Abstract
Processive transcription antitermination requires the assembly of the complete antitermination complex, which is initiated by the formation of the ternary NusB–NusE–BoxA RNA complex. We have elucidated the crystal structure of this complex, demonstrating that the BoxA RNA is composed of 8 nt that are recognized by the NusB–NusE heterodimer. Functional biologic and biophysical data support the structural observations and establish the relative significance of key protein–protein and protein–RNA interactions. Further crystallographic investigation of a NusB–NusE–dsRNA complex reveals a heretofore unobserved dsRNA binding site contiguous with the BoxA binding site. We propose that the observed dsRNA represents BoxB RNA, as both single-stranded BoxA and double-stranded BoxB components are present in the classical lambda antitermination site. Combining these data with known interactions amongst antitermination factors suggests a specific model for the assembly of the complete antitermination complex.
INTRODUCTION
Processive transcription antitermination, a system best characterized in bacteriophage lambda (λ), involves hijacking and structural modification of host RNA polymerase (RNAP) by a complex of proteins and RNA, enabling the regulation of transcription through downstream termination sites. During lytic infection, phage λ utilizes this process to control the timing of transcription of particular genes essential for lytic growth as the virus switches from early, to delayed-early, to late gene expression (1). Many other viruses, including the HIV provirus, use analogous systems of transcription control through the use of RNA regulatory elements (2).
The λ antitermination complex consists of phage N protein, N-utilization (nut) RNA control sequences BoxA and BoxB, and host proteins referred to as N-utilization substances A, B, E and G (NusA, NusB, NusE and NusG). NusE is also known as ribosomal protein S10 of the 30 S ribosome subunit (3). As such, this protein exhibits moonlighting behavior by playing roles in distinct complexes involved in transcription or translation in a mutually exclusive manner (4,5). BoxA and BoxB facilitate antitermination through several protein–RNA interactions. BoxA binds to the host NusB–NusE heterodimer (6,7) and the BoxB stem loop binds to the N protein (8–11). A spacer region, to which NusA binds (12), separates these two RNA segments. Although less well understood, a similar mechanism is employed by the bacterial host in the transcription control of ribosomal RNA operons (rrn) using the same Nus factors, other ribosomal proteins, a host-encoded BoxA RNA and a BoxB-like RNA (reviewed in 13).
The λ antitermination complex assembly likely begins with the association of NusB with BoxA as its sequence is transcribed. This is in concert with the formation of the NusB–NusE heterodimer, which facilitates and strengthens the binding to other transcription elongation components including N protein, BoxB, NusA, NusG and RNAP (14). Association of NusB, NusE and BoxA is therefore a regulatory event during the initial assembly of the complete antitermination complex (4,15), and has been chosen as a key element for our structural investigation. Moreover, the NusB–NusE heterodimer has been shown to bind BoxA with greater affinity than either protein alone (7,16,17). The ribosome binding loop of Escherichia coli (Ec) NusE/S10 was shown to be dispensable for λ transcription antitermination by genetic replacement of the loop with a serine (4). The crystal structure of the EcNusB–NusE complex was thus solved and UV cross-linking studies were used in an attempt to identify the amino acids responsible for binding of BoxA RNAs encoded in the Ec rrn operons (EcBoxA) (4). Those experiments provided a general indication of the BoxA binding region, but atomic details necessary to understand the protein–RNA interactions are still lacking, and have proven to be a major experimental hurdle. As an alternative approach, we used orthologous proteins from Aquifex aeolicus (Aa). Although phylogenetically divergent, the NusB and NusE proteins from these two bacteria are similar, sharing amino acid sequence identities of 28 and 53%, respectively (Figure 1A and B). Furthermore, we previously reported the solution structure of AaNusB and demonstrated similar protein–protein interactions at the NusB–NusE interface for the two bacterial systems (18). In this study, we have also replaced the ribosome binding loop of AaNusE with a single serine (Figure 1B) to improve solubility and provide a more globular structure conducive for crystallization.
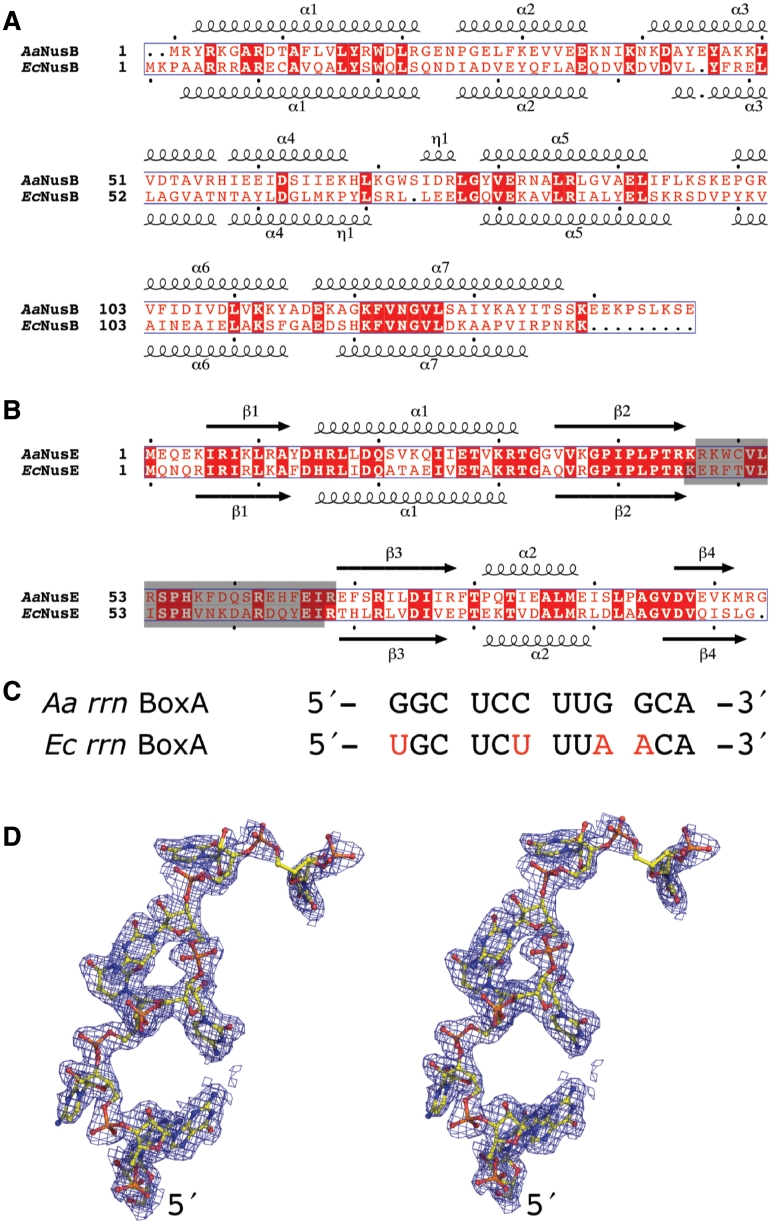
Figure 1.
Sequence and structure of NusB, NusE and rrn BoxA. (A and B) Structure-based sequence alignments of NusB and NusE from A. aeolicus (Swiss-Prot codes: O66530, O66430) and E. coli (Swiss-Prot codes: P0A780, P0A7R5). Residues shaded in gray were removed by genetic manipulation and replaced by a serine to form modified versions of EcNusE (4) and AaNusE (this work). (C) Sequence alignment of the rrn BoxA regions. Nucleotides in red denote positions that are different in the E. coli sequence relative to that of A. aeolicus. (D) Stereo representation of AaBoxA (chain R) as it exists in the crystal structure of AaNusB–NusE–BoxA, including electron density from a 2Fo−Fc omit map contoured at 1σ.
An Aa rrn BoxA (AaBoxA) was identified by sequence comparison with Ec rrn operons, and a synthetic 12-mer BoxA RNA (5′-GGC UCC UUG GCA-3′) was used in crystallization experiments (Figure 1C). This particular 12-mer RNA exists as a stable duplex at room temperature, with a melting temperature of ∼32°C (see Supplementary Data), and was therefore utilized as both single-stranded BoxA and generic double-stranded (ds) RNA by varying experimental temperatures. In this work, we outline the detailed RNA interactions of AaNusB–NusE with both BoxA and dsRNA, as well as the structure–functional significance of each. We also demonstrate by NMR and biophysical measurements that the AaNusB–BoxA interactions are equivalent in E. coli. Lastly, our genetic complementation assays for AaNusB in E. coli support the functional interpretation of the new crystal structures and demonstrate cross-species NusB–NusE–RNA interaction.
MATERIALS AND METHODS
Cloning, coexpression and purification of AaNusB-NusE
AaNusB was prepared as described (18). To coexpress the AaNusB–NusE complex, genes encoding AaNusE (residues 47–68 replaced with a single Ser) and AaNusB were cloned into suitable donor vectors, followed by multi-site recombination into a tagless destination vector (pDEST42) using Gateway® cloning technology (Invitrogen, Carlsbad, CA, USA). The proteins were coexpressed in E. coli and soluble lysate was incubated for 30 min at 90°C and clarified by centrifugation. The sample was purified by ion-exchange chromatography. Further details are provided in Supplementary Data.
Crystallization and structure determination
Crystals of AaNusB–NusE–BoxA were grown at 37°C. Crystals of AaNusB–NusE–dsRNA were grown at room temperature. All data were collected at the Advanced Photon Source and processed with HKL 3000 (19). Molecular replacements were performed using Phaser (20). Model building and refinement were carried out using the programs COOT (21) and PHENIX (22), respectively. All figures were generated using PyMol (23). Data, refinement statistics and other structural information are summarized in Table 1. Further details are provided in Supplementary Data.
Table 1.
X-ray diffraction data and refinement statistics
| AaNusB– NusE–BoxA | AaNusB– NusE–dsRNA | |
|---|---|---|
| Crystal | ||
| Space group | P21 | P212121 |
| Unit cell parameters | ||
| a (Å) | 40.5 | 76.8 |
| b (Å) | 113.0 | 79.9 |
| c (Å) | 65.8 | 106.6 |
| β (°) | 102.9 | 90 |
| Matthews coefficient (Å3/Da) | 2.4 | 2.6 |
| Data | Overall (last shell) | Overall/(last shell) |
| Resolution (Å) | 40.00–1.90 (1.97–1.90) | 50.00–2.20 (2.28–2.20) |
| Unique reflections | 42 602 (3354) | 32 257 (3160) |
| Redundancy | 5.0 (2.8) | 5.7 (5.5) |
| Completeness (%) | 94.2 (74.5) | 94.7 (94.5) |
| Rmergea | 0.102 (0.520) | 0.057 (0.542) |
| I/σ(I) | 19.3 (1.7) | 26.7 (3.1) |
| Refinementb | Overall/(last shell) | Overall/(last shell) |
| Resolution (Å) | 35.67–1.90 (1.95–1.90) | 33.64–2.20 (2.26–2.20) |
| Unique reflections | 42 498 (2150) | 32 201 (2634) |
| Completeness (%) | 93.9 (71) | 94.6 (94) |
| Data in the test set | 2160 (98) | 1659 (135) |
| Rwork | 0.169 (0.258) | 0.201 (0.273) |
| Rfree | 0.202 (0.292) | 0.239 (0.357) |
| Structure | ||
| Observed Electron Density | ||
| AaNusB (A) | 1–138 | 0–69, 73–134c |
| AaNusB (B) | 1–137 | 0–137c |
| AaNusE (J)d | 3–47, 69–102 | 4–47, 69–103 |
| AaNusE (K)d | 3–47, 69–103 | 4–47, 69–103 |
| AaBoxA (R) | 1–9e | 1–12f |
| AaBoxA (S) | 1–10 | 5–12 |
| Protein non-H atoms/B (Å2) | 4066/32.9 | 3524/64.7 |
| RNA non-H atoms/B (Å2) | 432/37.7 | 375/88.4 |
| Water oxygen atoms/B (Å2) | 240/40.3 | 98/56.9 |
| Other solvent atoms/B (Å2) | 78/56.4 | 35/91.5 |
| R.m.s.d. | ||
| Bond lengths (Å) | 0.007 | 0.004 |
| Bond angles (°) | 1.028 | 0.723 |
| Coordinate error (Å) | 0.23 | 0.33 |
| Ramachandran plot | ||
| Favored regions (%) | 99.3 | 99.0 |
| Disallowed regions (%) | 0.0 | 0.0 |
aRmerge = Σ|(I−<I>)|/Σ(I), where I is the observed intensity.
bRefinements included the use of TLS parameters.
cAmino acid residue 0 is an additional methionine beyond the N-terminal methionine of the sequence and is a cloning artifact.
dNusE residues 47–68 have been replaced by a serine as S47.
eNucleotide residue 9 contains atoms P, O1P, O2P and O5*, i.e. the 5′-phosphate, only.
fNucleotide residue 3 has a zero occupancy.
EcNusB and EcBoxA preparations and biophysical measurements
EcNusB proteins were cloned, expressed and purified using the methods previously described (18,24). NMR spectra were collected on Varian Inova spectrometers at either 600 or 800 MHz equipped with ChiliProbes. Sequential backbone assignments were made using standard triple resonance pulse sequences (25). Data were processed and analyzed using NMRPipe (26) and CCPNMR (27) and scripts therein. Sequential backbone and side-chain assignments of the EcNusB-BoxA complex were complete for ∼95% of the protein. Analytical ultracentrifugation was performed at 25 000 rpm, 20°C, using a Beckman XL-1 Optima analytical ultracentrifuge (Beckman Coulter, Inc., Brea, CA, USA). The protein–RNA interaction studies were performed using a VP-ITC Microcalorimeter (MicroCal LLC, Northampton, MA, USA) at 25 or 35°C. The integrated heat-of-interaction values were fit using the Origin 7.0-based ITC data analysis software (MicroCal). Further details are provided in Supplementary Data.
Functional assay for AaNusB complementation in E. coli
The entire nusB ORF (open reading frame) was deleted from the E. coli chromosome by recombineering as described (28). The Ec W3110 ΔnusB cells were transformed with plasmids expressing Aa or EcNusB proteins or with naked pET vector as a control. The transformed cells were then analyzed for cell growth at different temperatures. To assay for the NusB requirement in the growth of phage λ, the ΔnusB cells containing the AaNusB, EcNusB or control plasmids, were plated, spotted with serial phage dilutions and incubated at 42°C to observe the formation of λ plaques. Further details are provided in Supplementary Data.
RESULTS
The NusB–NusE heterodimer recognizes 8 nt of BoxA
Crystal data, refinement and structural details are provided in Table 1. Representative electron density is shown in Figure 1D. The crystal structure of AaNusB–NusE–BoxA contains two independent copies each of AaNusB, AaNusE and AaBoxA in the form of a dimer of ternary complexes (Supplementary Figure S1A), which can be superimposed with a root-mean-square deviation (r.m.s.d.) of 0.3 Å (Supplementary Figure S1B). Dynamic light-scattering experiments demonstrate that the interaction between the two complexes is both concentration and temperature dependent and that AaNusB–NusE is monomeric under native conditions. The four protein molecules are interconnected through two distinct NusB–NusE interfaces: one biologically relevant and previously observed in solution (18) and the other induced by dimerization. The data revealed electron density for eight and 10 out of the 12 nt for BoxA molecules R and S, respectively, but the 10th nucleobase of chain S is buried between symmetry-related AaNusB molecules in the crystal lattice and its position is thus considered an artifact of crystallization (Supplementary Figure S1C).
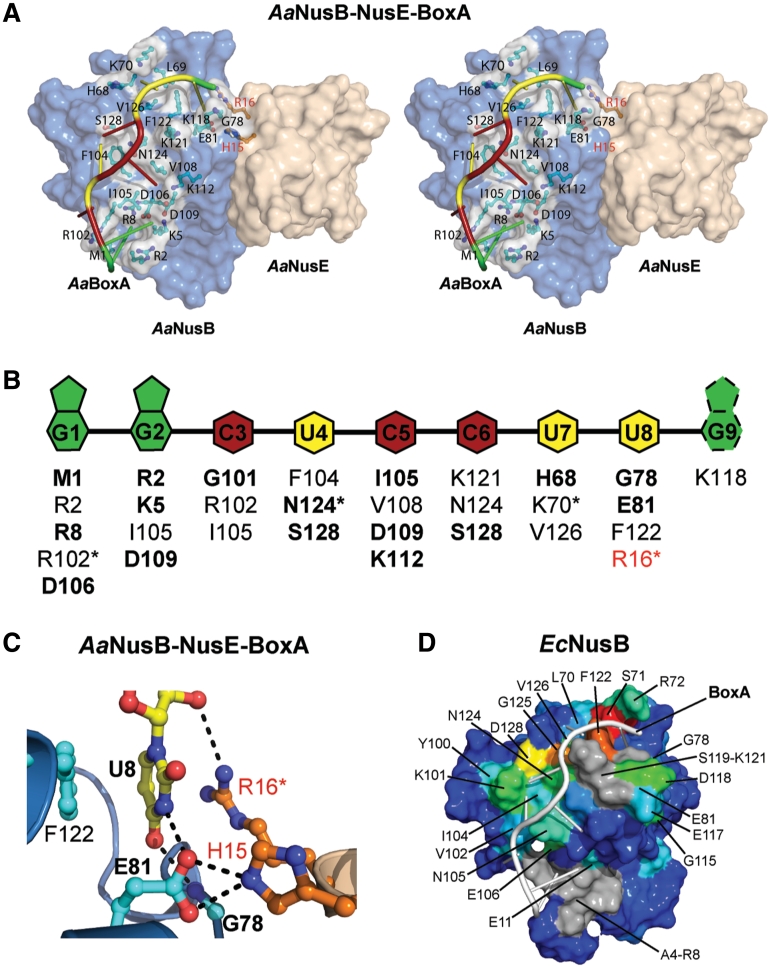
The AaBoxA RNA demonstrates extensive base-specific interactions with protein, beginning at the N-terminal amino group of AaNusB and ending at the AaNusB–NusE interface (Figure 2 and Supplementary Figure S2). These contacts include both hydrogen bonds and hydrophobic interactions with bases of AaBoxA nucleotides 1–8. The AaBoxA–protein interface comprises at least 30 interactions with an average of 3–4 interactions per nucleotide, but all protein–RNA interactions except those involving AaBoxA-U8 are specific to AaNusB alone (Figure 2B and C). The direct contribution to AaBoxA binding by AaNusE is observed in the AaBoxA-U8 nucleotide binding site formed by the AaNusB–NusE protein–protein interface (Figure 2A and C). Within this binding pocket, AaBoxA-U8 is held tightly in place through hydrophobic stacking interactions with both faces of the uracil ring, provided by AaNusB-F122 and AaNusE-R16. The guanidinium group of R16 also donates a hydrogen bond directly to the 2′-O of the AaBoxA-U8 ribose. Aside from spatial constraints, base-specificity of AaBoxA-U8 is achieved through two hydrogen bonds: one, which exists between the backbone NH of AaNusB-G78 and O4 of the uracil base and the other between endocyclic N3 and AaNusB-E81. AaNusE-H15 donates a hydrogen bond that is bifurcated by the carboxylate oxygen atoms of AaNusB-E81, thereby stabilizing the E81 side chain in a conformation favorable for AaBoxA-U8 recognition. Together, these contributions by AaNusE enhance AaBoxA affinity and specificity. AaNusB-K118 also binds to this region of AaBoxA through ionic interaction with the phosphate group of AaBoxA-G9 (Supplementary Figure S2). However, information pertaining to the ribose and nucleobase moieties of AaBoxA-G9 is inconclusive due to crystallographic disorder as well as differences between the two complexes of AaNusB–NusE–BoxA observed in the crystal structure (Supplementary Figures S1 and S2). NMR data of a complex of EcNusB and EcBoxA also reveal interaction between EcBoxA nucleotide 9 and amino acid residues near EcNusB-118, yet these data do not reveal a single interacting conformation (Supplementary Figure S3E). We believe, therefore, that the relevant BoxA nucleotides do not extend beyond position 8.
Figure 2.
The BoxA binding site is highly sequence-specific. (A) Surface representation in stereo of the AaNusB–NusE–BoxA ternary complex illustrating all protein-BoxA interactions. AaNusB-contributing residues are shown in cyan and AaNusE residues in orange as ball-and-stick models. AaBoxA nucleotide bases are color-coded by base type according to the schematic in panel B. (B) Schematic of interactions between AaBoxA and AaNusB–NusE residues as observed in the crystal structure. Amino acid residues in bold are involved in nucleobase specificity. Asterisks denote residues interacting with 2′-OH groups of RNA. Residues labeled in red belong to AaNusE. The base of AaBoxA-G9 is shown with dotted lines to indicate that it is disordered. (C) Details of AaBoxA-U8 interactions with AaNusB–NusE at the heterodimer interface. (D) Surface representation of the chemical shift map of EcNusB upon binding of EcBoxA, presented on the solution structure of EcNusB (PDB ID: 1EY1) with EcBoxA (white) positioned as in the AaNusB–NusE–BoxA structure. The residues are colored according to the magnitude of Δδ (EcNusB subtracted from EcNusB–BoxA) ranging from ∼0 ppm (blue) to 1.2 ppm (red) (Supplementary Figure S3C). Differences between the chemical-shift-mapped EcBoxA binding site of EcNusB, relative to that observed in the Aa crystal structure, are primarily at the N-terminus. Residues A4-R8 (gray) are disordered in the free EcNusB protein and thus no Δδ could be calculated. However, the N-terminal resonances are observed in the EcNusB–BoxA complex, suggesting that this region is stabilized by interactions with EcBoxA. Similarly, exchange-broadened residues S119-K121, also for which no Δδ could be calculated, are shown in gray.
The NusB-NusE interface is conserved across A. aeolicus and E. coli
The comparison between the crystal structures of AaNusB–NusE–BoxA (this work) and EcNusB–NusE (4) shows that the r.m.s.d. for the NusB components, the NusE components and the NusB–NusE complexes are 1.5, 0.8 and 1.7 Å, respectively. The structures show that 6 out of 11 NusB and all 10 NusE residues involved in NusB–NusE heterodimer formation are conserved between the two species, with buried surface areas of 1620 and 1626 Å2 for the Aa and Ec complexes, respectively. The NusB–NusE interface is dominated by hydrophobic interactions and assisted by additional electrostatic interactions between the two proteins. Almost all interactions can be complemented in a cross-species NusB–NusE complex, indicating that the formation of such a complex is possible. Our genetic complementation experiments of AaNusB in E. coli show that an AaNusB–EcNusE complex is indeed formed in vivo (vide infra).
NMR chemical-shift mapping reveals a similar BoxA binding site for EcNusB
The BoxA binding site on EcNusB was determined using NMR chemical-shift differences (Δδ) between the free (29) and EcBoxA-bound states of EcNusB (Figure 2D and Supplementary Figure S3C). The Δδ shift map reveals a contiguous EcBoxA binding surface spanning part of helix α6 (K101-E106), residues E117 and D118 in the loop connecting helices α6 and α7, and most of helix α7 (S119-D128) (Figures 1A and 2D). The largest Δδ are observed near F122, which emphasizes the significance of this conserved amino acid for BoxA recognition. Additionally, all amino acid residues in the EcNusB loop containing Y69-G78 show changes in chemical shift upon EcBoxA binding.
The orientation of EcBoxA on the EcNusB surface was determined using Δδ values obtained upon binding of EcBoxA mutants U1C and A9C. EcNusB-D118N was employed in these NMR experiments as this mutant binds EcBoxA with higher affinity in the absence of EcNusE (30). In the D118N/BoxA-U1C spectrum, small Δδ (∼0.1 ppm) are observed for residues L74, E81, K101, N105, I134 and K139, with the largest Δδ for K101 and K139 (Supplementary Figure S3D). In the D118N/BoxA-A9C spectrum, small Δδ (∼0.1 ppm) are observed for I27, R72, E106, A116, D117 and N118 NH’s (Supplementary Figure S3E). These data indicate that the 3′-end of EcBoxA is near the N-terminus of EcNusB helix α6, with nucleotides 8 and 9 near the A116-D118 loop. The EcBoxA binding site on EcNusB, as well as the orientation of EcBoxA on the EcNusB surface, is consistent with the AaNusB–NusE–BoxA crystal structure (Figure 2A and D).
NusB-F122 is important for both cellular and λN-mediated antitermination
NusB is involved in both cellular and phage λ functions in E. coli, being conditionally required for cell growth at low temperatures and λ growth at 42°C (31). Therefore, EcΔnusB cells possess a characteristic cold-sensitive growth phenotype at temperatures below 30°C. Compared to wild-type (wt) Ec cells (nusB+), the EcΔnusB cells grow slower at 37°C, but at a comparable rate at 42°C. However, the EcΔnusB cells do not support formation of wt λ (λ+) plaques during phage infection at 42°C. These two functions of NusB can be efficiently restored through trans-expression of wt EcNusB from a plasmid and is the molecular basis for genetic complementation (28).
The BoxA binding site on NusB is a region of high phylogenetic conservation, particularly along its C-terminal α-helix (α7) (29). As seen in the AaNusB–NusE–BoxA structure, the semi-exposed and conserved aromatic residue in α7, NusB-F122, exhibits ring stacking with the U8 nucleobase (Figure 2C). In order to test its importance for NusB function, EcNusB-F122 was mutated to aspartic acid and analyzed in vivo for its ability to complement EcΔnusB cells in the support of E. coli and phage λ growth (Supplementary Table S1).
The EcnusB-F122D mutant is defective as compared to EcnusB+, which is demonstrated by its cold-sensitivity and inability to promote λ growth (Supplementary Table S1). The mutant protein is correctly folded, as shown by minimal differences between the NMR spectra (Supplementary Figure S4A and B) and similar retention times on a size exclusion column as wt EcNusB. The wt EcNusB binds to EcBoxA in a 1:1 complex with a Kd = 2 µM (Supplementary Table S2). However, EcNusB-F122D did not detectably bind EcBoxA as measured by either ITC or NMR when present in a 1 : 1 ratio. It remains to be determined how this single point mutation, which would potentially result in a minimal loss of protein–RNA interactions, can abolish binding of BoxA. Nevertheless, it is the only known EcnusB missense mutation resulting in defects in both cellular and phage λ antitermination activities. Two previously known missense mutations, EcnusB5 (32) and EcnusB101 (33–35) show effects mostly in phage λ antitermination activities, and the effect of EcnusB101 is only observed in conjunction with EcnusA or EcnusE mutations, being otherwise neutral (33).
Complementation by AaNusB in E. coli is BoxA-sequence dependent
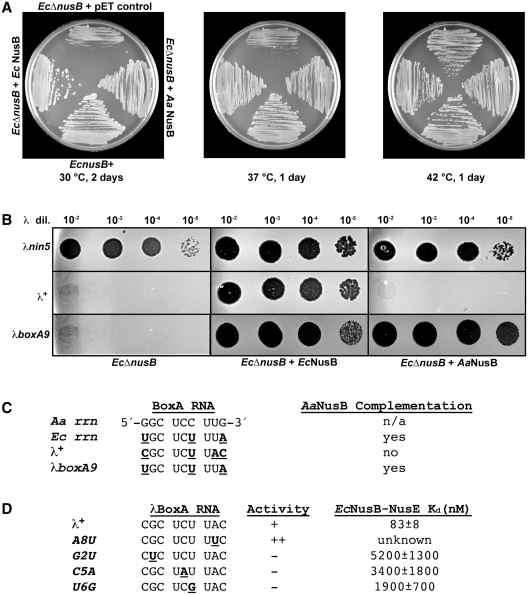
The functional homology between the Aa and EcNusB proteins has been assessed in a genetic complementation assay using EcΔnusB cells. Once again, the cellular and λ growth defects of the EcΔnusB cells can be compensated by the trans-expression of functional EcNusB from a plasmid (28) (Figure 3A and B). Unlike cells transformed with the control vector, cells transformed with the EcNusB plasmid grow normally at 30 and 37°C. Under the same growth conditions, the expression of AaNusB from plasmid DNA also restores the growth of EcΔnusB cells at 30 and 37°C. AaNusB is therefore capable of complementing EcNusB in the context of the protein’s cellular functions, most likely through productive interactions with EcBoxA and EcNusE during rRNA synthesis. This interpretation was validated using ITC, which revealed that EcBoxA binds to AaNusB (Supplementary Table S2).
Figure 3.
Complementation by AaNusB in E. coli is BoxA-sequence dependent. (A) Growth complementation assay of EcΔnusB cells with NusB expressed from plasmid DNA. The EcΔnusB cells were transformed with plasmids expressing EcNusB (pAB47), AaNusB (pKM772) or with the control pET vector. The growth of transformants was measured by incubating cells on L medium containing 50 µg/ml ampicillin at 30°C for 2 days, or at 37 or 42°C for 1 day (Supplementary Table S1). (B) Plaque assays demonstrating the complementation of nusB-deficient λ growth by trans-expression of Ec or AaNusB from plasmid DNA. By contrast to phage λ+, λnin5 does not require NusB for phage growth and was thus used as a positive control for plaque formation. A mutant derivative of λ, λboxA9, has three base changes that create a BoxA RNA that is identical to EcBoxA. It gains the ability to plaque and illustrates that complementation by AaNusB is dependent on specific NusB-BoxA interactions. (C) Comparison of BoxA RNA sequences for Aa, Ec, λ+ and λboxA9. Nucleotides in underlined boldface indicate differences in BoxA sequences relative to that of AaBoxA. (D) Other λ derivatives with boxA mutations that affect λN-dependent antitermination activity (indicated in underlined boldface) and their reported Kd values (30). Plaquing ability of λ carrying these mutations is shown. The ++ indicates the enhanced ability to make plaques by the A8U mutant (52).
Contrasting results for genetic complementation are observed in the λ-related activities of EcNusB. Here, λnin5 (36) is used as a control, as it is known to form plaques even in the absence of EcNusB (37). The λ+ infection of EcΔnusB cells expressing EcNusB from a plasmid generates plaques, whereas, in cells expressing AaNusB from a plasmid, no plaques are generated at 42°C (Figure 3B). By contrast, AaNusB is capable of supporting growth of a phage λ mutant, λboxA9, whose BoxA is identical to EcBoxA (Figure 3C), thus indicating that the AaNusB complementation is BoxA-sequence dependent.
The crystal structure of AaNusB–NusE–BoxA provides a molecular basis for the differences observed for AaNusB complementation of Ec cellular versus λN-mediated functions. The rrn BoxA sequences of E. coli and A. aeolicus differ at positions 1, 6 and 9 (Figure 3C). Structural analysis suggests that two of these differences are likely to be insignificant. A BoxA-C6U substitution would have minimal effects on binding, as the interaction between O2 of the pyrimidine base with AaNusB-S128 would be conserved (Supplementary Figure S2). A substitution from N4 to O4 on the pyrimidine ring would result in the loss of a hydrogen bond with phosphate O2 of AaBoxA-C5, and require slight changes in atomic position to avoid electrostatic repulsion between the two oxygen atoms. Similarly, a BoxA-G9A substitution would likely be tolerated, as this nucleotide is a purine in both cases and AaBoxA-G9 exhibits no clear base-specific interactions with NusB or NusE proteins. Therefore, the only difference in the rrn BoxA sequences of E. coli and A. aeolicus that would have structural implications for binding is at position 1. As observed in the crystal structure, AaBoxA-G1 participates in several hydrogen bonds with AaNusB that would be lost in the case of EcBoxA-U1 (Figure 2 and Supplementary Figure S2). Nevertheless, the sum of these changes has very little effect on AaNusB binding and function in E. coli (Figure 3A). The greater differences between Aa and λBoxA sequences have greater structural implications, by which the latter is incapable of binding to AaNusB (Supplementary Table S2). Once again, a BoxA-C6U substitution would likely have little or no effect (vide supra). However, a transversion substitution, BoxA-G1C, when accompanied by two other transversion substitutions, BoxA-U8A and G9C (Figure 3C), would lead to the disruption of several hydrogen bonds, resulting in significantly reduced binding energy between λBoxA and the AaNusB–EcNusE complex. AaNusB residues in the BoxA-binding site that are not conserved in EcNusB may cause further disruption of RNA–protein interactions.
Taken together, the major structural difference at position 1 of EcBoxA is tolerated by AaNusB–EcNusE with respect to binding and cellular growth, whereas the added differences at positions 8 and 9 of λBoxA are not tolerated. How EcNusB–NusE tolerates the transversion substitutions at positions 8 and 9 of λBoxA remains to be established. However, previous studies have shown a weaker EcNusB–NusE binding affinity for λBoxA relative to EcBoxA (15,17,30), most likely caused by forcing a bulkier adenine base into the EcBoxA-U8 binding pocket shaped by the EcNusB–NusE interface. This is further supported by the fact that the EcBoxA- and λBoxA binding affinities for NusB alone are virtually the same (Supplementary Table S2). Hence, the absence of the binding pocket results in less nucleotide discrimination.
Structural bases for observed mutant phenotypes
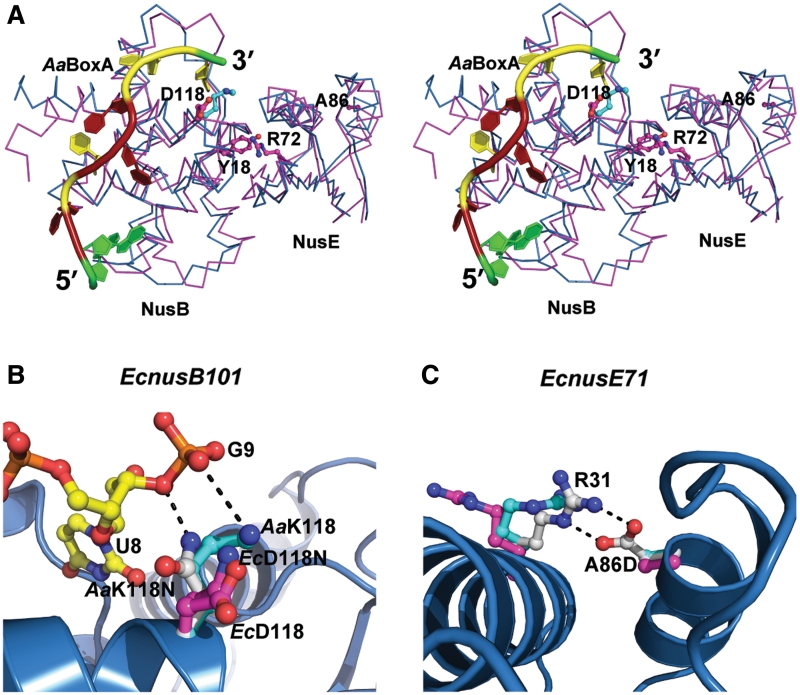
Previous studies have identified mutations in both EcNusB and EcNusE, as well as mutations in λBoxA that affect the efficiency of antitermination. Two of these mutants, EcnusB5 (32) and EcnusE100 (38), whose single point mutations were identified as EcNusB-Y18D (35) and EcNusE-R72G, respectively, likely result in the disruption of the EcNusB–NusE heterodimer, as both of these amino acid side chains are buried deep within the protein–protein interface (4,18). The distant location of these residues in relation to the BoxA binding site suggests that they have little effect on BoxA binding (Figure 4A). Indeed, EcNusB-Y18D maintains EcBoxA-binding affinity relative to wt EcNusB and is otherwise a folded, monomeric protein (Supplementary Table S2 and Supplementary Figure S4C).
Figure 4.
Structural bases for EcNusB and EcNusE mutant phenotypes. (A) Stereoscopic view shows the Cα-trace superposition of EcNusB–NusE (PDB ID: 3D3B, in magenta) and AaNusB–NusE–BoxA (this work, in cyan). Also shown are BoxA as a cartoon (colored as in Figure 2) and side chains of amino acid residues whose mutational effects have been previously reported, as ball-and-sticks. (B and C) Structural bases for the EcnusB101 and EcnusE71 mutants through side-chain modeling. The AaNusB–NusE–BoxA (this work), EcNusB–NusE (PDB ID: 3D3B), and EcNusB101–NusE (PDB ID: 3IMQ) structures are superimposed. For AaNusB–NusE–BoxA, polypeptide backbones are shown as cartoons (helices as spirals, strands as arrows and loops as tubes) in blue, and nucleotides and amino acid side chains as ball-and-sticks in atomic colors (N in blue, O in red, P in orange, C of RNA in yellow and C of protein in cyan). For EcNusB–NusE and EcNusB101–NusE, polypeptide backbones are not shown for clarity and amino acid side chains are shown as ball-and-sticks but C in magenta. Modeled amino acid side chains are shown as ball-and-sticks but C in white. Potential interactions are shown as dashed lines in black.
The structural bases for EcNusB-D118N and EcNusE-A86D phenotypes (35), caused respectively by the EcnusB101 (33–35) and EcnusE71 (3) mutations, are much less obvious. EcNusB-D118N and EcNusB-D118K (30) are compensatory mutants that rescue λ growth defects induced by a mutation in EcNusA (EcnusA1), most likely by enhancing λBoxA affinity for EcNusB-NusE via interaction with asparagine or lysine side chains, which is not possible in the case of the native aspartic acid. Indeed, the crystal structure of AaNusB–NusE–BoxA, which naturally contains a lysine at AaNusB position 118, exhibits contacts between AaNusB-K118 and AaBoxA-G9. Likewise, modeling of AaNusB-K118N reveals favorable interaction potential between the asparagine side chain and the phosphate group of AaBoxA-G9 (Figure 4A and B).
By contrast, the addition of these few interactions provided by lysine or asparagine at this position, do not significantly improve binding to EcBoxA. Previous in vitro studies using fluorescence anisotropy titrations showed that the EcNusB-D118N and D118K mutants have enhanced EcNusB–NusE binding affinity for λBoxA but not for EcBoxA (30). Further supporting evidence is provided by experiments with λimm21 (35), whose BoxA RNA is identical to EcBoxA, demonstrating that EcNusB-D118N rescues the growth of phage λ, but not λimm21, in E. coli cells carrying the nusA1 mutation. We propose, therefore, that the selective binding enhancement of the EcNusB-D118K and D118N mutants for λBoxA results from the transversion substitutions at positions 8 and 9 (Figure 3C), in which the additional interactions with BoxA generated by these rescue mutations are of greater impact in the case of λBoxA and are required to rescue phage growth. However, further structural investigation of RNA–protein interactions is necessary, particularly for λBoxA, in order to support this claim.
The EcNusE-A86D mutation, which is far from both the heterodimer interface and the BoxA binding site (Figure 4A), most likely plays another role. An aspartate in this position can be modeled within hydrogen-bonding distance of a conserved arginine (NusE–R31) that is solvent exposed (Figure 4C). In the structure of AaNusB–NusE–BoxA, NusE–R31 forms crystal contacts with the second copy of AaNusB. This may indicate that EcNusE-R31 is involved in the binding of another protein in the antitermination complex, possibly RNAP (39), and that its interaction with EcNusE-A86D may weaken or preclude its native function. The introduction of a negative charge at this position may also directly disrupt interactions electrostatically. However, EcNusE-A86D, which inhibits growth of λ+, does not inhibit growth of λimm21 or Ec host cells (35). This suggests that any negative impact posed by EcNusE-A86D is compensated by the greater BoxA affinities in the λimm21 and E. coli cellular antitermination systems. Whereas, in the case of λ+, EcNusE-A86D necessitates a compensatory mutation (EcnusB101, EcNusB-D118N) in order to achieve the required λBoxA affinity.
Several phenotypes resulting from mutations in λBoxA have also been observed (Figure 3D). The mutation, λBoxA-A8U (39), actually enhances λN-mediated antitermination. Indeed, BoxA-U8, present in both Aa and EcBoxA sequences, binds tightly in the pocket formed by the NusB–NusE interface in the crystal structure of AaNusB–NusE–BoxA, and exhibits key base-specific interactions that are likely to be absent in the case of adenine (Figure 2C). Other mutations in λBoxA, including G2U, C5A and U6G, inhibit λN-mediated antitermination and have greatly reduced affinity for the EcNusB–NusE heterodimer (30), suggesting that transversion substitutions are not tolerated at λBoxA positions 2, 5 or 6 (Figure 3D). It is also evident from the crystal structure of AaNusB–NusE–BoxA that these conserved nucleotides, G2, C5 and C6 (AaBoxA) or G2, C5 and U6 (Ec and λBoxA), play significant roles in both cellular and λN-mediated antitermination activities, due to their extensive and specific interactions with NusB residues (Figure 2 and Supplementary Figure S2).
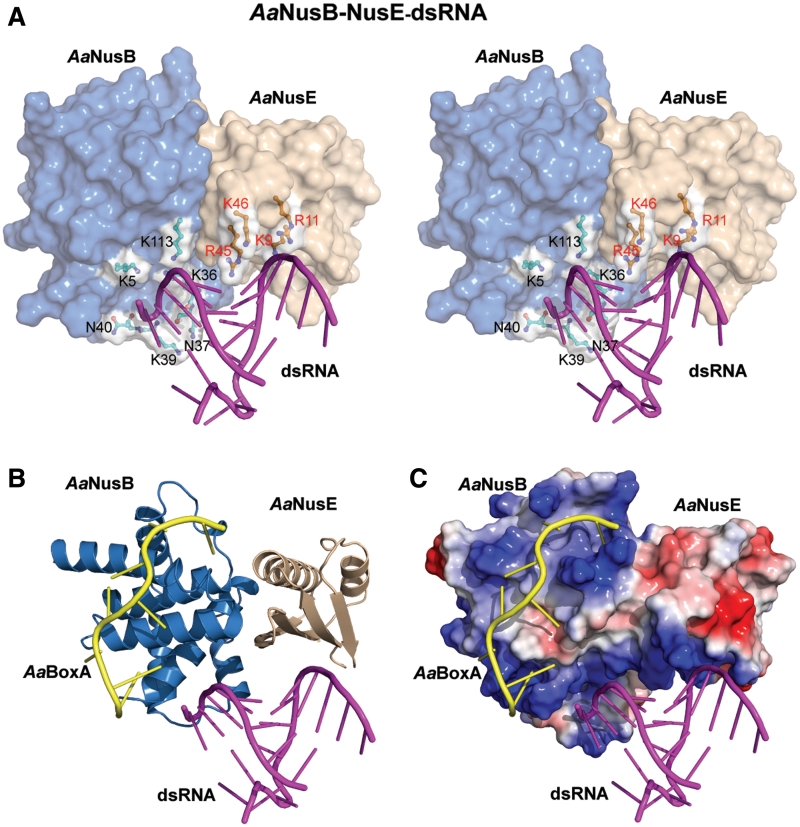
The AaNusB-NusE heterodimer forms a contiguous binding site for dsRNA
As aforementioned, the 12-mer RNA used in crystallization experiments forms a stable duplex at temperatures below 32°C. Room-temperature crystallization of the A. aeolicus ternary complex (Supplementary Figure S5) led to the discovery of a novel binding site for dsRNA, which is generated by the surface of the NusB–NusE heterodimer (Figure 5A), and is positioned directly adjacent to the BoxA binding site (details in Discussion). Although not as numerous as the interactions with BoxA, the strength of dsRNA binding by NusB–NusE is achieved through several electrostatic interactions between dsRNA backbone atoms and positively-charged protein side chains. The structure reveals six residues of AaNusB (K5, K36, N37, K39, N40 and K113) and four residues of AaNusE (K9, R11, R45 and K46) that contribute to the binding of dsRNA. As virtually all protein-dsRNA contacts are limited to the RNA backbone, the mode of binding is primarily sequence-independent (Figure 5A), with the exception of AaNusE-R45, which has additional interactions with a base pair. The four AaNusE residues are conserved in E. coli (Figure 1) and at least three out of the four (R11, R45 and K46) contribute to NusE/S10-dsRNA interaction within the 30 S ribosome subunit (PDB codes 2AVY and 3KIQ). Some of the dsRNA-binding residues of NusB, on the other hand, demonstrate weaker conservation, particularly D39, D42 and S113 (EcNusB). This may indicate a smaller, yet still relevant, contribution of EcNusB in the binding of dsRNA by EcNusB–NusE. Our NMR experiments indicate that EcNusB–dsRNA interaction is more dependent on the high-affinity interaction with EcBoxA (Supplementary Figure S6).
Figure 5.
Binding of dsRNA by the NusB–NusE heterodimer. AaNusB molecules are colored in blue, AaNusE molecules in wheat and dsRNA in magenta. Orientations in all panels are identical to that in Figure 2A. (A) Crystal structure of the AaNusB–NusE–dsRNA ternary complex in stereo. Residues involved in dsRNA binding are depicted as ball-and-sticks with white surface. (B and C) Superposition of the crystal structures of AaNusB–NusE–BoxA and AaNusB–NusE–dsRNA, illustrating the contiguous binding regions for AaBoxA and dsRNA. In panel C, the protein surface representation is colored by vacuum-electrostatic potential (red, negative; blue, positive), which depicts the AaBoxA and dsRNA binding sites as a contiguous positively-charged surface.
DISCUSSION
The phenomena of transcription termination and antitermination are best characterized in the λN-mediated system (40–42). Hence, the discussion will be focused on the λN-mediated transcription antitermination (For a discussion on other systems, see Supplementary Data). The λN protein, whose interaction with RNAP is facilitated by host proteins (NusA, NusB, NusE and NusG), binds to the λnut site in the nascent RNA (43). How λN assists RNAP to read through terminators remains unclear (44), but its site of action appears to be in proximity to the RNA:DNA hybrid of the transcription elongation complex (45). As mentioned previously, the assembly of the complete antitermination complex is required for processive antitermination and begins with the formation of the NusB–NusE–BoxA complex.
We have determined the crystal structure of the NusB–NusE–BoxA ternary complex from A. aeolicus, indentifying key protein–RNA interactions at atomic resolution. The equivalence of the EcBoxA and AaBoxA binding sites enables detailed interpretation of these findings with respect to the biologic functions of the NusB and NusE proteins. The structure shows that the BoxA sequence comprises 8 nt. The extensive interaction between NusB and BoxA illustrates not only the strength of binding energy (Supplementary Table S2), but also the significance of nucleotide-sequence specificity. The direct contribution of NusE to the binding of BoxA is limited, suggesting that the importance of NusE in the initiation of transcription antitermination is demonstrated by additional interactions with other antitermination complex components.
The NusB–NusE complex binds an additional RNA component not previously observed. We have shown that a binding site for dsRNA is generated by the formation of the NusB–NusE heterodimer, which is contiguous with the 5′-end of the BoxA binding site (Figure 5B and C). The binding of dsRNA involves primarily RNA-backbone interactions, indicating that this dsRNA binding site is independent of sequence, provided that canonical A-form RNA secondary structure is maintained. Nonetheless, some sequence specificity may be imposed by the conserved NusE-R45 to discriminate generic dsRNA species from the physiological ligand of the NusB–NusE heterodimer.
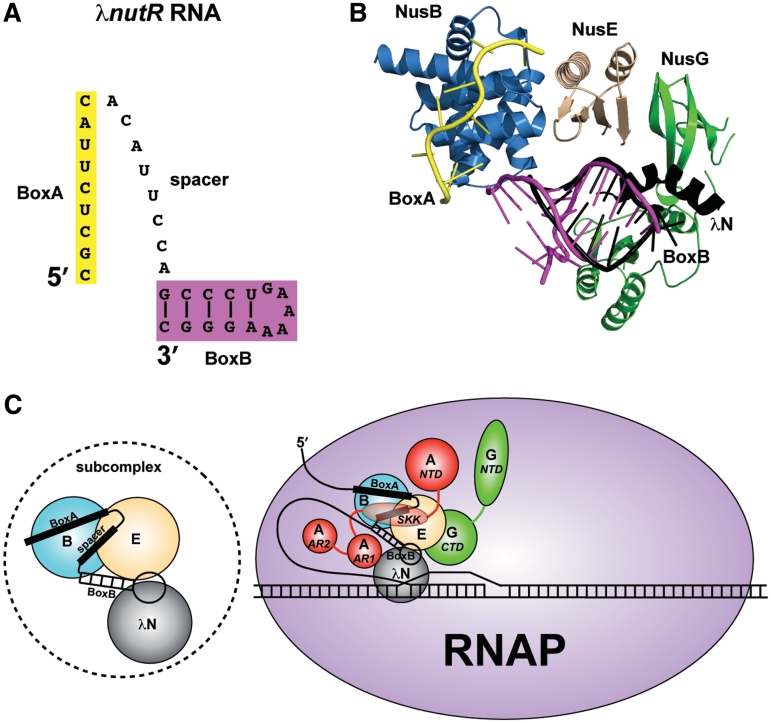
The only defined dsRNA component involved in λN-mediated antitermination, particularly within proximity to BoxA, is the stem-loop structure formed by BoxB (Figure 6A). We propose, therefore, that the dsRNA observed in these crystal structures may provide an accurate representation of the BoxB stem-loop and that NusB and NusE cooperatively form contiguous binding surfaces for both BoxA and BoxB components (Figures 5C and 6B). As previously discussed, the conserved NusE residues demonstrate similar dsRNA interactions within the ribosome. Thus, the dual functionality of NusE may involve, not only protein–protein interactions, but also conserved protein-dsRNA interactions: with rRNA in translation, with BoxB in antitermination.
Figure 6.
Models for the λN-mediated antitermination subcomplex and complete complex. (A) Representation of the λnutR RNA showing the possible spatial relationship between BoxA (yellow) and BoxB (magenta) components. (B) Superimposed components of the antitermination complex with known structures, including AaNusB–NusE–BoxA and AaNusB–NusE–dsRNA (this work), the N-terminal α-helix of λN protein bound to BoxB (PDB ID: 1QFQ, black), the C-terminal domain of EcNusG in its complex with EcNusE (PDB ID: 2KVQ, green), and the N-terminal domain of EcNusG (PDB ID: 2K06, green). The N- and C-terminal domains of EcNusG were positioned relative to one another by superimposing them with the N- and C-terminal domains, respectively, of AaNusG (PDB ID: 1NPP). (C) Left: the NusB–NusE–BoxA–spacer–BoxB–λN subcomplex; Right: updated model for the complete λN-mediated antitermination complex, illustrating all known interactions. Nus factors B (blue) and E (wheat) cooperatively bind BoxA and BoxB (this work). λN protein (gray) binds BoxB (8–11) and also possibly NusE (this work). λN protein binds RNAP near the transcription bubble (45). NusA (red) stabilizes these interactions by interacting with the λnut spacer RNA between BoxA and BoxB (12), and with λN protein (51) through NusA SKK (S1-KH1-KH2) and AR1 domains, respectively, while anchoring the complex to RNAP through the NusA NTD and AR2 domains (48–50). NusG (green) further stabilizes the complex through its CTD interaction with NusE (5), also serving as an anchor to RNAP through its NTD (53).
It is known that the BoxB stem loop binds to phage λN protein (8–11). To address the implications of this model in the λ system, the complexes presented in this work were structurally aligned with an NMR structure of the BoxB stem-loop bound to the N-terminal α-helix of λN protein (PDB ID: 1QFQ) (11). The BoxB structure aligns with the dsRNA observed in this work with near precision, and positions the λN α-helix within close proximity to AaNusE without steric clash (Figure 6B). Previous studies have shown that the presence of NusB and NusE increases the efficiency of a λN-like protein (Nun) to modify transcription elongation (46), which, by this model, would result from the formation of a stable NusB–NusE–BoxA–BoxB complex that enables more effective recruitment of λN/Nun protein.
It is indicated that NusG promotes anchoring of a subcomplex, which we suggest to include NusB, NusE, BoxA, BoxB and λN protein (Figure 6C, left), to RNAP through its interactions with NusE (5). Superposition of previous structures of NusG (47) and the NusE–NusG complex (5), with the combined crystal structures presented in this work, is shown in Figure 6B. Association of NusA with RNAP (48–50) aids in the recruitment of the subcomplex through NusA interactions with λN protein (51) and the λnut spacer RNA (12), which is located between BoxA and BoxB. Together, the network of interactions implies a highly compact quaternary structure for the complete antitermination complex (Figure 6C, right). These findings demonstrate the complexity of structural interplay among various components of the antitermination machinery, and provide a framework for studying the underlying molecular mechanisms.
ACCESSION NUMBERS
The coordinates and structure factors for the AaNusB-NusE–BoxA and AaNusB–NusE–dsRNA complexes have been deposited in the Protein Data Bank as PDB entries 3R2C and 3R2D. The sequential backbone and side-chain assignments of the EcNusB–BoxA complex have been deposited in the Biological Magnetic Resonance Bank as BMRB entry 17526.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research; a Trans NIH/FDA Intramural Biodefense Program Grant from National Institute of Allergy and Infectious Diseases (to D.L.C.) and federal funds from the National Cancer Institute, National Institutes of Health (N01-CO-12400). Funding for open access charge: The National Institutes of Health Intramural Research Program.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jennifer Briggs and Aaren King for EcNusB mutant protein preparation; Mikhail Kashlev, Yun-Xing Wang and Alexander Wlodawer for discussion and Paul Wingfield for analytical ultracentrifugation data. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
REFERENCES
- 1.Friedman DI, Court DL. Transcription antitermination: the lambda paradigm updated. Mol. Microbiol. 1995;18:191–200. doi: 10.1111/j.1365-2958.1995.mmi_18020191.x. [DOI] [PubMed] [Google Scholar]
- 2.Brigati C, Giacca M, Noonan DM, Albini A. HIV Tat, its TARgets and the control of viral gene expression. FEMS Microbiol. Lett. 2003;220:57–65. doi: 10.1016/S0378-1097(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 3.Friedman DI, Schauer AT, Baumann MR, Baron LS, Adhya SL. Evidence that ribosomal protein S10 participates in control of transcription termination. Proc. Natl Acad. Sci. USA. 1981;78:1115–1118. doi: 10.1073/pnas.78.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo X, Hsiao HH, Bubunenko M, Weber G, Court DL, Gottesman ME, Urlaub H, Wahl MC. Structural and functional analysis of the E. coli NusB-S10 transcription antitermination complex. Mol. Cell. 2008;32:791–802. doi: 10.1016/j.molcel.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch P. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–504. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 6.Mason SW, Li J, Greenblatt J. Direct interaction between two Escherichia coli transcription antitermination factors, NusB and ribosomal protein S10. J. Mol. Biol. 1992;223:55–66. doi: 10.1016/0022-2836(92)90715-v. [DOI] [PubMed] [Google Scholar]
- 7.Luttgen H, Robelek R, Muhlberger R, Diercks T, Schuster SC, Kohler P, Kessler H, Bacher A, Richter G. Transcriptional regulation by antitermination. Interaction of RNA with NusB protein and NusB/NusE protein complex of Escherichia coli. J. Mol. Biol. 2002;316:875–885. doi: 10.1006/jmbi.2001.5388. [DOI] [PubMed] [Google Scholar]
- 8.Legault P, Li J, Mogridge J, Kay LE, Greenblatt J. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell. 1998;93:289–299. doi: 10.1016/s0092-8674(00)81579-2. [DOI] [PubMed] [Google Scholar]
- 9.Mogridge J, Legault P, Li J, Van Oene MD, Kay LE, Greenblatt J. Independent ligand-induced folding of the RNA-binding domain and two functionally distinct antitermination regions in the phage lambda N protein. Mol. Cell. 1998;1:265–275. doi: 10.1016/s1097-2765(00)80027-1. [DOI] [PubMed] [Google Scholar]
- 10.Cilley CD, Williamson JR. Analysis of bacteriophage N protein and peptide binding to boxB RNA using polyacrylamide gel coelectrophoresis (PACE) RNA. 1997;3:57–67. [PMC free article] [PubMed] [Google Scholar]
- 11.Scharpf M, Sticht H, Schweimer K, Boehm M, Hoffmann S, Rosch P. Antitermination in bacteriophage lambda. The structure of the N36 peptide-boxB RNA complex. Eur. J. Biochem. 2000;267:2397–2408. doi: 10.1046/j.1432-1327.2000.01251.x. [DOI] [PubMed] [Google Scholar]
- 12.Prasch S, Jurk M, Washburn RS, Gottesman ME, Wohrl BM, Rosch P. RNA-binding specificity of E. coli NusA. Nucleic Acids Res. 2009;37:4736–4742. doi: 10.1093/nar/gkp452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan EA. Antitermination mechanisms in rRNA operons of Escherichia coli. J. Bacteriol. 1986;168:1–5. doi: 10.1128/jb.168.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeVito J, Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc. Natl Acad. Sci. USA. 1994;91:8660–8664. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greive SJ, Lins AF, von Hippel PH. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J. Biol. Chem. 2005;280:36397–36408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- 16.Mogridge J, Mah TF, Greenblatt J. Involvement of boxA nucleotides in the formation of a stable ribonucleoprotein complex containing the bacteriophage lambda N protein. J. Biol. Chem. 1998;273:4143–4148. doi: 10.1074/jbc.273.7.4143. [DOI] [PubMed] [Google Scholar]
- 17.Nodwell JR, Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–268. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 18.Das R, Loss S, Li J, Waugh DS, Tarasov S, Wingfield PT, Byrd RA, Altieri AS. Structural biophysics of the NusB:NusE antitermination complex. J. Mol. Biol. 2008;376:705–720. doi: 10.1016/j.jmb.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 23.DeLano WL. Delano Scientific. CA: San Carlos; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 24.Altieri AS, Mazzulla MJ, Zhou HJ, Costantino N, Court DL, Byrd RA. Sequential assignments and secondary structure of the RNA-binding transcriptional regulator NusB. FEBS Lett. 1997;415:221–226. doi: 10.1016/s0014-5793(97)01128-9. [DOI] [PubMed] [Google Scholar]
- 25.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Mag. Res. Sp. 1999;34:93–158. [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe - a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct. Funct. Genet. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 28.Bubunenko M, Baker T, Court DL. Essentiality of ribosomal and transcription antitermination proteins analyzed by systematic gene replacement in Escherichia coli. J. Bacteriol. 2007;189:2844–2853. doi: 10.1128/JB.01713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altieri AS, Mazzulla MJ, Horita DA, Coats RH, Wingfield PT, Das A, Court DL, Byrd RA. The structure of the transcriptional antiterminator NusB from Escherichia coli. Nat. Struct. Biol. 2000;7:470–474. doi: 10.1038/75869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burmann BM, Luo X, Rosch P, Wahl MC, Gottesman ME. Fine tuning of the E. coli NusB:NusE complex affinity to BoxA RNA is required for processive antitermination. Nucleic Acids Res. 2010;38:314–326. doi: 10.1093/nar/gkp736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swindle J, Ajioka J, Dawson D, Myers R, Carroll D, Georgopoulos C. The nucleotide sequence of the Escherichia coli K12 nusB (groNB) gene. Nucleic Acids Res. 1984;12:4977–4985. doi: 10.1093/nar/12.12.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman DI, Baumann M, Baron LS. Cooperative effects of bacterial mutations affecting lambda N gene expression. I. Isolation and characterization of a nusB mutant. Virology. 1976;73:119–127. doi: 10.1016/0042-6822(76)90066-0. [DOI] [PubMed] [Google Scholar]
- 33.Ward DF, DeLong A, Gottesman ME. Escherichia coli nusB mutations that suppress nusA1 exhibit lambda N specificity. J. Mol. Biol. 1983;168:73–85. doi: 10.1016/s0022-2836(83)80323-4. [DOI] [PubMed] [Google Scholar]
- 34.Patterson TA, Zhang Z, Baker T, Johnson LL, Friedman DI, Court DL. Bacteriophage lambda N-dependent transcription antitermination. Competition for an RNA site may regulate antitermination. J. Mol. Biol. 1994;236:217–228. doi: 10.1006/jmbi.1994.1131. [DOI] [PubMed] [Google Scholar]
- 35.Court DL, Patterson TA, Baker T, Costantino N, Mao X, Friedman DI. Structural and functional analyses of the transcription-translation proteins NusB and NusE. J. Bacteriol. 1995;177:2589–2591. doi: 10.1128/jb.177.9.2589-2591.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Court D, Sato K. Studies of novel transducing variants of lambda - dispensability of genes N and Q. Virology. 1969;39:348–352. doi: 10.1016/0042-6822(69)90060-9. [DOI] [PubMed] [Google Scholar]
- 37.Friedman DI, Schauer AT, Olson ER, Carver DL, Eades LJ, Bigelow B. Proteins and nucleic acid sequences involved in regulation of gene expression by the bacteriophage X N transcription antitermination function. Microbiology. 1985:271–276. [Google Scholar]
- 38.Robledo R, Atkinson BL, Gottesman ME. Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. J. Mol. Biol. 1991;220:613–619. doi: 10.1016/0022-2836(91)90104-e. [DOI] [PubMed] [Google Scholar]
- 39.Mason SW, Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991;5:1504–1512. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- 40.Roberts JW. Termination factor for RNA synthesis. Nature. 1969;224:1168–1174. doi: 10.1038/2241168a0. [DOI] [PubMed] [Google Scholar]
- 41.Krebs JE, Goldstein ES, Lewin B, Kilpatrick ST. Lewin’s Essential Genes. 2nd edn. Massachusetts: Jones and Bartlett Publishers, Sudbury; 2010. Antitermination may be a regulated event; pp. 287–291. [Google Scholar]
- 42.Weisberg RA, Gottesman ME. Processive antitermination. J. Bacteriol. 1999;181:359–367. doi: 10.1128/jb.181.2.359-367.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 44.Vieu E, Rahmouni AR. Dual role of boxB RNA motif in the mechanisms of termination/antitermination at the lambda tR1 terminator revealed in vivo. J. Mol. Biol. 2004;339:1077–1087. doi: 10.1016/j.jmb.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 45.Cheeran A, Kolli NR, Sen R. The site of action of the antiterminator protein N from the lambdoid phage H-19B. J. Biol. Chem. 2007;282:30997–31007. doi: 10.1074/jbc.M704864200. [DOI] [PubMed] [Google Scholar]
- 46.Hung SC, Gottesman ME. Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J. Mol. Biol. 1995;247:428–442. doi: 10.1006/jmbi.1994.0151. [DOI] [PubMed] [Google Scholar]
- 47.Knowlton JR, Bubunenko M, Andrykovitch M, Guo W, Routzahn KM, Waugh DS, Court DL, Ji X. A spring-loaded state of NusG in its functional cycle is suggested by X-ray crystallography and supported by site-directed mutants. Biochemistry. 2003;42:2275–2281. doi: 10.1021/bi0272508. [DOI] [PubMed] [Google Scholar]
- 48.Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J. Mol. Biol. 1991;220:307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 49.Gill SC, Yager TD, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. II. Physical properties and self-association states. J. Mol. Biol. 1991;220:325–333. doi: 10.1016/0022-2836(91)90016-y. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Hanna MM. NusA changes the conformation of Escherichia coli RNA polymerase at the binding site for the 3′ end of the nascent RNA. J. Bacteriol. 1994;176:1787–1789. doi: 10.1128/jb.176.6.1787-1789.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonin I, Muhlberger R, Bourenkov GP, Huber R, Bacher A, Richter G, Wahl MC. Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda. Proc. Natl Acad. Sci. USA. 2004;101:13762–13767. doi: 10.1073/pnas.0405883101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedman DI, Olson ER. Evidence that a nucleotide sequence, “boxA,” is involved in the action of the NusA protein. Cell. 1983;34:143–149. doi: 10.1016/0092-8674(83)90144-7. [DOI] [PubMed] [Google Scholar]
- 53.Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J. Mol. Biol. 2009;391:341–358. doi: 10.1016/j.jmb.2009.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.