associated with the repression of transcription in a cell type-specific manner (5,12–17). PRC2 is an important regulator in many cell types, both embryonic and adult, including embryonic stem (ES) cells and neural, epidermal and haematopoietic stem cells (18–20). In ES cells, PRC2 cooperates with Oct4, SOX2 and NANOG to silence lineage-specific genes and to preserve the pluripotent state (12,13). H3K27me3 occurs together with the activating mark H3K4me3 in regions referred to as bivalent domains (21,22). Bivalent domains consist of nucleosomes containing both modifications simultaneously, although localization on neighbouring nucleosomes can also occur (22–24). These domains often occur in the promoter of lineage specific transcription factors, and are thought to keep the genes poised to respond to developmental cues (25,26). As well as being found on individual genes, H3K27me3 can occur in large domains that spread over hundreds of kilobases (17,27). In some cases, these extended domains are associated with gene families, including the Hox gene clusters. The mechanisms that govern the targeting of PRC2 and the distribution of H3K27me3 remain poorly understood.
In this work, we investigated the distribution of H3K27me3, both within genes and on adjacent regulatory elements. To this end, we developed new approaches to visualize ChIP-seq data and to classify genes by their H3K27me3 profile. We found differences both in the placement of H3K27me3 and the spread of the signal; H3K27me3 can either be deposited in distinct peaks over a small number of nucleosomes, or can spread to blanket an entire locus. We identified three distinct H3K27me3 enrichment profiles that were strongly correlated with transcriptional activity. These distinct profiles were observed in four different cell types. Although H3K27me3 was generally found on repressed genes, we identified a set of genes that carry H3K27me3 in their promoters and yet remain highly expressed. Finally, we investigated the relationship between patterns of H3K27me3, H3K4me3 and H3K36me3, which led to the identification of an H3K27me3 profile that is enriched for bivalent genes.
MATERIALS AND METHODS
Cell culture
Mouse C57BL/6 Bruce 4 (B4) ES cells (mECSs) were adapted to culture without helper fibroblasts and expanded using standard techniques. mESCs were cultured in DMEM (Invitrogen) with 15% (v/v) FCS (batch tested for ES cell culture), 100 μg/ml streptomycin, 100 IU/ml penicillin, β-mercaptoethanol (Sigma), non-essential amino acids, Glutamax (Gibco) and recombinant mLIF 103 IU/ml (Millipore). To prepare chromatin for ChIP, mESCs were grown to ∼80% confluence and fixed with buffered formaldehyde (1%) for 10 min at room temperature.
G1ME cells were grown in alpha-MEM (Invitrogen) supplemented with 20% (v/v) FCS, 100 μg/ml streptomycin, 100 IU/ml penicillin and 70 ng/ml recombinant thrombopoietin (28). For ChIP studies, G1ME cells were grown at 5 × 105 cells/ml and were fixed with 1% formaldehyde for 20 min at room temperature.
Chromatin immunoprecipitation and antibodies
Chromatin was prepared from fixed mouse ES cells and G1ME cells and was sonicated with a Bandelin sonicator (30% amplitude, 15–25 30-s bursts with a 2 min reprieve) to produce fragments from 200 to 1000 bp, with peak signal between 200 and 500 bp. Approximately 2 × 107 cell equivalents were used for each immunoprecipitation. Of the sample, 1.7% was removed for use as an input control. ChIP was performed as described previously (Lee, 2006), using antibodies towards H3K27Me3 (07-449, Millipore), phosphorylated RNA polymerase C-terminal domain (ab5131, Abcam) or a control rabbit IgG (ab46540).
Sequencing
Samples were processed for sequencing at the Australian Genome Research Facility. ChIP-enriched DNA fragments were size selected on an agarose gel to enrich for fragments 200 bp in size, linkers were then added and the library was amplified using polymerase chain reaction (PCR). Each library was loaded on to an individual lane of a Genome Analyser II next-generation sequencing platform (Illumina). All samples were processed using the standard 36 bp single-end protocol, except for the G1ME RNApol-II library that was run at a later date using a 75 bp single-end protocol. Total read counts and unique read counts are presented in Supplementary Table S2.
In addition to the in house ChIP-seq data, previously published data sets were sourced from the gene expression omnibus (GEO). Raw sequencing reads were extracted from GSE12241: mouse embryonic fibroblasts (MEF) H3K4me3 (GSM307608), MEF H3K27me3 (GSM307609), MEF H3K36me3 (GSM307610), MEF whole-cell extract (Input, GSM307612), ES H3K4me3 (GSM307618), ES H3K27me3 (GSM307619), ES H3K36me3 (GSM307620), ES RNApol-II (GSM307623), ES whole-cell extract (Input, GSM307625), NP H3K4me3 (GSM307613), Neural Progenitor (NP) H3K27me3 (GSM307614), NP H3K36me3 (GSM307615) and NP whole-cell extract (Input, GSM307617). The publicly available ES H3K27me3, RNApol-II and Input data were compared with our in house data. Where individual samples were run on multiple lanes the data was pooled for analysis. All data (both in-house and public) was processed in the same way.
ChIP-QPCR
ChIP-QPCR was performed using SYBR Green I master mix (Roche). Primers for β-actin, HoxA11 and Oct4 were described previously (29,30). Primers that targeted the promoter, TSS and gene body regions of PDE8A, SCUBE2, DNMT3A, FNIP1 and RTN4 were also used (see Supplementary Table S1 for primer sequences). Standard curves were generated for each amplicon using purified mouse genomic DNA (Clontech). Absolute quantification was performed and enrichment expressed as a fraction of the whole-cell extract control. Each experiment was performed in triplicate.
Microarrays
Gene expression was assessed in G1ME cells with GeneChip Mouse Genome 430 2.0 microarrays (Affymetrix). For ES, MEF and NP cells, we used expression data from the same microarray platform (GEO GSE8024) (5). There were three technical replicates for ES and NP cells and two for G1ME and MEFs. All microarrays were background corrected and quantile normalized using the gcrma bioconductor R package (31). In order to integrate the expression data with our mapped sequencing data, we first converted all Affymetrix probe IDs into ENSEMBL gene identifiers. For genes with multiple probe sets, the average probe set value was taken to represent the expression level of the gene.
Mapping and processing
All reads were mapped to version 9 of the mouse reference genome using bowtie version 0.11.3 (32) with the default settings and output to SAM format (33). Any reads that appeared on the same strand at the same location more than five times were discarded to remove PCR artefacts that appear as large spikes in the data. While the value of 5 is an arbitrary choice, other values (such as 1 or 10) were tried and did not alter our overall conclusions. Mapped sequencing data and processed microarray data can be downloaded from the GEO under accession number GSE27970.
Identification of marked genes
We used version 58 of the ENSEMBL mouse reference and defined a gene as being the region between the 5′- and 3′-end of the longest ENSEMBL transcript, plus a 3 kb promoter region. In order to identify genes marked by a particular protein complex (histone modification or RNApol-II in our case), we used two separate methods to capture different sized domains of marking. MACS version 1.4.0alpha2 (34) was used with the default parameters and the appropriate input as control to identify regions of the genome that were significantly enriched for the relevant protein complex. A gene was called as bound if one of the MACS identified peaks overlapped an ENSEMBL gene. We supplemented this test with a targeted Poisson test of broad enrichment. Specifically, we counted the number of reads within each gene, in each of the different experimental conditions. We then used a Poisson exact test (35) to obtain a P-value for each gene being higher in the ChIP than the Input control. P-values were multiple hypothesis testing corrected using Benjamini Hochberg FDR (36). Furthermore, we calculated the log ‘fold-change’ of the gene as log2 (Xc/Xi), where X is the number of reads mapping to the gene in the ChIP sample (c) or the input sample (i). Any gene with an FDR < 0.001 and a log2 fold change >1 (2-fold) was called as marked. This Poisson test allowed us to identify genes with significantly broad marking, which was missed by the MACS test. However, it should be noted that the Poisson test suffers from gene length bias in a similar way to RNA-seq data (37). Our final list of marked genes was taken to be the union of MACS and Poisson identified genes (see Table 1 for a summary).
Table 1.
Number of genes that are significantly enriched for the specified mark in different cell types using the MACS test, Poisson test or both
| Cell type | ChIP type | Source | MACS only | Poisson only | Called in both | All bound |
|---|---|---|---|---|---|---|
| ES | H3K27me3 | WEHI | 2298 | 660 | 2613 | 5571 |
| RNApol II | WEHI | 8055 | 155 | 6118 | 14328 | |
| H3K4me3 | Mikkelsen | 6436 | 536 | 9705 | 16677 | |
| H3K36me3 | Mikkelsen | 2639 | 533 | 6088 | 9260 | |
| G1ME | H3K27me3 | WEHI | 1574 | 1969 | 2840 | 6386 |
| RNApol II | WEHI | 6949 | 170 | 5411 | 12530 | |
| MEF | H3K27me3 | Mikkelsen | 249 | 2939 | 47 | 3235 |
| H3K4me3 | Mikkelsen | 5863 | 1754 | 6192 | 13809 | |
| H3K36me3 | Mikkelsen | 3008 | 606 | 6973 | 10587 | |
| NP | H3K27me3 | Mikkelsen | 856 | 377 | 480 | 1713 |
| H3K4me3 | Mikkelsen | 8176 | 245 | 3447 | 11868 | |
| H3K36me3 | Mikkelsen | 2866 | 532 | 1818 | 5216 |
The numbers in the ‘MACS only’ and ‘Poisson only’ columns are the number of genes found as marked with that method but not the other, i.e. they are exclusive to MACS or Poisson.
Normalization and visualization
For each sample, a genome-wide coverage track was generated where the value at each base represents the number of 5′-ends of reads that overlap that base, normalized by the total library size. To facilitate the investigation of the ChIP-seq data, we used two key visualization techniques; the TSS plot and the average scaled enrichment (ASE) plot. Both visualize the ChIP profile of a group of genes, either around the TSS or across the length of the gene respectively. The TSS plot is created by taking a fixed length (in bp) around the TSS, with all genes in the same orientation (5′ to 3′ open reading frame), and then averaging the signal across all the selected genes.
To create the ASE plots, we first selected a resolution or sampling frequency. This is the number of evenly spaced points that will be taken to represent a gene. For all plots in this manuscript, a sampling frequency of 1000 points/gene was used. As there are genes that exceed the sampling frequency, the raw coverage track was smoothed by taking a sliding window across the genome and averaging the number of reads within each bin. The window size is set to the maximum gene size divided by the gene sampling frequency to ensure data at every base pair of every gene was used in creating the smoothed signal. Throughout this article, the genome was smoothed using a sliding window of size 2258 bp when creating ASE plots. Finally, the genes were arranged from 5′ to 3′ and are averaged at each sampling point.
Classification scheme
The ChIP-seq profile of each marked gene was tested to see if it met criteria sufficient for it to be called a member of the TSS, Promoter or Broad class. Each gene was broken into three regions (see Supplementary Figure S1A): (i) The TSS region, which was 1100 bp long and was anchored around the TSS, and includes 100 bp upstream and 1000 bp downstream of the TSS; (ii) The promoter region, which extended from 100 bp upstream to 3000 bp upstream of the TSS; and (iii) The broad region, which encompassed 1000 bp downstream of the TSS to the end of the gene (30). Genes shorter than 5000 bp were excluded from consideration, as they were too small for broad binding to be reliably differentiated from a TSS profile [c.f. Ref. (10) who exclude genes shorter than 8000 bp].
The number of reads per base pair (coverage) in each of the three defined regions was calculated. Peaks in each region were then determined using a 200 bp sliding window to capture the local density around a point. Each gene was allocated to the subclass corresponding to the region with the highest coverage, provided it also satisfied the following criteria: (i) if the highest coverage was in the promoter region, then it must also have a peak in the promoter region that was >25% higher than any other peak in the gene; (ii) if the gene had the highest coverage in the TSS region, then it must also have a peak in the TSS region that is >25% higher than any other peak in the gene; and (iii) if the gene had the highest coverage in the broad region, then the signal at each point must be above the mean coverage in >35% of the region. This removed high average coverage genes that contained a large peak in the body, rather than a sustained level of enrichment across the gene (see Supplementary Figure S1B).
If none of the three criteria was met, then the gene was considered for the broad category if the average in the broad region was >90% of the other regions. This additional step captures genes that had a slightly lower signal on average, but were still broadly marked. If none of these criteria were satisfied, then the gene was not assigned to a subclass. R code has been included which can perform this classification on any data set and can be downloaded from the online supplementary materials.
k-means clustering
K-means clustering used the gene length scaled signal for each gene across the entire body of the gene, plus a 20% of gene length upstream. The signal was capped at the 97th percentile of the combined signal from all genes, to prevent extreme spikes in enrichment dominating the clustering. K-means clustering was then performed in R with Euclidean distance similarity metric. In most cases k (the number of clusters) was set to five but other values of k were also explored. After the clusters were identified, the genes were sorted by their classification into broad, promoter, TSS or none and plotted in a heatmap. The log2 expression values from the microarrays were also plotted.
R code to generate plots and perform classification
The scripts used to perform the analyses in this article and generate the plots are included in Supplementary Data. We also provide visualization functions in the Repitools R package (38).
RESULTS
Data generation and quality control
PRC2 binding has been mapped in a variety of cell types, including mouse ES cells, fibroblasts, NPs and cancer cell lines (5,12–17,21,22,39). To investigate the role of PRC2 during blood cell differentiation we used ChIP-seq to study the distribution of H3K27me3 and RNA polymerase II (RNApol-II) in the haemopoietic cell line G1ME (28). To gain insight into lineage-specific functions of PRC2, we also examined the distribution in mouse ES cells. ChIP samples plus input DNA from the two cell types were sequenced, yielding an average of 8 million uniquely mapped reads per condition (Supplementary Table S2). In addition, we made use of previously published ChIP-seq data sets for H3K27me3, H3K4me3 and H3K36me3 in ES cells, NP cells and MEFs (5) (Supplementary Table S2). All reads were mapped against version mm9 of the mouse genome using bowtie (32).
Initially, we assessed the quality of our data by comparing the distribution of H3K27me3 in ES cells with previously published data (5). We found the number of reads in 5 kb bins across the genome between the two experiments was highly correlated after normalizing for the total number of mapped reads (P = 0.92, Figure 1A), indicating good concordance between the two data sets. Furthermore, we generated standard quality control metrics on both our newly generated and publicly available data using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Our data, produced with newer sequencing technology, consistently showed higher quality (Supplementary Figure S2). Consistent with previous studies, we found that H3K27me3 is highly enriched in genic regions in both ES cells and G1ME cells, with 41 and 45% of total reads falling within a gene and 3 kb promoter region with these regions constituting <2% of the genome.
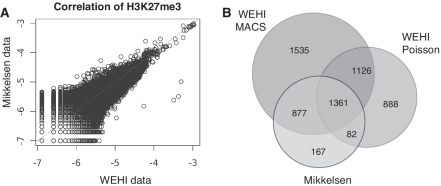
Figure 1.
A comparison of H3K27me3 ChIP-seq data sets from ES cells. (A) The number of mapped reads in H3K27me3 data was assessed in 5 kb intervals across the entire genome in ES cells for both our data (WEHI) and the public data (Mikkelsen). The number of reads in each interval was expressed as a proportion of total mapped reads. The data are plotted on a log2 scale and show a strong positive correlation (Pearson correlation co-efficient 0.92). (B) A Venn diagram showing the number of genes identified as marked with H3K27me3 in our data using different calling methods (MACS or Poisson). We recovered 93.3% of genes previously characterized as marked, as well as 3549 new genes. The majority of genes identified by Mikkelsen et al. (2007) were identified in our data using MACS (90%), whereas a much smaller proportion was identified using the Poisson test (58%). This difference may indicate a bias towards shorter domains of H3K27me3 in the genes defined by Mikkelsen et al. (2007).
To identify genes that were significantly enriched for H3K27me3, we used MACS (34). The MACS algorithm is designed to detect transcription factor binding events, which typically produce short peaks with strong signal intensity. In order to avoid a bias towards specific binding profiles, we designed an additional test to look for broad enrichment across the entire gene region. Specifically, the number of reads coming from each gene region was compared to an input control using a Poisson test (see ‘Materials and Methods’ section). When these methods were applied to identify genes marked with H3K27me3 in ES cells, 2298 genes were identified exclusively using MACS, 660 genes were identified exclusively using the Poisson test and 2613 genes (46.9%) were identified by both methods (Table 1 and Figure 1B). As expected, we found those genes identified as marked by MACS had a much shorter domain of H3K27me3 than genes identified as marked by the Poisson test (Supplementary Figure S3). The same methods were applied to find enriched genes in all other ChIP-seq experiments used in this study.
Our analysis recovered the vast majority of the genes that were previously identified as PRC2 targets by Mikkelsen et al. (5) in ES cells (93.3%), and identified an additional 3549 H3K27me3 marked genes (Figure 1B). Using the same approach, we identified 6567 genes that were enriched for H3K27me3 in G1ME cells (Table 1). The fraction of genes detected by the MACS or Poisson method varied widely with cell type, indicating differences in the prevalence of broad binding domains.
Visualizing gene-wide enrichment profiles
To assess the distribution of each mark of interest, we first calculated the average coverage around the TSS across all marked genes. In addition, we examined the pattern of enrichment across the entire gene length in more detail by utilizing an ASE plot. The ASE plot provides the enrichment profile for a set of genes by scaling each gene to a common length and then averaging the signal appropriately. These plotting functions are available in the Repitools package in R (38) (see ‘Materials and Methods’ section for a more detailed description and Supplementary Data for R code).
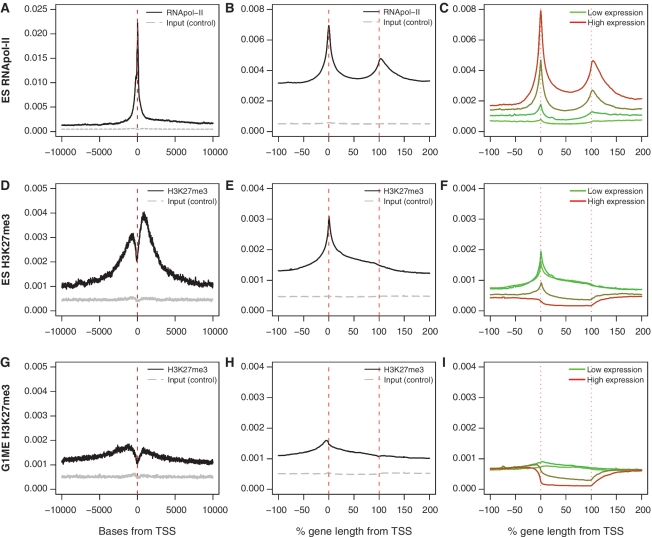
When we assessed the distribution of RNApol-II with a TSS centered plot, we observed a sharp peak of enrichment at the TSS, which extends over a 2 kb interval. Additionally, the signal was stronger downstream of the TSS, in the gene body, than in the upstream region (Figure 2A). By considering the structure of each gene, the ASE plot revealed additional details about RNApol-II occupancy. In the ASE plot, RNApol-II enrichment is seen at the TSS and across the gene body, with a second peak at the end of the gene, which may indicate stalling of the polymerase during termination (Figure 2B). Stratifying genes based on expression level clearly showed that RNApol-II occupancy increases proportionally with expression level (Figure 2C). The binding profile of RNApol-II is also highly consistent across cell types (Supplementary Figure S4B, F, N and R).
Figure 2.
TSS centered, ASE and expression stratified ASE plots of H3K27me3 and RNApol-II. Comparison of the TSS centered averaged plots (A, D and G) with the ASE plots (B, E and H) for genes marked by RNApol-II in ES cells and H3K27me3 in ES cells and G1MEs. There is information contained in the scaled version that is not observed from the TSS centered view. (C, F and I) ASE plots of all genes stratified by expression. The average level is lower as these include ChIP-seq data for all genes on the expression array, not just marked genes.
The distribution of H3K27me3 was examined in ES cells and G1ME cells. In ES cells, there was strong enrichment of H3K27me3 around the TSS (Figure 2D), whereas the signal was much lower in G1ME cells (Figure 2G). A common feature of the TSS centered plots is a sharp dip in H3K27me3 around the TSS, corresponding to the position of the nucleosome-depleted zone (40). The nucleosome-depleted zone was not evident in ASE plots of H3K27me3 signal, due to smoothing applied to the data. The ASE plots demonstrate that H3K27me3 is distributed over the entire length of genes, but we noticed stark differences in the enrichment profile between the cell types. In ES cells, the strongest peak in enrichment occurred at the TSS (Figure 2E), whereas in G1ME cells the peak was shifted upstream of the TSS (Figure 2H). Stratifying the genes based on expression level confirmed that H3K27me3 is highly enriched at repressed genes; however, the shape of the H3K27me3 profile also changed with expression level, suggesting that the distribution of H3K27me3 across the gene may be important for regulating transcription (Figure 2F and I).
Genes marked with H3K27me3 have distinct enrichment profiles
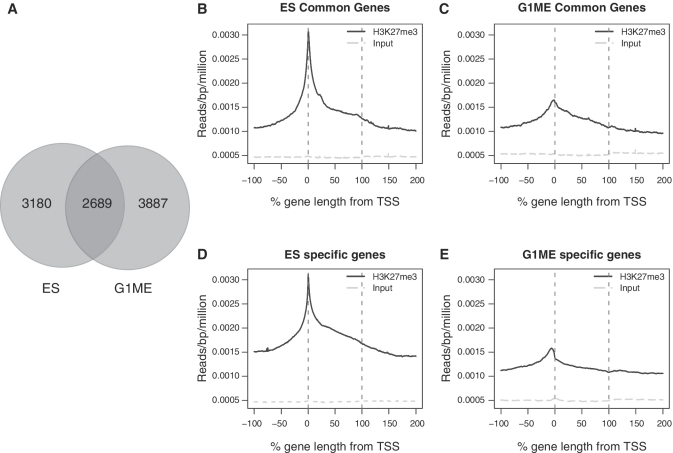
Despite the differences in H3K27me3 enrichment profiles between ES cells and G1MEs, the genes marked in each cell type show a large degree of overlap (Figure 3A). However, the cell type specificity of the enrichment profiles remained even for those genes marked in common (Figure 3B and C), indicating that the pattern of PRC2 marking differs depending on the precise cellular and developmental context. In addition, profiles for genes marked by H3K27me3 uniquely for each cell type showed striking differences between the two cell types (Figure 3D and E). The most prominent difference was an obvious peak in the promoter region of the G1ME-specific genes compared to ES cells with a stronger signal at the TSS.
Figure 3.
Profiles of genes exclusive and commonly marked by H3K27me3 in ES and G1ME cells. (A) The Venn diagram shows the overlap between the marked genes in ES cells and G1MEs. (B and C) ASE plots of H3K27me3 signal for the 2689 genes that are enriched for H3K27me3 in ES and G1ME. In each plot, the red lines denote the boundaries of the gene and the level of signal in the input control is plotted (grey line). (D and E) ASE plots of H3K27me3 signal for cell type-specific genes (the number of genes differs for each cell line: ES cells = 3180, G1MEs = 3887).
We hypothesized that these distinct enrichment profiles were the result of many genes in each cell type with a similar pattern of enrichment. To test this prediction, we identified three general patterns of enrichment seen in the ASE plots that we believed to be important and distinct. First, there were genes with an abundance of H3K27me3 around the TSS, mostly clearly seen in the ES cells (Figure 3B and D). Second, there were genes that showed a broad enrichment across the entire length of the gene. Third, there appeared to be a class of genes that were strongly enriched for H3K27me3 upstream of the TSS, that were most obvious in G1ME cells (Figures 2H and 3E).
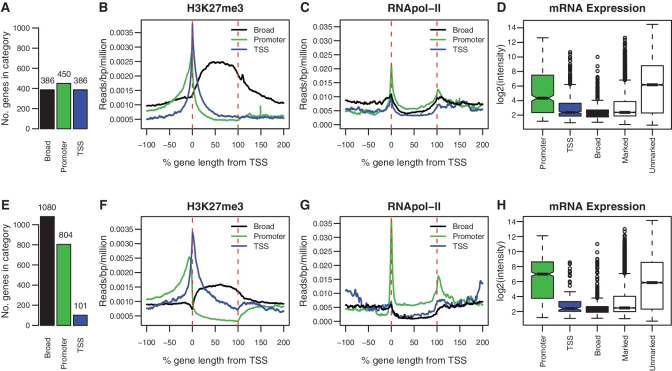
We developed a set of conservative criteria to identify individual genes that had patterns of H3K27me3 that fall into one of the three classes that we call broad, TSS and promoter (see ‘Materials and Methods’ section). These criteria were used to classify all genes enriched for H3K27me3 in our ES cell and G1ME data. We found that a sizeable fraction of genes could be robustly and unambiguously assigned to one of the three classes we identified. We classified 21% of enriched genes from ES cells and 30% from G1ME cells into our three profile groups (see Supplementary Table S3 for the complete list of marked genes and profile classifications). In both cell types, we find evidence for all three classes of enrichment, but the proportions of genes in each class are very different (Figure 4A and E). Figure 4 shows the ASE plot for ES cells and G1MEs, separated into each of the three classes. As we would expect from looking at the ASE plots of all marked genes (Figure 3), ES cells contain a high proportion of TSS genes, while G1ME cells contain a high proportion of broad genes and promoter genes.
Figure 4.
Classification of profiles. ASE plots for each H3K27me3 enrichment profile are shown for ES cells (B) and G1ME cells (F). The number of genes classified into each enrichment profile is shown in the adjoining bar plots (A and E). ASE plots of RNApol-II for each class of genes in ES (C) and G1ME cells (G). Promoter genes show strong enrichment for RNApol-II, while TSS and broad genes do not. Box plots of expression levels are shown for each class of gene in ES cells (D) and G1ME cells (H). Genes classified with the promoter profile show high levels of expression, whereas genes with the broad profile have the lowest expression levels. Genes classified as TSS have intermediate expression levels, but are still repressed relative to the average of all genes on the array.
In addition to showing the expected enrichment features that were used to classify genes as broad, TSS and promoter, Figure 4 also revealed additional features specific to each class and persistent across the cell types. Specifically, the TSS genes show the expected peak in enrichment around the TSS with little or no enrichment across the body of the gene. In contrast, the broad genes show strong enrichment in the gene body that slowly tapers to background beyond both the start and end of transcription. This suggests that for some of these genes the H3K27me3 domain extends well into the intergenic region, consistent with the observation that H3K27me3 can mark the genome in large blocks (27). Finally, the promoter genes show strong enrichment of H3K27me3 upstream of the TSS, but also show a depletion of H3K27me3 below background levels, across the body of the gene. Depletion of the H3K27me3 mark is also seen in highly expressed genes (Figure 2F and I), which suggested that these genes might be actively transcribed.
Validation of H3K27me3 enrichment profiles
To validate our classification of H3K27me3 marked genes into three distinct profiles (TSS, promoter and broad), we selected a number of genes in each category and confirmed their enrichment profile using ChIP-qPCR. Specifically, we showed high levels of enrichment for H3K27me3 in the promoter of DNMT3A, FNIP1 and RTN4 in G1ME cells, and low or absent levels of H3K27me3 at the TSS and in the gene body for these genes. Furthermore, we selected two genes, SCUBE2 and PDE8A, which had a TSS profile of H3K27me3 enrichment in ES cells and a broad profile in G1ME cells and confirmed this difference using two qPCR amplicons. All experiments were performed in triplicate and we observed good concordance between ChIP-Seq and ChIP-qPCR. This verified the distribution of H3K27me3 observed in the ChIP-Seq data and confirmed that the H3K27me3 enrichment profile differs depending on the cellular context (Supplementary Figure S5).
To test the broader applicability of our classifications, we extended our analysis to include publicly available H3K27me3 ChIP-Seq data (5) for ES, MEF and NP cells and identified promoter, TSS and broad genes in each case (Supplementary Figure S6). We classified a significant fraction of marked genes as promoter, TSS or broad in all three cell types (20% ES, 26% MEF, 21% NP). Despite being able to identify genes in each class in each cell type, the overall number of marked genes identified was higher in our newly generated data compared with the public data sets. This is likely a consequence of the technology driven improvement in data quality, with higher resolution giving more power to identify marked genes and to classify enrichment profiles.
H3K27me3 in promoters is associated with active transcription
In order to test the functional properties of the different profile classes (promoter, TSS and broad), we assessed the mRNA expression level of each class. For each of the different classes of genes, we calculated box plots of their corresponding expression values (Figure 4D and H). A consistent trend was seen in ES and G1ME cells. First, broadly marked genes were the most lowly expressed set of genes identified. In both cell types, broad genes were more lowly expressed than the average marked gene (P < 5.1 × 10−12, <2.2 × 10−16, Mann–Whitney test for ES cells and G1ME cells, respectively). Second, the TSS class of genes had expression levels consistent with the average of the marked genes and were repressed relative to the average gene on the array (P < 2.2 × 10−16, 1.1 × 10−7). In contrast to broad and TSS genes, the promoter genes were highly expressed relative to the average marked gene. Indeed, for G1MEs the expression level of the promoter genes was significantly higher than the average of all unmarked genes on the array (P < 1.39 × 10−9). Despite having H3K27me3 in their promoters these genes are not repressed, instead they appear to be expressed at high levels.
The degree of RNApol-II enrichment for the various classes was consistent with the findings from the expression data (Figure 4C and G). That is, the promoter genes have a high level of RNApol-II binding, whereas the broad genes and the TSS class show little evidence of RNApol-II being present. Finally, by taking all marked genes and plotting the 5% most highly expressed genes and the 5% most lowly expressed genes, we were able to recover the broad and promoter profiles without using our classification criteria (Supplementary Figure S7).
To address whether H3K27me3 was contributing to the high level of expression, we examined the expression level of promoter genes in Suz12 knockout ES cells (41) and in Suz12 knockdown G1ME cells, which have impaired PRC2. On the whole, we observed modest expression changes in H3K27me3 modified genes, and found no consistent change in promoter genes between the two cell types (Supplementary Figure S10). Two major caveats influence our interpretation of this data: first, the expression data and ChIP-Seq data were not obtained from the same cells (the Suz12 knockout ES cell data was from published microarrays) and second, we did not assess the level of H3K27me3 on the promoters after inhibition of PRC2. Therefore, additional experiments are required to determine whether H3K27me3 in the promoter is having an activating role in transcription or performing an alternate function.
K-means clustering confirms classification
Clustering approaches have been used to separate distinct enrichment profiles for the H3K4me2 mark in T cells (10). To further explore the classifications of our H3K27me3 profiles, we performed k-means clustering for all genes longer than 5 kb. Figure 5 shows the results of clustering genes using five groups in G1ME cells. The largest cluster corresponds to genes without any enrichment of H3K27me3. Other clusters can be clearly associated with our defined classes. Specifically, there is one cluster of promoter genes, a cluster of broadly marked genes with a high average enrichment and a cluster of broadly marked genes with a low average enrichment. These results support our classification scheme and illustrate the conservative nature of our criteria. Our classification scheme only identifies the most robust examples in each class; applying k-mean clustering assigned a larger number of genes to each cluster. Our classification approach was able to reliably identify smaller populations of enrichment profiles, such as the TSS profile, which do not form a separate cluster in G1MEs. Similarly, when applying clustering to ES cells the TSS class was reliably identified but the promoter class was not. Thus, while clustering does well at identifying the most prevalent classes, it cannot reliably identify less common profiles (Supplementary Figure S8A–D). R code is provided to produce cluster plots using the Repitools package (38) (see Supplementary Data).
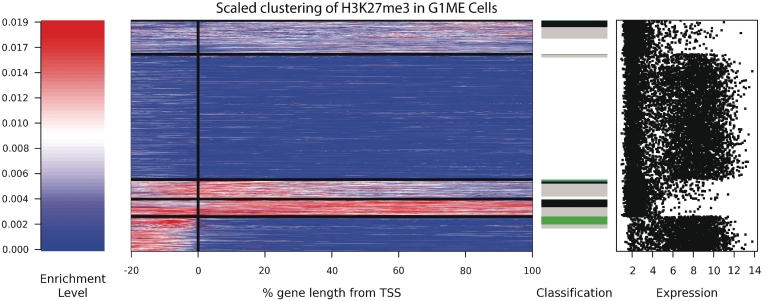
Figure 5.
K-means clustering of genic H3K27me3 profiles in G1ME cells. The signal intensity is shown as a spectrogram, with red reflecting a high enrichment signal and blue reflecting no signal. All genes were scaled to have the same length, and position relative to the TSS is shown in percentage terms. Genes were sorted first by cluster, then by classification (black: broad; green: promoter; blue: TSS; grey: marked but unclassified). The expression level of all genes is shown on the far right. Additional cluster profiles are provided for the other cell types (Supplementary Figure S8).
The promoter profile is conserved between cell types
We plotted the profiles of those genes marked as promoter, TSS and broad in G1MEs in the other cell types (ES, MEF, NP Supplementary Figure S9A–C). We found that the genes that were classified as broad or TSS in G1MEs generally show a TSS profile in other cell types. In contrast, the genes identified in the promoter class showed the same promoter profile in the other cell types. These promoter genes are also highly expressed in all cell types, indicating that they have a stable enrichment profiles and expression pattern between cell types (Supplementary Figure S9D–F).
To explore additional factors that correlate with promoter genes, we first identified genes with any CpG islands in the 3 kb upstream of the TSS using the UCSC track (http://genome.ucsc.edu/cgi-bin/hgTables). Roughly 80% of promoter genes had a CpG island within 3 kb upstream of their TSS, compared to the genome-wide average of 37%. Genes with CpG islands in their promoter have previously been associated with housekeeping genes (5), strengthening the case for promoter genes being ubiquitously expressed.
It remained possible that the H3K27me3 signal in the promoter was not a direct regulatory mechanism, but instead reflected the spread of methylation from a neighbouring gene. This may be the case for a proportion of the promoter genes, as we found that genes directly upstream of the promoter genes have a higher level of H3K27me3 compared to the genes downstream of the promoter genes, or the flanking genes of the broad class (Supplementary Figure S11). Nevertheless, only 35% of promoter genes were downstream of a gene classified as marked, which suggests that the promoter profile also occurs independently.
To gain insight into the categories of genes represented within the promoter class, we performed Gene Ontology and KEGG pathway analysis (42,43) (clustered results shown in Supplementary Table 4, KEGG pathways in Supplementary Table S5). This analysis indicated that promoter genes are enriched for genes involved in cell signalling, including many genes involved in the Ras pathway. However classic Polycomb target genes, including the Hox genes, and genes involved in development, pattern specification and organ development, were represented in the broad class (Supplementary Table S3).
Bivalent genes are TSS class genes
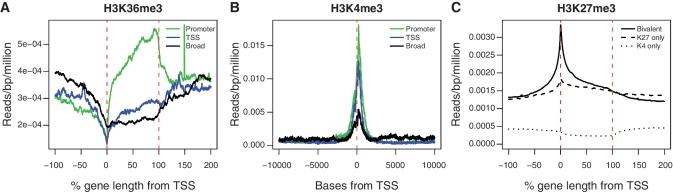
To further characterize the three classes of H3K27me3 marked genes, we investigated the relationship between the H3K27me3 profile and the presence of other histone modifications. Both H3K4me3 and H3K36me3, a mark associated with transcriptional elongation, have been studied in ES cells, NP cells and in MEFs (5). This data were processed in the same way as the H3K27me3 and RNApol-II data and marked genes were identified (Table 1). ASE plots of the genes marked by H3K4me3 in ES cells, NP cells and MEFs (Supplementary Figure S4O, T and W) indicate that the vast majority of H3K4me3 signal is localized to the region around the TSS, confirming previously published findings (44). In contrast, ASE plots show that H3K36me3 is almost always present across the entire gene, which is also consistent with earlier studies (9) (Supplementary Figure S4P, U and X). Therefore we used TSS centered plot to examine H3K4me3 and ASE plots for H3K36me3. Finally, we confirmed that H3K36me3 and H3K4me3 were more prevalent on highly expressed genes by stratifying both marks by mRNA expression (Supplementary Figures S12 and S13).
We found that the promoter genes (defined by the H3K27me3 mark) were enriched for both H3K36me3 and H3K4me3, consistent with these genes being actively transcribed. In contrast, the broad class of genes showed little enrichment for either of these modifications (Figure 6A and B). The TSS class of genes were strongly enriched for H3K4me3, but showed little H3K36me3 signal or RNApol-II binding. These observations support the idea that the TSS class contains many bivalent genes, which are poised to respond to regulatory cues.
Figure 6.
The relationship between H3K4me3, H3K36me3 and H3K27me3 enrichment in ES cells. H3K36me3 enrichment across the gene (A) and H3K4me3 enrichment around the TSS (B) for the promoter, TSS and broad classes of genes in ES cells. ASE plots of the H3K27me3 signal in ES cells, where genes have been separated based on being called marked by H3K27me3 and H3K4me3 (C). The solid line is the profile for bivalent genes (marked by both H3K27me3 and H3K4me3). The dashed line corresponds to genes marked with only H3K27me3 and the dotted line is the H3K27me3 profile for genes marked with only H3K4me3.
Next, we calculated the ASE for all genes designated as doubly marked (those that possess both the H3K27me3 and H3K4me3 marks), and for genes marked with H3K27me3 or H3K4me3 alone (Figure 6C). The H3K27me3 profile for doubly marked genes is strikingly similar to the profile of the TSS class of genes, whereas genes marked only with H3K27me3 show a broad enrichment profile. This provides further evidence that bivalent genes predominantly have a small domain of H3K27me3 (22). As expected, genes that were classified as H3K4me3-only showed negligible levels of H3K27me3.
Of the 5538 genes marked by H3K27me3 in ES cells, 4737 (86%) showed evidence of marking by H3K4me3. TSS and promoter genes were over represented among doubly marked genes, with 94% of TSS genes being doubly marked (P = 3 × 10−7, two-sided Fisher's exact test) and 96% of promoter genes being doubly marked (P = 2 × 10−9). The TSS genes are likely bivalent genes, with the H3K27me3 mark and the H3K4me3 mark overlapping, whereas the promoter genes are unlikely to have overlapping profiles. In contrast, broad genes were under-represented in this set with only 75% being doubly marked (P = 8 × 10−12 c.f. 86% of all H3K27me3 marked genes being doubly marked).
DISCUSSION
The chromatin landscape is highly complex. Understanding the functional role of a modification requires a detailed analysis of its distribution, its relationship to other epigenetic factors (chromatin context), and correlation with functional properties of modified genes. We have utilized ChIP-seq data to explore the relationship between the pattern of H3K27me3 enrichment and gene expression. Three H3K27me3 enrichment profiles were identified: enrichment in the promoter, enrichment at the TSS and broad marking across the full length of the gene. All classes were present in four different cell types, but the proportions differed. These results provide new insights into transcriptional control mediated by PRC2, and call attention to the way in which we process and utilize ChIP-Seq data.
In this study, we employed several analytical and visualization tools to assess the distribution of H3K27me3. In particular, we used an averaged scaled enrichment (ASE) plot, which shows the average signal over the body of a gene, compared enrichment profiles between distinct cell types, stratified genes based on their expression level and assessed the interaction of H3K27me3 with other histone modifications and RNApol-II. We observed dramatic differences between the average H3K27me3 profile in ES and G1ME cells, and by focusing on the differences we were able to identify three predominant enrichment profiles. Comparable results were obtained when we used the k-means algorithm to cluster genes based on their H3K27me3 enrichment profile. Similar clustering methods were used previously to assess the distribution of many histone modifications, including H3K27me3, in CD4+ T cells (45). Hon et al. (45) confirmed a strong association between H3K27me3 and transcriptional repression, but they did not identify genes that carry H3K27me3 specifically in the promoter region. We have seen that the proportion of marked genes in each class can vary dramatically between cell types, which may explain why the promoter class was not identified in CD4+ T cells. Indeed, our results suggest that clustering tends to miss profiles that contain small numbers of genes. Additionally, the k-means algorithm requires the number of clusters to be selected prior to running the analysis and it uses a randomly chosen ‘seed’ gene to form each cluster, altering these variables can produce different results. Each visualization tool possesses distinct advantages and multiple tools should be used to interpret ChIP-Seq data. Although the ASE plot includes some structural landmarks, including the TSS, transcriptional end site and a loosely defined promoter, it still misses internal structures, such as introns and exons. Because of this limitation, the ASE plot did not provide sufficient resolution to identify a modification specifically enriched on exons as has previously been identified for H3K36me3 (9). One possible extension of the ASE plot would be to scale the first exon and intron to the same length.
By combining gene expression data, RNApol-II, H3K36me3 and H3K4me3 ChIP-seq data with our classification scheme, we have been able to show that each of the promoter, TSS and broad classes of H3K27me3 is associated with a distinct transcriptional outcome. The promoter genes are highly expressed despite possessing significant enrichment of H3K27me3 in the promoter. In both ES and G1ME cells, promoter genes have a depletion of the repressive mark H3K27me3 across the body of the gene and a significant level of RNApol-II binding. These genes also show significant enrichment of H3K4me3 and H3K36me3 and higher than average mRNA expression. In contrast, the broad genes are strongly repressed. They have enrichment for H3K27me3 across the entire gene that can extend into the flanking regions and have little to no RNApol-II, H3K36me3 or H3K4me3. Finally, genes in the TSS class lack significant RNApol-II or H3K36me3 binding and have lower mRNA expression levels than the average gene, although not as low as the broad class. Many of the TSS genes are likely to be bivalent, having a peak in both H3K27me3 and H3K4me3 around the TSS, although we lack the sequential ChIP data needed to confirm co-occupancy. These findings support the generally accepted view that bivalent genes are ‘poised’, meaning that they are not yet committed to either activation or repression.
While our analysis identified many genes that carry H3K27me3 in their promoter, the precise role of the modification in this context remains unclear. The vast majority of promoter genes possess CpG islands upstream of their TSS. Several groups have noticed strong association between PRC2 binding sites and GC-rich sequence elements and it is thought that these sequences play a key role in recruiting PRC2 (5,13,14,39). It was surprising to find that, in contrast to the promoter class, the frequency of CpG islands in the broad class was only 30%, which is comparable to the genome-wide frequency. This suggests that alternative mechanisms may be employed to recruit PRC2 to promoter and broad genes. Although promoter genes are highly expressed, it remains possible that H3K27me3 has a repressive function at these sites and acts to moderate expression levels. Another alternative is that deposition of H3K27me3 in the promoter occurs at regulatory elements. H3K27me3 has previously been found in the promoter of repressed Polycomb target genes that express small RNAs, which act as recruitment signals for PRC2 (46); however, in this context H3K27me3 also occurs downstream of the TSS where it acts to block RNApol-II extension. If H3K27me3 does block RNApol-II extension, then marking in the promoter may provide a way to guide alternative promoter use or prevent inappropriate transcription in the opposite direction. It is not possible to address these questions using standard microarray expression analysis alone. A detailed analysis of gene expression by RNA sequencing, including promoter usage, splicing patterns and transcriptional orientation will be required to resolve this issue.
In this study, we focused on understanding how differences in the distribution of H3K27me3 impact on gene expression; however, there are many functional properties of genes that could also be considered, including promoter usage, alternative splicing, antisense transcription and replication timing. Many new insights into the biology of PRC2 will come as we continue to map more chromatin modifications and uncover new mechanisms that influence transcription. It will be important to integrate these new data to gain a better understanding of the mechanisms that govern the distribution of PRC2 and that regulate its activity. Our study demonstrates that it is important to consider the precise pattern of H3K27me3 enrichment on genes. Many Polycomb group proteins are involved in cancer and it will be interesting to see whether H3K27me3 enrichment profiles are also altered in disease.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council (NHMRC) fellowship (406676 to M.E.B.); Australian Research Council QEII fellowship (DP1096092 to M.E.B.). I.J.M. funded by NHMRC grant 575581. Funding for open access charge: NHMRC grant 490037.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mark Robinson, Aaron Statham and Dario Strbenac for incorporating the plotting functions into the Repitools package. We thank Kristian Helin's group for sharing microarray data and Kylie Greig, Manasa Ramakrishna and Mark Robinson for helpful discussion and comments on the article.
REFERENCES
- 1.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 2.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 4.Mendenhall EM, Bernstein BE. Chromatin state maps: new technologies, new insights. Curr. Opin. Genet. Dev. 2008;18:109–115. doi: 10.1016/j.gde.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hon G, Ren B, Wang W. ChromaSig: a probabilistic approach to finding common chromatin signatures in the human genome. PLoS Comput. Biol. 2008;4:e1000201. doi: 10.1371/journal.pcbi.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pekowska A, Benoukraf T, Ferrier P, Spicuglia S. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res. 2010;20:1493–502. doi: 10.1101/gr.109389.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat. Rev. Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 13.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 16.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 17.Squazzo SL, O'Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majewski IJ, Blewitt ME, de Graaf CA, McManus EJ, Bahlo M, Hilton AA, Hyland CD, Smyth GK, Corbin JE, Metcalf D, et al. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 2008;6:e93. doi: 10.1371/journal.pbio.0060093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell. Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 23.Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O'Neill L, Azuara V. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137:2483–2492. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzarella L, Jorgensen HF, Soza-Ried J, Terry AV, Pearson S, Lacaud G, Kouskoff V, Merkenschlager M, Fisher AG. ES cell-derived hemangioblasts remain epigenetically plastic and require PRC1 to prevent neural gene expression. Blood. 2010;117:83–7. doi: 10.1182/blood-2010-03-273128. [DOI] [PubMed] [Google Scholar]
- 25.Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr. Opin. Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 27.Pauler FM, Sloane MA, Huang R, Regha K, Koerner MV, Tamir I, Sommer A, Aszodi A, Jenuwein T, Barlow DP. H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res. 2009;19:221–233. doi: 10.1101/gr.080861.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stachura DL, Chou ST, Weiss MJ. Early block to erythromegakaryocytic development conferred by loss of transcription factor GATA-1. Blood. 2006;107:87–97. doi: 10.1182/blood-2005-07-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kidder BL, Yang J, Palmer S. Stat3 and c-Myc genome-wide promoter occupancy in embryonic stem cells. PLoS ONE. 2008;3:e3932. doi: 10.1371/journal.pone.0003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee AP, Koh EG, Tay A, Brenner S, Venkatesh B. Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proc. Natl Acad. Sci. USA. 2006;103:6994–6999. doi: 10.1073/pnas.0601492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909. [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23:2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 37.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol. Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Statham AL, Strbenac D, Coolen MW, Stirzaker C, Clark SJ, Robinson MD. Repitools: an R package for the analysis of enrichment-based epigenomic data. Bioinformatics. 2010;26:1662–1663. doi: 10.1093/bioinformatics/btq247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Schmid CD, Bucher P. ChIP-Seq data reveal nucleosome architecture of human promoters. Cell. 2007;131:831–832; author reply 832–833. doi: 10.1016/j.cell.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 45.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum. Mol. Genet. 2009;18:R195–201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.