Abstract
The development of new methods for gene addition to mammalian genomes is necessary to overcome the limitations of conventional genetic engineering strategies. Although a variety of DNA-modifying enzymes have been used to directly catalyze the integration of plasmid DNA into mammalian genomes, there is still an unmet need for enzymes that target a single specific chromosomal site. We recently engineered zinc-finger recombinase (ZFR) fusion proteins that integrate plasmid DNA into a synthetic target site in the human genome with exceptional specificity. In this study, we present a two-step method for utilizing these enzymes in any cell type at randomly-distributed target site locations. The piggyBac transposase was used to insert recombinase target sites throughout the genomes of human and mouse cell lines. The ZFR efficiently and specifically integrated a transfected plasmid into these genomic target sites and into multiple transposons within a single cell. Plasmid integration was dependent on recombinase activity and the presence of recombinase target sites. This work demonstrates the potential for broad applicability of the ZFR technology in genome engineering, synthetic biology and gene therapy.
INTRODUCTION
Technologies for introducing gene sequences into mammalian cells are central to numerous applications in medicine, biopharmaceutical production and mechanistic studies of gene function. Similarly, the burgeoning fields of synthetic biology and metabolic engineering are founded on complex genetic engineering of cell systems. Ideally, these applications would involve the addition of genes to specific sites in the genome that facilitate desirable gene expression characteristics and minimize aberrant effects on the cell. However, current methods for chromosomal gene addition use viral delivery vehicles or DNA-modifying enzymes that integrate DNA sequences semi-randomly in the billions of base pairs of mammalian genomes. This approach has the potential to disrupt endogenous gene sequences that leads to unpredictable consequences on cell activity (1). Additionally, isogenic cell lines must be clonally derived after gene addition to ensure robust and uniform levels of gene expression across the cell population. Methods for reproducibly integrating genes at specific genomic target sites would overcome these challenges and enable robust genome manipulation for diverse fields of biotechnology and biological research.
Retroviral and lentiviral vectors are the conventional delivery vehicles for gene addition to mammalian genomes. These vectors integrate semi-randomly into the genome with a preference for promoters or intragenic regions of actively transcribed genes (2,3). In several gene therapy clinical trials, integration of the strong viral promoters nearby proto-oncogenes has led to gene deregulation and clonal expansions (4–6). Consequently, these vectors are not useful for applications that require targeted gene addition. The piggyBac and Sleeping Beauty transposon systems have both been used to integrate genes into mammalian genomes in vitro and in vivo (7–9). Because the transposon systems do not contain the strong viral promoters, it is anticipated that activation of nearby oncogenes will be unlikely. Additionally, Sleeping Beauty, and to a lesser extent piggyBac, appear to integrate more randomly into the genome than γ-retroviral vectors and do not show as strong a preference for integration into genes (8,10,11). However, transposition into an oncogene or tumor suppressor and subsequent insertional mutagenesis has been demonstrated in genetic screens that are designed to favor these events (12,13). Although a few studies have attempted to direct gene transposition by these enzymes to genomic target sites by the fusion of sequence-specific DNA-binding proteins, the majority of gene addition events in these systems are still random (14,15).
In contrast to the random addition of genes by viral vectors and transposases, the Cre and Flp recombinases catalyze the exchange of DNA strands between loxP and FRT sequences, respectively (16,17). Cre and Flp have both been used to target plasmid integration into loxP or FRT sites that have been pre-introduced into mammalian cell genomes (17,18). The efficiency of Flp-mediated plasmid integration is comparable to random plasmid integration (17,19). Therefore additional selection or screening steps are necessary to ensure site-specific integration. Cre expression is toxic to mammalian cells (20,21) and can lead to chromosomal rearrangements by reacting with off-target pseudo-loxP sites present in the human genome (22,23). Similarly, the phage-derived integrase phiC31 catalyzes the integration of a transfected plasmid into pseudo-recognition sites in the human genome (24,25). Thorough characterization of the pseudo-recognition sites in human cells identified over 100 distinct genomic integration sites, with a slight preference for integration into intragenic regions (26). Chromosomal rearrangements have also been observed in human cells following phiC31 expression (27,28). Although recent in vivo safety studies suggest that phiC31 expression does not lead to oncogenic insertional mutagenesis (29), there remains a clear need for enzymes with strict DNA-binding domains that recognize unique sites within mammalian genomes.
The Cys2-His2 zinc-finger domain is the most common DNA-binding motif in the human proteome. A single zinc finger contains ∼30 amino acids and typically functions by binding three consecutive base pairs of DNA via interactions of a single amino acid side chain per base pair (30). The specificity of particular zinc fingers for the 64 possible nucleotide triplets has been examined extensively through site-directed mutagenesis, rational design and the selection of large combinatorial libraries (31–33). The modular structure of the zinc-finger motif permits the fusion of several domains in series, allowing for the recognition and targeting of extended sequences in multiples of 3 nucleotides (34). It is now possible to design synthetic zinc-finger proteins to bind practically any target site in the human genome (35,36).
These targeted DNA-binding proteins can be fused to enzymatic domains to direct enzyme activity to specific sites in the genome. This approach has been most prominently exemplified by the development of zinc-finger nucleases (ZFNs), in which the synthetic zinc-finger protein is fused to the catalytic domain of the FokI restriction endonuclease (37). When expressed within mammalian cells, ZFNs cleave DNA to create a double-strand break at a targeted genomic locus (37). This DNA cleavage stimulates DNA repair pathways and increases the efficiency of homologous recombination at the site by several orders of magnitude, which otherwise occurs below background levels of random plasmid integration in human cells. This method has been used to incorporate gene sequences at specific locations in the genomes of cells from a variety of species, including human cell lines and embryonic and adult stem cells (37). However, the potential for off-target DNA cleavage, the induction of the DNA-damage response pathway and the associated genotoxicity that has been often observed with these enzymes remain concerns for this method (37–39).
Inspired by the success of the ZFN technology, we have recently developed zinc-finger recombinases (ZFRs) to autonomously perform precise gene addition to the human genome without cleaving genomic DNA and activating the DNA damage response pathway (40). ZFRs are a fusion of a synthetic zinc-finger protein and the catalytic domain of a serine recombinase (41,42). For the chromosomal integration of plasmid DNA, the designed zinc-finger domain binds to specific target sites in the genome and the plasmid, and the recombinase domain catalyzes the exchange of DNA strands (40). In the original demonstration of this approach, a single model recombinase target site was introduced into a specific, but unknown, chromosomal location in the human HEK-293 cells using the Flp-InTM cell lines and reagents from Invitrogen. We demonstrated that ZFRs could target plasmid integration into this site with >98% specificity (40). This specific integration occurred only if the correct target site was present and the DNA-binding domain of the ZFR contained at least three zinc-finger motifs. In the current study, we sought to evaluate the general applicability of this system by investigating ZFR-mediated plasmid integration into target sites in diverse regions of the genome and in a variety of cell lines. The piggyBac transposon system was used to distribute ZFR recognition sites randomly throughout the genome in a population of cells. Genomic integration of a transfected plasmid was dependent on both an active ZFR and the presence of ZFR target sites in the genome, demonstrating the stringent selectivity of these enzymes. ZFR and piggyBac activity were both dependent on cell type. Clonal analysis showed that the vast majority of stably transfected cells contained integration events at the intended target site, and in some instances the ZFR was able to integrate plasmids into multiple transposon target sites within the same cell. These results support the broad utility of the ZFR technology and suggest possibilities for engineering of isogenic cell lines and stem cell-based gene therapies.
MATERIALS AND METHODS
Plasmids
The plasmids pTpB and pCMV-pB were provided by Matthew H. Wilson (8). The pTpB plasmid contains the piggyBac transposon, which carries a kanR/neoR cassette driven by the SV40 promoter and a p15A origin of replication for propagation in Escherichia coli. pCMV-pB carries the expression cassette for the piggyBac transposase. For our piggyBac expression vector, the piggyBac transposase was PCR amplified and inserted into a pcDNA3.1-Zeocin ZFR expression plasmid (40) in which the ZFR had been removed by SfiI digestion. The 44-bp ZFR target site was added to pTpB by PCR amplification of a 1109-bp fragment containing the C.20G ZFR target sequence and a neighboring promoterless EGFP transgene from the pcDNA5/FRT-EGFP-C.20G plasmid (40) and ligation into the BamHI site in pTpB downstream of the kanR/neoR cassette in the piggyBac transposon. The luciferase-encoding piggyBac transposon was created by removing the kanR/neoR cassette in pTpB by digestion with BglII and SacII and ligation of a CMV promoter driving the luciferase gene into these sites. The ZFR expression plasmid pcDNA3.1-Zeocin-GinC4 and the C.20G-Puro ZFR donor plasmid carrying the puroR cassette have been described previously (40). The luciferase-encoding ZFR donor plasmid was created by removing the puroR gene from the C.20G-Puro donor plasmid (40) by digestion with HindIII and NheI and ligating the luciferase gene into these sites, under the control of the SV40 promoter. All vector sequences were confirmed by DNA sequencing.
Transposition and plasmid integration
All cell lines were cultured in DMEM, 10% fetal bovine serum, 100 U/ml penicillin G sodium and 100 μg/ml streptomycin sulfate in a humidified 5% CO2 atmosphere at 37°C. All cell culture media components and reagents were obtained from Invitrogen unless otherwise noted. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Twenty-four hours prior to transfection, 100 000 cells were plated into 24-well plates. For piggyBac-mediated transposition, cells were transfected with 250 ng of pTpB and 250 ng of pcDNA3.1-Zeocin-piggyBac or control pcDNA3.1-Zeocin plasmid (no insert). For ZFR-mediated donor plasmid integration, cells were transfected with 50 ng donor plasmid and 500 ng of control pcDNA3.1-Zeocin plasmid (no insert), pcDNA3.1-Zeocin-ZFRS9A, or pcDNA3.1-Zeocin-ZFR.
For colony-counting assays, 10% of the transfected cell population was moved to one well of a six-well plate at 3 days post-transfection. The following day, cell culture media was exchanged with media containing 800 μg/ml G418 sulfate and/or 2 μg/ml puromycin as appropriate. Approximately 14 days later, cells were stained with crystal violet solution and colony number was determined by automated counting using a GelDoc XR imaging system with Quantity One 1-D analysis software (Bio-Rad). Reported colony numbers have been multiplied by a factor of ten to account for the initial 1:10 cell division. For luciferase assays, cells were continuously cultured in the absence of selection with passaging every 3–5 days. At each passaging, cell samples were harvested and frozen. At the conclusion of the experiment, samples were thawed and assayed for luciferase activity with the Luciferase Reporter Assay System (Promega) according the manufacturer’s instructions using a Veritas Microplate Luminometer (Turner Biosystems) equipped with injectors.
Genomic DNA isolation and analysis
Genomic DNA was purified with QIAamp DNA Blood kits (Qiagen) from G418R/puroR polyclonal cell populations or monoclonal populations derived by limiting dilution. Genomic DNA was used as template in PCR reactions with primer combinations that amplified the unmodified ZFR target site (pTpB-prim1 & pTpB-prim2), the integration of the donor plasmid into the ZFR target site in the forward orientation (donor-prim1 & pTpB-prim2), or the integration of the donor plasmid into the ZFR target site in the reverse orientation (donor-prim1 & pTpB-prim1). Primer binding sites are indicated in Figure 1A and C. Primer sequences are: pTpB-prim1: 5′-TTTTCCGGGACGCCGGCTGG-3′; pTpB-prim2: 5′-CTTGTCGGCCATGATATAGACG-3′; donor-prim1: 5′-TGACGTCAATGACGGTAAATGG-3′.
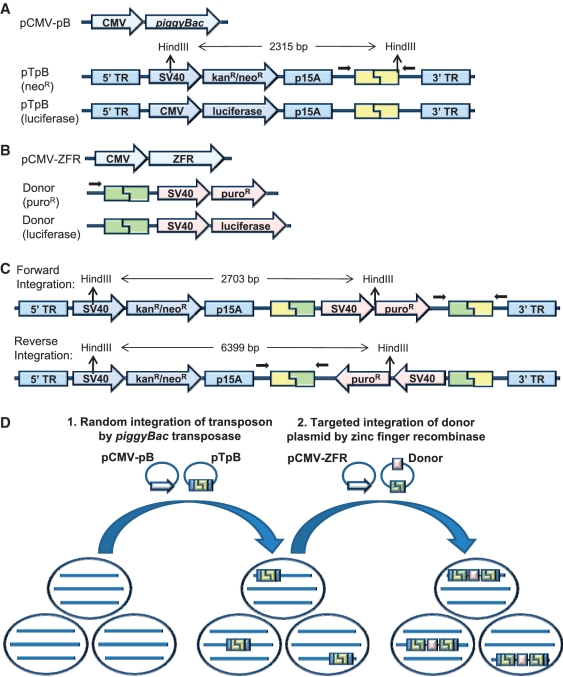
Figure 1.
(A) Schematic of plasmids used for piggyBac-mediated transposition. (B) Schematic of plasmids used for ZFR-mediated integration of the donor plasmid. (C) Schematic of predicted results of targeted donor plasmid integration into the piggyBac transposon in forward or reverse orientations. Small black arrows indicate the positions of PCR primers used for detection of targeted integration. HindIII restriction sites and fragment sizes indicate expected results of Southern blot analysis with a probe against kanR/neoR. (D) Experimental design. Cell lines were first co-transfected with a plasmid encoding the piggyBac transposase (pCMV-pB) and a plasmid carrying a piggyBac transposon that contains a ZFR target site and neoR/kanR cassette (pTpB). Random transposition was used to generate a polyclonal cell population with ZFR target sites distributed throughout the genome. Following selection of neoR cells, these populations were co-transfected with a ZFR expression plasmid (pCMV-ZFR) and a donor plasmid containing a ZFR recognition site and puroR cassette to assess targeted plasmid integration into the piggyBac transposon by the ZFR.
Southern blots were performed using standard procedures. Briefly, 1 μg of genomic DNA was digested with HindIII (Figure 1A and C) for 4 h. Digested DNA was separated on a 1% agarose gel by electrophoresis. The gel was washed and blotted onto a membrane (GeneScreen Plus, Perkin Elmer) overnight using an upward capillary transfer. The membrane was crosslinked with UV light for 30 s and dried on filter paper. An 800-bp probe of the neoR gene in the pTpB plasmid was generated by PCR with the primers 5′-ATGATTGAACAAGATGGATTGC-3′ and 5′-TCGTCAAGAAGGCGATAGAA-3′ and labeled with 32P using the PrimeIT II kit (Stratagene) according to manufacturer’s instructions. The blot was pre-hybridized in MiracleHyb buffer (Stratagene). Following overnight hybridization with the radiolabeled probe at 65°C in a rotary hybridization oven, blots were washed twice in solution A (2× SSC, 0.1% SDS) and twice in solution B (0.01× SSC, 0.1% SDS) for 15 min at 50°C. Labeled blots were then exposed to a phosphorimager cassette (Molecular Dynamics) for 14 days and visualized on a Storm840 phosphorimager (GE Healthcare).
Plasmid rescue
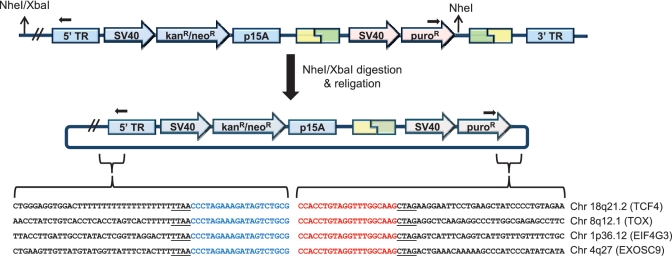
Genomic DNA (10 μg) was digested with NheI and XbaI, which have compatible 5′ CTAG overhangs. NheI cut inside the donor plasmid, and NheI or XbaI were expected to cut within the neighboring genomic DNA outside of the 5′-end of the piggyBac transposon. Digested DNA was purified by ethanol precipitation and ligated in dilute conditions of 300 μl total ligation volume to favor self-ligation. The resulting plasmid contained the kanR gene and p15A replication of origin from the pTpB transposon. The DNA was concentrated by ethanol precipitation, resuspended in water and electroporated into XL1 Blue E. coli and plated onto kanamycin-containing LB agar plates. Colonies were screened by colony PCR for plasmids that contained the donor plasmid sequence integrated into the piggyBac transposon (∼10% of all colonies), rather than only the transposon. Plasmid DNA from positive colonies was purified by miniprep (Qiagen) and sequenced with primers extending from both the 5′ terminal inverted repeat (TR) of the transposon and the puroR gene within the donor plasmid (Figure 6).
Figure 6.
Mapping the chromosomal locations of donor plasmid integrations into the transposon target site. Genomic DNA of the polyclonal G418R/puroR HEK293 cell populations was digested with NheI and XbaI, circularized by ligation and transformed into E. coli. Purified plasmid was sequenced by reverse and forward primers that bind to the transposon and donor plasmid, respectively, to ensure recovery of correctly targeted transposons. The recovered sequences were intragenic, which is a favored location for piggyBac transposition (8). Gene names and chromosomal locations are indicated to the right of the corresponding sequencing results. The positions of sequencing primers are indicated with black arrows. The underlined TTAA and CTAG correspond to the site of piggyBac transposition and the NheI/XbaI compatible 5′ overhang, respectively.
Statistics
Data are presented from representative experiments as the mean of triplicate samples ± standard error of the mean (mean ± SEM). Statistical analyses for colony number assays included two-sided, two-sample Student’s t-test assuming equal variances (Figure 2A) or two-way ANOVA accounting for both cell type and treatment (Figure 3A). Statistical analyses for luciferase assays included two-way ANOVA accounting for both time and treatment, for which only P-values with respect to treatment were reported (Figures 2B and 3B). In order to make the variance independent of the mean, statistical analysis of luciferase assays was performed following logarithmic transformation of the raw data. Analyses were performed with Microsoft Office Excel 2007 with α = 0.05.
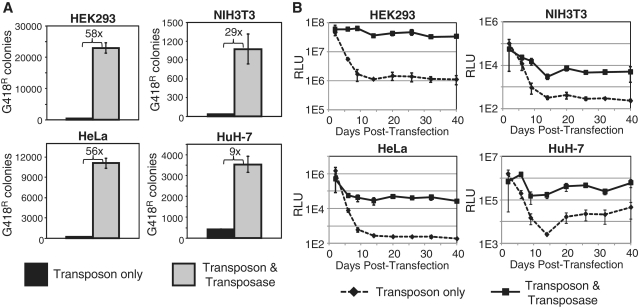
Figure 2.
PiggyBac-mediated gene transposition into the genome of human and mouse cell lines. Cells were co-transfected with the transposon vector and either empty expression plasmid or transposase expression plasmid. (A) Stable chromosomal integration of a piggyBac transposon carrying a neoR cassette was measured as the number G418R cell colonies. Fold increase in colony number is indicated in each graph (n = 3, mean ± SEM; HEK293 P = 0.0002; NIH3T3 P = 0.01; HeLa P = 0.0001; HuH-7 P = 0.001). (B) Stable chromosomal integration of a piggyBac transposon carrying a luciferase gene was measured as luciferase activity [relative luminescence units (RLU)] in an unselected cell population (n = 3, mean ± SEM; HEK293 P = 5E-19, NIH3T3 P = 1E-9, HeLa P = 1E-14, HuH-7 P = 6E-8).
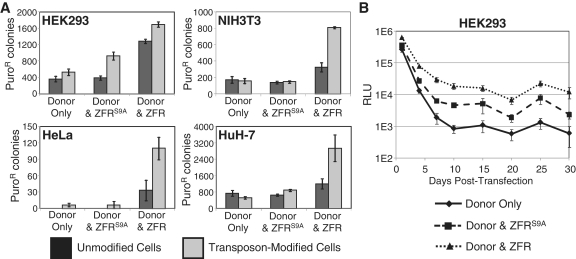
Figure 3.
ZFR-mediated plasmid integration into the genome of human and mouse cell lines. Unmodified parental cell lines or G418R transposon-modified polyclonal cell populations were co-transfected with a donor plasmid and control (empty), ZFRS9A, or ZFR expression plasmid. (A) Stable chromosomal integration of a donor plasmid carrying a puroR cassette was measured as the number of puroR cell colonies (n = 3, mean ± SEM). P-values for each cell type with respect to expression vector and unmodified vs. transposon modified cells are: HEK293 P = 1E-8, P = 4E-5; NIH3T3 P = 2E-8, P = 7E-5; HeLa P = 9E-5, P = 9E-3; HuH-7 P = 6E-4, P = 0.03. (B) Stable chromosomal integration of a donor plasmid carrying a luciferase gene was measured as luciferase activity in an unselected cell population (n = 3, mean ± SEM; P = 4E-19).
RESULTS
In order to create a heterogeneous population of polyclonal cells carrying ZFR target sites, the piggyBac transposon system was used to randomly distribute target sites throughout the genomes of several cell lines (Figure 1). The C.20G ZFR target site (40) was added to the piggyBac transposon in the pTpB plasmid, which also contains a kanamycin/neomycin resistance (kanR/neoR) cassette and a p15A origin of replication for propagation in E. coli (8). This modified transposon was co-transfected with control plasmid DNA or a plasmid encoding the piggyBac transposase into four cell lines: human embryonic kidney (HEK293) cells, human epithelial cervical cancer (HeLa) cells, human hepatocarcinoma (HuH-7) cells and NIH3T3 mouse embryonic fibroblasts. Cells in which the transposon was stably integrated into the genome were neomycin resistant and selected by growth in the presence of antibiotic G418. The number of G418-resistant (G418R) cell colonies was used as a measure of transposase activity (Figure 2A). The presence of the transposase significantly increased the number of stable integrants, ranging from 9-fold in HuH-7 cells to 58-fold in HEK293 cells. The level of background integration of plasmid DNA, the fold-increase of G418R colonies upon the addition of transposase, and the total number of G418R colonies varied greatly among cell lines, as noted previously (43).
To monitor stable transposition in the absence of selective pressure, the neoR gene in the piggyBac transposon was replaced with a luciferase gene. This plasmid was co-transfected into each cell line with control plasmid DNA or the transposase expression plasmid and samples were harvested over a time course of 40 days to monitor stable luciferase gene expression (Figure 2B). In the absence of transposase, luciferase levels dropped precipitously over 10–15 days post-transfection as a result of episomal plasmid degradation and dilution during cell division. Luciferase activity stabilized at a low level after this period. In contrast, addition of the transposase resulted in sustained gene expression at significantly higher levels for the duration of the experiment in all cell lines.
PiggyBac transposes semi-randomly into mammalian genomes at TTAA sequences (8). Consequently the G418R cell populations from the samples transfected with both transposase and transposon were heterogeneous; each original cell incorporated one or more transposons into different locations in its genome (44). Therefore these polyclonal cell populations were used to assess the ability of the ZFR to integrate plasmid DNA into different genomic loci in various cell types (Figure 1). The ZFR used for this study, GinC4, is a fusion of the catalytic domain of the Gin invertase and a four-finger designed zinc-finger protein (40). For this experiment, both the parental cell lines and the G418R transposon-modified cell lines (Figure 2A) were co-transfected with a donor plasmid containing both a ZFR target site and a puromycin-resistance (puroR) cassette and either control DNA, a plasmid that expresses a catalytically inactive ZFR (ZFRS9A), or a plasmid that expresses the ZFR. At 3 days post-transfection, the cells were moved to puromycin-containing media for 14 days and the level of plasmid integration into the genome was measured as the number of puroR cell colonies (Figure 3A). The addition of active ZFR to the transposon-modified cells increased the level of plasmid integration relative to cells without ZFR, cells with inactive ZFR and cells with ZFR but without transposon target sites. Similar to piggyBac-mediated transposition, the level of background plasmid integration, the fold-increase of puroR colonies upon the addition of ZFR, and the total number of puroR colonies varied greatly among cell lines. The differences in plasmid integration levels between the parental cell lines and the transposon-modified cell lines when both were treated with donor plasmid and active ZFR was particularly notable in the NIH3T3, HeLa and HuH-7 cell lines. The selectivity of the ZFR was such that in the absence of the appropriate 44-bp target site, the level of ZFR-mediated recombination into any of the >3 billion base pairs of the human or mouse genomes was not significantly higher than background plasmid integration. It is unclear why there was a measurable increase in plasmid integration in the HEK293 cell line upon the addition of active ZFR despite the absence of transposon target sites, although there was a significant increase when the transposon was present. It is also noteworthy that binding of the catalytically inactive ZFRS9A to the target sites with intact zinc-finger proteins led to a moderate increase in colony numbers in HEK293 cells but did not have any effect on plasmid integration in the other three cell types. Importantly, we have previously validated that the ZFRS9A mutant is catalytically inactive in a high-throughput excision assay in E. coli (45). Therefore the mechanism for ZFRS9A-mediated stable transfection in HEK293 cells is unknown, although it is clear that none of the plasmid was correctly targeted to the transposon (Figure 4). It is possible that binding of the ZFR to its target site on the plasmid may modify its stability in the absence of any catalytic activity.
Figure 4.
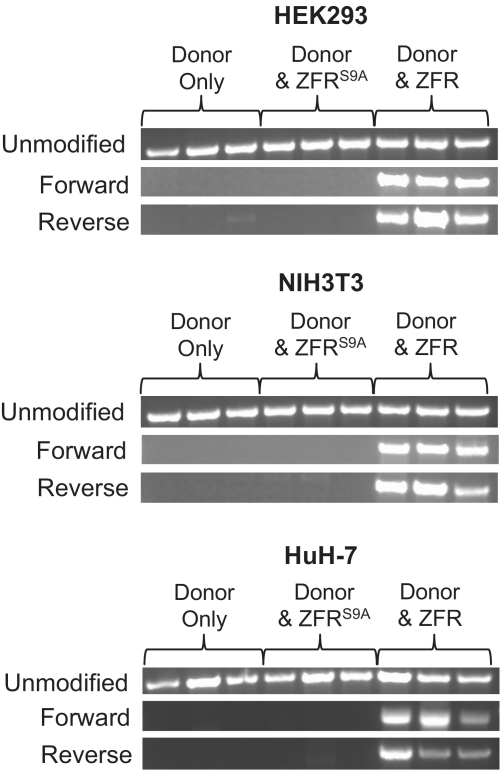
Specificity of ZFR-mediated plasmid integration. Genomic DNA from polyclonal G418R/puroR cell populations that had been transfected with donor plasmid and control (empty), ZFRS9A, or ZFR expression plasmid was analyzed by PCR. PCR primer combinations amplified either the unmodified ZFR target site on the piggyBac transposon or the integration of the donor plasmid at this site in either forward or reverse orientations. Targeted plasmid integration was detected only in samples treated with the ZFR.
In order to measure the time course of gene expression following ZFR-mediated plasmid integration in the absence of the selective pressure of antibiotics, the puroR gene in the donor plasmid was replaced with the luciferase gene. G418R HEK293 cells were co-transfected with the luciferase-encoding donor plasmid and either control DNA, the ZFRS9A expression plasmid, or the active ZFR expression plasmid. Cell samples were collected at various time points up to 30 days post-transfection and assayed for luciferase activity (Figure 3B). Cells treated with donor plasmid and ZFR showed >10-fold increases in the levels of stable gene expression relative to cells treated with donor plasmid only. The expression of the inactive ZFRS9A led to an intermediate level of stable luciferase expression in HEK293 cells similar to the colony formation assay (Figure 3A).
In order to determine whether the plasmid integration events mediated by the ZFR were correctly targeted to the transposon, genomic DNA was purified from the polyclonal puroR cell populations and assayed by PCR (Figure 4). PCR primers were used that amplify the unmodified transposon or the ZFR donor integrated into the transposon in either forward or reverse orientations (Figure 1A and C) (40). Although these cell populations were polyclonal with respect to transposon integration sites, the PCR amplification should occur independently of chromosomal location. The PCR products corresponding to the correct plasmid integration events in forward and reverse orientations were present only in samples containing both the donor and an active ZFR. Unmodified transposons were detected in all cell populations, which is not surprising given that it is possible for >15 transposition events to occur in a single cell (44) and cells would be selected in this assay if the plasmid integrated into at least one of these transposons. Although most samples from ZFR-treated HeLa cells were positive for site-specific plasmid integration, results were inconsistent due to low numbers of puroR colonies (Figure 3A) and overgrowth of the cell population by small numbers of rapidly expanding colonies. Therefore results obtained from experiments with HeLa cells are not shown.
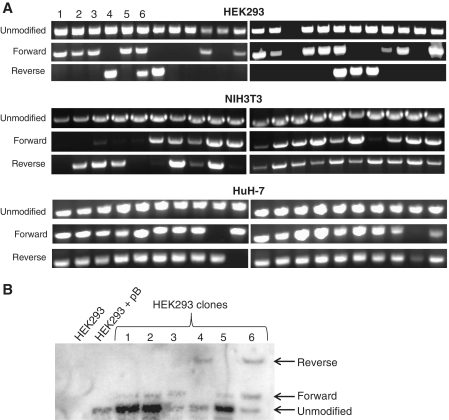
Although the genomic PCR of the polyclonal population indicated that site-specific integration events were present within the sample, how frequently these events occurred was unknown. Therefore monoclonal cell populations were derived from single cells of the puroR cultures that had been treated with donor and active ZFR to determine the overall level of specificity in the population (Figure 5). The genomic DNA from these clonal populations was analyzed by PCR for targeted integration events (Figure 5A). Except for one aberrant HEK293 clone, all of the clonal samples contained transposons that had not been modified by plasmid integration, which could be predicted by previous studies showing multiple transpositions per cell (44). Of 24 HEK293 clones, only five clones had no targeted integration events, including the single clone in which the unmodified transposon was not detectable. These clones represent cells in which the puroR donor plasmid integrated at an off-target site. Thirteen clones had only forward integrations, four clones had only reverse integrations, and two clones contained both reverse and forward integrations. All 20 of the HuH-7 clones and 19 of the 20 NIH3T3 clones contained targeted integration events, and the vast majority of these (17/20 NIH3T3 and 17/20 HuH-7 clones) contained both forward and reverse integrations. The PCR products from selected samples were sequenced to confirm the accurate junction of the transposon and donor plasmid. Southern blots of the genomic DNA from a subset of the HEK293 clones verified the PCR results and validated the PCR-based method for detecting clones with forward and/or reverse integration orientations (Figure 5B).
Figure 5.
Clonal analysis of ZFR-mediated plasmid integration events. Single cell colonies were isolated by limiting dilution from polyclonal G418R/puroR cell populations that had been transfected with donor plasmid and ZFR expression plasmid. (A) Genomic PCR was used to detect the unmodified ZFR target site on the piggyBac transposon and/or the integration of the donor plasmid at this site in either forward or reverse orientations. (B) Southern blot of unmodified HEK293 cells, G418R HEK293 cells containing the piggyBac transposon, and the first six G418R/puroR clonal HEK293 cell populations was performed with a probe against the neoR cassette. Distinct bands correspond to the unmodified transposon and forward and reverse orientations of the correctly targeted donor plasmid integration (Figure 1A and C) and coincide with the PCR results in (A).
The chromosomal locations of representative ZFR-mediated integration events were determined to confirm that donor plasmid integration occurred at multiple chromosomal locations (Figure 6). A plasmid rescue assay was performed in which the genomic DNA of the polyclonal puroR/G418R ZFR-treated HEK293 cells was digested with the restriction enzymes NheI, which cuts inside the donor plasmid and XbaI, which is expected to cut in the chromosomal DNA near the transposon integration site. Self-ligation of the digestion products resulted in a new plasmid that contained sequences from the piggyBac transposon, donor plasmid and flanking chromosomal DNA. This plasmid can be transformed and propagated in E. coli because of the kanR cassette and p15A origin of replication within the transposon. Plasmids containing the donor fragment were distinguished from non-targeted transposons by colony PCR and sequenced with transposon- and donor-specific primers to determine chromosomal location. Representative sequences show that the piggyBac target sites were on different chromosomes (Figure 6). The recovered events occurred within genes, which is a preferred region for piggyBac transposition (8). These results show that the ZFR was active at several chromosomal locations in human cells.
DISCUSSION
This study demonstrates the ability of ZFRs to direct the targeted chromosomal integration of a plasmid at specific recognition sites distributed throughout human and mouse genomes in several cell types. As measured by the fraction of stable transfectants containing successfully targeted integration events (Figure 5A), ZFR specificity is significantly greater than any of the enzyme- or viral-mediated vector integration strategies described to date. In our previous study using an isogenic HEK293 cell line with a ZFR target site at a single accessible locus, we observed targeted integration in 55 of 56 clones (40). We have now observed similarly high levels of specificity into polyclonal targets in 19/24, 19/20 and 20/20 clones for HEK293, NIH3T3 and HuH-7 cells, respectively (Figure 5A), as well as detectable site-specific integration in HeLa cells (Figure 4). For comparison, a previous study which inserted the 39-bp target site of the phiC31 integrase into the genome of HEK293 cells showed only 14/96 clones contained site-specific integration events (24), with the other 82 clones likely containing integration events at any of the other >100 genomic sites that are also recognized by this enzyme (26).
The high frequency of targeted integration events in the clonal cell populations (Figure 5) does not eliminate the possibility that off-target integration events also occurred in these cells. However, given that only ∼1% of the total transfected cell population contains an integration event (40), we expect it is unlikely that high levels of ZFR-mediated off-target integration are occurring. Furthermore, ZFR activity increased ∼3-fold when the transposon target site was pre-introduced in the genome of NIH3T3, HeLa and HuH-7 cells (Figure 3A). Plasmid integration by the ZFR in the absence of transposon target sites was only marginally higher than background plasmid integration in these cells (Figure 3A), suggesting that many off-target integration events in our experiments were the result of random plasmid integration that was not ZFR-mediated. Nevertheless, additional studies dedicated to thoroughly and quantitatively characterizing potential off-target recombination and ZFR-DNA interactions are necessary to understand the full potential of this technology and its values relative to methods that enhance gene targeting by homologous recombination (37,46,47).
Commercial systems are available for cell line engineering by targeted plasmid integration. For example, the Flp-InTM and Jump-InTM systems marketed by Invitrogen make use of the Flp recombinase and phiC31 integrase, respectively. As described above, Flp activity is not significantly greater than background levels of plasmid integration and phiC31 does not have the intrinsic sequence specificity to recognize a single site in mammalian genomes. Therefore these commercial systems account for this lack of activity and specificity by dividing a single expression cassette for an antibiotic resistance gene between the genomic target site and the donor plasmid such that only plasmid integration at the target site will reconstitute this cassette (48,49). In contrast, we readily recovered cells with targeted integration events by selecting for any plasmid incorporation into the genome by incorporating the complete puroR cassette on the donor plasmid. The ability to target multiple sites within the same cell is further evidence of the high level of ZFR efficiency and specificity.
The two-step strategy for genome engineering described here (Figure 1D) will be directly useful for a variety of applications in stem cell-based gene therapy, cell line engineering, genetic engineering for biopharmaceutical production and animal transgenesis. For example, the polyclonal population of transposon-modified cells can be screened for individual clones in which the ZFR target site has been integrated into a favorable genomic locus, as was recently demonstrated in the creation of induced pluripotent stem cells with the piggyBac transposon system (50) and the screening of lentiviral safe harbor integration sites (51). The favorable locus may be defined by the degree of interactions with endogenous genes, the distance to neighboring genes, lack of silencing effects, high levels of gene expression, or other factors (51). The ZFR could then be used to integrate transgenes specifically at this locus in the clonal transposon-modified population. This two-step approach is advantageous because many different transgenes can be added to the transposon-modified cell source in parallel, all of which would be targeted to the same favorable locus. An analogous method has been proposed for using piggyBac and Cre recombinase for the genomic analysis of transgenic mice (52). A similar approach has also been used with phage integrases to engineer the genomes of cell lines, embryonic stem cells and whole organisms (49,53). However all of these systems require additional levels of selection to compensate for insufficient enzyme specificity, as discussed above. Further investigation of ZFR activity in primary cells, embryonic stem cells, induced pluripotent stem cells and animals will elucidate the advantages and limitations of ZFRs in these settings.
Several other studies have investigated targeted gene addition by fusing engineered zinc-finger proteins or other targeted DNA-binding proteins to transposases (14,15) and retroviral integrases (54,55). Although these strategies have been successful in directing integration and transposition, the vast majority of integration events occur at locations other than the target locus. Importantly, these fusion proteins were not engineered to abrogate the integration activity of the parent enzyme. Consequently, any rare targeted integration events occur amidst many more semi-random events. This approach to protein engineering is in contrast to our development of ZFRs, in which the serine recombinase catalytic domain is only active when fused to a DNA-binding protein (40). We expect that this modular structure and function of ZFRs contributes substantially to their precise specificity.
Ultimately, we envision the design of ZFRs that specifically target endogenous genome sequences, enabling the direct integration of plasmid DNA into any natural locus. The ZFR used in this study is composed of a fusion between a designed four-finger zinc-finger protein and a hyperactive catalytic domain of the Gin invertase from bacteriophage Mu (40). Synthetic zinc-finger proteins can be engineered to target almost any DNA sequence (35,36). However, the Gin catalytic domain retains a strict sequence specificity for its native recognition sequence in the bacteriophage genome, thus preventing the reprogramming of ZFR activity solely through exchanging the zinc-finger DNA-binding domain (40,42). To facilitate the use of ZFRs at diverse target sequences, we have used directed evolution to alter the sequence specificity of ZFR catalytic domains (42,45). In particular, we have recently described a structure-guided approach to directed evolution that facilitates the engineering of domains with high levels of activity exclusively on new targets (56). We anticipate that this work will lead to the development of ZFRs that integrate plasmids efficiently and specifically at any genomic site of interest. This work has the potential to transform customized genome engineering into a facile and routine procedure.
FUNDING
National Institutes of Health (R01GM065059 and R21CA126664 to C.F.B.); Skaggs Institute for Chemical Biology and a National Institutes of Health Postdoctoral Fellowship (F32CA125910 to C.A.G.). Funding for open access charge: Grants and internal funding.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The plasmids pTpB and pCMV-pB were provided by Matthew H. Wilson.
REFERENCES
- 1.Baum C, Kustikova O, Modlich U, Li Z, Fehse B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum. Gene Ther. 2006;17:253–263. doi: 10.1089/hum.2006.17.253. [DOI] [PubMed] [Google Scholar]
- 2.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 3.Bushman F, Lewinski M, Ciuffi A, Barr S, Leipzig J, Hannenhalli S, Hoffmann C. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 2005;3:848–858. doi: 10.1038/nrmicro1263. [DOI] [PubMed] [Google Scholar]
- 4.Fischer A, Hacein-Bey-Abina S, Cavazzana-Calvo M. 20 years of gene therapy for SCID. Nat. Immunol. 2010;11:457–460. doi: 10.1038/ni0610-457. [DOI] [PubMed] [Google Scholar]
- 5.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, Glimm H, Kuhlcke K, Schilz A, Kunkel H, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 6.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VandenDriessche T, Ivics Z, Izsvak Z, Chuah MK. Emerging potential of transposons for gene therapy and generation of induced pluripotent stem cells. Blood. 2009;114:1461–1468. doi: 10.1182/blood-2009-04-210427. [DOI] [PubMed] [Google Scholar]
- 8.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 9.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 10.Vigdal TJ, Kaufman CD, Izsvak Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 2002;323:441–452. doi: 10.1016/s0022-2836(02)00991-9. [DOI] [PubMed] [Google Scholar]
- 11.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 13.Rad R, Rad L, Wang W, Cadinanos J, Vassiliou G, Rice S, Campos LS, Yusa K, Banerjee R, Li MA, et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivics Z, Katzer A, Stuwe EE, Fiedler D, Knespel S, Izsvak Z. Targeted Sleeping Beauty transposition in human cells. Mol. Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 15.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl Acad. Sci. USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 18.Sauer B, Henderson N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990;2:441–449. [PubMed] [Google Scholar]
- 19.Logie C, Stewart AF. Ligand-regulated site-specific recombination. Proc. Natl Acad. Sci. USA. 1995;92:5940–5944. doi: 10.1073/pnas.92.13.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl Acad. Sci. USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naiche LA, Papaioannou VE. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 2007;45:768–775. doi: 10.1002/dvg.20353. [DOI] [PubMed] [Google Scholar]
- 22.Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc. Natl Acad. Sci. USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol. Cell. Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calos MP. The phiC31 integrase system for gene therapy. Curr. Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- 26.Chalberg TW, Portlock JL, Olivares EC, Thyagarajan B, Kirby PJ, Hillman RT, Hoelters J, Calos MP. Integration specificity of phage phiC31 integrase in the human genome. J. Mol. Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 27.Ehrhardt A, Engler JA, Xu H, Cherry AM, Kay MA. Molecular analysis of chromosomal rearrangements in mammalian cells after phiC31-mediated integration. Hum. Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Jeppesen I, Nielsen K, Jensen TG. Phi c31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- 29.Woodard LE, Keravala A, Jung WE, Wapinski OL, Yang Q, Felsher DW, Calos MP. Impact of hydrodynamic injection and phiC31 integrase on tumor latency in a mouse model of MYC-induced hepatocellular carcinoma. PLoS ONE. 2010;5:e11367. doi: 10.1371/journal.pone.0011367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 31.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 32.Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 33.Dreier B, Fuller RP, Segal DJ, Lund CV, Blancafort P, Huber A, Koksch B, Barbas CF., 3rd Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 34.Beerli RR, Segal DJ, Dreier B, Barbas CF., 3rd Toward controlling gene expression at will: specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl Acad. Sci. USA. 1998;95:14628–14633. doi: 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandell JG, Barbas CF., 3rd Zinc Finger Tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 38.Cornu TI, Thibodeau-Beganny S, Guhl E, Alwin S, Eichtinger M, Joung JK, Cathomen T. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol. Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 39.Guo J, Gaj T, Barbas CF., 3rd Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J. Mol. Biol. 2010;400:96–107. doi: 10.1016/j.jmb.2010.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordley RM, Gersbach CA, Barbas CF., 3rd Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc. Natl Acad. Sci. USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordley RM, Smith JD, Graslund T, Barbas CF., 3rd Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gersbach CA, Gaj T, Gordley RM, Barbas CF., 3rd Directed evolution of recombinase specificity by split gene reassembly. Nucleic Acids Res. 2010;38:4198–4206. doi: 10.1093/nar/gkq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendrie PC, Russell DW. Gene targeting with viral vectors. Mol. Ther. 2005;12:9–17. doi: 10.1016/j.ymthe.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 48.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol. Adv. 2005;23:431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Lieu PT, Machleidt T, Thyagarajan B, Fontes A, Frey E, Fuerstenau-Sharp M, Thompson DV, Swamilingiah GM, Derebail SS, Piper D, et al. Generation of site-specific retargeting platform cell lines for drug discovery using phiC31 and R4 integrases. J. Biomol. Screen. 2009;14:1207–1215. doi: 10.1177/1087057109348941. [DOI] [PubMed] [Google Scholar]
- 50.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A, et al. Genomic safe harbors permit high beta-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu S, Ying G, Wu Q, Capecchi MR. Toward simpler and faster genome-wide mutagenesis in mice. Nat. Genet. 2007;39:922–930. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- 53.Huang J, Zhou W, Dong W, Watson AM, Hong Y. From the cover: directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc. Natl Acad. Sci. USA. 2009;106:8284–8289. doi: 10.1073/pnas.0900641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan W, Dong Z, Wilkinson TA, Barbas CF, 3rd, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim KI, Klimczak R, Yu JH, Schaffer DV. Specific insertions of zinc finger domains into Gag-Pol yield engineered retroviral vectors with selective integration properties. Proc. Natl Acad. Sci. USA. 2010;107:12475–12480. doi: 10.1073/pnas.1001402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF., III Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc. Natl Acad. Sci. USA. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]