Abstract
In plant, primary transcripts (pri-miRNAs) transcribed from miRNA genes by RNA polymerase II are first processed into stem-loop pre-miRNAs and further chopped into ∼21 nt long miRNAs by RNase III-like enzyme DCL1. SERRATE (SE) protein is an essential component for miRNA processing by assisting DCL1 for accurate cleavage. Here we report the crystal structure of Arabidopsis SE core (residues 194–543) at 2.7 Å. SE core adopts the ‘walking man-like’ topology with N-terminal α helices, C-terminal non-canonical zinc-finger domain and novel Middle domain resembling the leading leg, the lagging leg and the body, respectively. Pull-down assay shows that SE core provides the platform for HYL1 and DCL1 binding, whereas in vitro RNA binding and in vivo mutant rescue experiments suggest that the non-canonical zinc-finger domain coupled with C-terminal tail binds miRNA precursors. SE presumably works as a scaffold-like protein capable of binding both protein and RNA to guide the positioning of miRNA precursor toward DCL1 catalytic site within miRNA processing machinery in plant.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs of ∼21 nt in length and participate in a wide variety of physiological cellular processes by regulating the gene expression at a sequence-specific manner (1). Primary microRNA (pri-miRNA) is transcribed by RNA polymerase II to form a hairpin-like secondary structure and subsequently processed into precursor microRNA (pre-miRNA) and then into miRNA/miRNA* duplex with 2 nt overhangs at 3′-terminus by ribonuclease III family protein Dicer (2,3). The mature miRNA is loaded into RNA induced silencing complex (RISC) and paired with target mRNAs for mRNA cleavage or translational inhibition (4,5).
In Arabidopsis, pri-miRNA is processed to pre-miRNA then miRNA/miRNA* duplex within the nucleus by DICER-LIKE1 (DCL1), assisted by dsRNA binding protein HYPONASTIC LEAVES1 (HYL1) and C2H2 zinc-finger protein SERRATE (SE) (6). Interestingly, DCL1 requires presence of both HYL1 and SE to secure its dicing accuracy up to 80% in vitro (6).
Arabidopsis SE controls leaf development, meristem activity and inflorescence architecture. Se-1 mutant plants display a pleiotropic phenotype and show defects on early phase of leaf development (7). SE is predicted to harbor four domains including a single C2H2-type zinc-finger domain near its C-terminus and multiple bipartite nuclear localization motifs at its N-terminus (Figure 1A). SE homolog genes are found in plants, fungi and animals, although the overall similarity is modest (8). The mutation of a SE-like gene in zebrafish displays an embryo-lethal phenotype, which suggests SE gene may have crucial developmental functions in animals (9). More recently, SE orthologue Ars2 was reported to play an essential role in miRNA-mediated silencing in flies and in mammals (10,11).
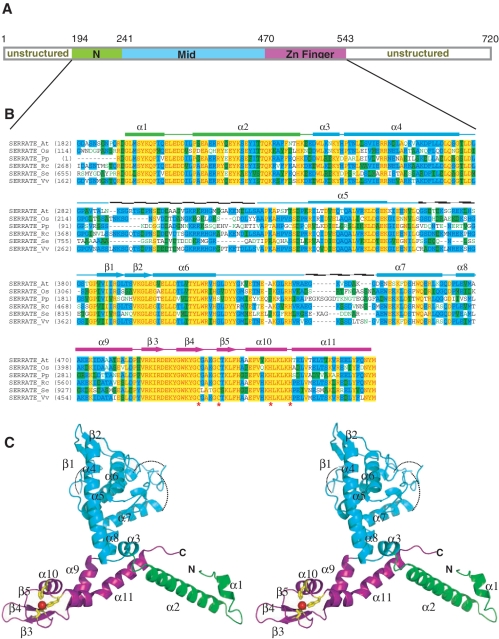
Figure 1.
Domain architecture, sequence alignment and overall structure of Arabidopsis SE core. (A) Schematic of the domain borders in Arabidopsis SE core, N- and C-terminal unstructured regions are indicated. (B) Sequence alignment and secondary structure of SE core. The aligned sequences are in the order of Arabidopsis, Oryza, Physcomitrella, Ricinus, Selaginella and Vitis. The secondary structure diagram for Arabidopsis SE is shown on the top. The α-helices are indicated as bricks, β-strands are indicated as arrows. Three domains are colored in green, cyan and magenta, respectively from N- to C-terminus. Conserved residues are shaded in cyan (80% similarity) and green (60% similarity), whereas essentially invariant residues are shaded in yellow. The critical residues for zinc ion coordination (C500, C505, H518 and H523) are indicated by asterisks beneath the residues. (C) Stereoview ribbon representation of SERRATE core showing the N-terminal domain (green), the middle domain (cyan) and the zinc-finger domain (magenta).
To initiate the structural efforts to investigate the molecular principles of SE-assisted, DCL1-mediated pre-miRNA dicing in Arabidopsis, we report here the crystal structure of Arabidopsis SE core (residues 194–543) at 2.7 Å, which displays a ‘walking man-like’ topology. Our pull-down assays shows that SE core provides the platform for HYL1 and DCL1 binding, whereas in vitro RNA binding and in vivo mutant rescue experiments suggests that the conserved non-canonical zinc-finger domain coupled with C-terminal tail recognizes structural features of miRNA precursors.
MATERIALS AND METHODS
Construction of Escherichia coli expression vectors and protein purification
SE core (residues 194–543) was cloned from cDNA in frame into pET28b vector (Novagen) to be expressed with a carboxy-terminal hexahistidine-tag. SE middle domain (residues 248–470) was cloned into pET28b and pGEX6p-1 vectors to bear carboxy-terminal hexahistidine (cHis) and amino-terminal glutathione-s-transferase (nGST) tags, respectively. HYL1 dsRBD1 (residues 15–84), HYL1 dsRBD2 (residues 100–172) and HYL1 dsRBD1+2 (residues 15–172) were cloned from cDNA into pGEX6p-1 vector with nGST. Cloned likewise were the fragments of DCL1, namely DCL1 DUF283 (residues 836–942), DCL1 PAZ (residues 1176–1353) and DCL1 RBD1+2 (residues 1732–1909) with nGST. All the recombinant proteins were expressed in E. coli (BL21/DE3 strain) overnight at 20°C induced by 0.4 mM isopropyl β-d-thiogalactoside (IPTG). The cHis-fused proteins were purified with Ni2+ affinity column followed by HiLoad Superdex S-75 26/60 column, whereas the nGST-fused proteins were purified with glutathione Sepharose resin (GE Healthcare) followed by HiLoad Superdex S-75 26/60 column. Non-tagged HYL1 RBD1 and RBD2 were obtained by subsequent cleavage of nGST by PreScission enzyme.
Crystallization and data collection
Crystals of SE core were grown by hanging drop vapor diffusion at 20°C. Typically, a 2.0 μl hanging drop contained 1.0 μl of protein (15 mg/ml) mixed with 1.0 μl of reservoir containing 13.5% PEG 4000, 0.7 M potassium formate and 100 mM Tris (pH 7.4) and equilibrated over 1 ml of reservoir solution. These crystals grew to a maximum size of 0.35 × 0.2 × 0.2 mm over the course of 2 days. For data collection, crystals were flash frozen (100 K) in the above reservoir solution supplemented with 30% glycerol. A total of 360 frames per wavelength of 1° oscillation were collected for each crystal on two wavelengths near the zinc edge (1.2814 and 1.2818 Å, respectively) and one wavelength at 1.1 Å and subsequently processed by HKL2000 (www.hkl-xray.com). The crystals belong to space group P212121, with unit cell dimensions a = 50.97 Å, b = 80.66 Å, c = 112.67 Å and α = β = γ = 90° with one molecule per asymmetric unit (Supplementary Table S1).
Structure determination
The crystal structure of SE core was determined by multiple anomalous dispersion method (MAD) based on the anomalous scattering signals from the bound zinc atom using SOLVE/RESOLVE (www.solve.lanl.gov). The initial phase was further improved by density modification assuming a solvent content of ∼53% using the SHARP program (www.globalphasing.com). The model was built by using the program O (http://xray.bmc.uu.se/alwyn) and refined using REFMAC/CCP4 (www.ccp4.ac.uk). The R-free set contained 5% of the reflections chosen at random. The model comprises residues 194–551 (including C-terminal eight extra residues from the vector). Disordered regions, including loop segments 290–324, 366–380 and 435–444, were not included in the model.
Electrophoretic mobility shift assay
Pre-miR164C (miRBase accession MI0001087) was transcribed in vitro containing digoxygenin-labelled uracile (Roche), using RiboMAX Large Scale RNA Production Systems-SP6 and T7 (Promega). The oligo ribonucleotides were incubated at room temperature with different concentrations of the protein in the buffer containing 10 mM Tris, pH 7.5, 50 mM KCl, 1 mM DTT, 10 mM MgCl2, 0.1% NP-40 and 5% glycerol in 20 µl reaction. Following 20 min incubation, the samples were immediately loaded onto 4% native polyacrylamide gel with non-denaturating dye. The resolved nucleic acids were electro-blotted onto Hybond-N+ (GE healthcare) and cross-linked. Blocking, detection and washing of the membrane were performed according to the instruction of DIG GelShift Kit 2nd Generation (Roche).
For electrophoretic mobility shift assay (EMSA) using biotin-labelled pre-miR164c, native pre-miR164c was transcribed in vitro and labelled with EZ-Link Psoralen-PEO3-Biotin (PIERCE). The binding reaction, separation and blotting were performed as above. Blocking, detection and washing of the membrane were performed using Chemiluminescent Nucleic Acid Detection Module Kit (PIERCE).
In vitro GST pull-down assay
GST tagged DCL1 fragments were bound to glutathione sepharose beads (GE healthcare) in binding buffer containing 25 mM Tris, pH 7.4, 1 mM EDTA, 0.01% NP-40 and 2 M NaCl, and incubated with His-tagged SE core protein overnight at 4°C. The bound SE core was washed ten times in the binding buffer and eluted by boiling in SDS–PAGE loading dye. Twenty microlitres of eluents were resolved on SDS–PAGE gel to be immunoblotted with anti-SE core polyclonal antibody. GST-fused SE Mid and NT domains were bound to glutathione sepharose beads (GE healthcare) in the binding buffer of a lower salt content to pull-down non-tagged HYL1 RBD1 and HYL1 RBD2 domains. The bound proteins were detected likewise using anti-HYL1 polyclonal antibody.
Generation of transgenic plants
The se-1 mutant and all transgenic lines used are in the Col-0 background. Sterilized seeds were germinated on MS medium with 1% sucrose. After 3 days of incubation in darkness at 4°C, seeds were incubated at 22°C under long day light condition. Plant were transformed by Agrobacterium (EHA105 strain)-mediated infiltration using the floral dip method (12,13). Seeds from infiltrated plants were selected on MS nutrient medium containing Basta (10 mg/l) and Carbenicillin (100 mg/ml). 35S::6Myc-SE, 35S::3HA-SE, 35S::6Myc-SE core, 35S::6Myc-SE core+C, 35S::6Myc-SE core+N and 35S::6Myc-SE ZF+C transgenes were constructed using pBA002-6myc binary vectors. Constructs were transferred into to se-1 mutant to generate se-1/35S::6Myc-SE, se-1/35S::3HA-SE, se-1/35S::6Myc-SE core, se-1/35S::6Myc-SE core+C, se-1/35S::6Myc-SE core+N, se-1/35S::6Myc-SE ZF+C transgenic lines.
Northern blot analysis
Total RNA was prepared by Trizol (Invitrogen), resolved on 15% TBE-Urea gel, electro-blotted onto Hybond-N+ (GE healthcare) and cross-linked. Biotinylated oligo deoxyribonucleotides complementary to miRNA sequences were purchased from 1st Base (Singapore) to be used as probe. The probe was hybridized with the membrane in ULTRAhybTM (Ambion) overnight at 42°C, washed and detected using LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific Inc., USA) according to the manufacturers’ protocols.
Real-time reverse transcription–PCR
Total RNA was isolated from the aerial parts of wild-type, se-1 and transgenic seedlings using Trizol reagent (Invitrogen). Reverse transcription was using First Strand cDNA Synthesis Kit (Fermentas Inc, USA) or Reverse Transcriptase AMV (Roche). PCR was carried out in the presence of the double-stranded DNA-specific dye Maxima SYBR Green (Fermentas Inc, USA). Amplification was monitored in real time with the 7500 Real Time PCR System (Applied Biosystems, Warrington, UK).
RESULTS
Overall structure of Arabidopsis SE core
Arabidopsis SE core adopts a ‘walking man-like’ topology with N-terminal two α-helices resembling the leading leg, the Middle α-helix dominant domain resembling the body and the C-terminal non-canonical C2H2 zinc-finger domain featuring Helix-kink-Helix (HkH) motif resembling the lagging leg (Figure 1).
The green-colored N-terminal domain (residues 195–240) consists of a short α-helix (α1) followed by an orthogonally oriented long α-helix (α2) that shares the fold of N-terminal half of histone H3 core (Z-score 4.4, r.m.s.d. 2.0 Å, 43 Cα) (Supplementary Figure S1A) (14). The N-terminal domain is connected to the Middle domain through a short α-helix (α3). The cyan-colored Middle domain (residues 241–469) displays a novel fold dissimilar to any solved structure hitherto, according to Dali (www2.embl-ebi.ac.uk/dali). The Middle domain consists of loosely packed three α-helices (α5–α7) oriented orthogonally against a central ∼30° bending long α-helix (α4). A pair of anti-parallel short β-strands (β1, β2) is embedded within a partially disordered long loop connecting α5 to α6, whereas α6 is connected to α7 by another partially disordered long loop. The Middle domain is followed by the C-terminal C2H2 zinc-finger domain via a short α-helix (α8). The presence of three disordered loops (residues 290–324, 366–380 and 435–444) in Middle domain implicates its role in protein–protein interaction with SE partners and/or substrates.
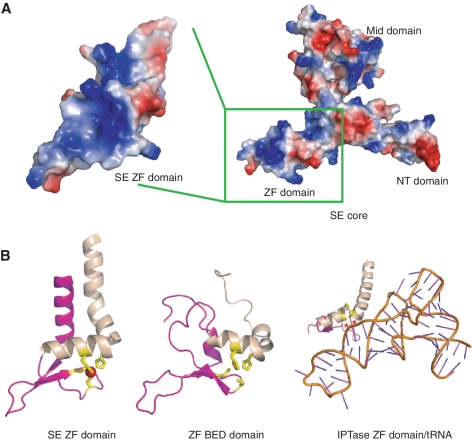
The magenta-colored C-terminal non-canonical C2H2 zinc-finger domain (residues 471–543) adopts a variant C2H2 zinc-finger fold with β-β-β-α architecture (β3–β5 and α10) flanked by one additional α-helix (α9) at its N-terminus and one additional atypical kinked long α-helix at its C-terminus (α11) (Figure 2B, left panel). The central β-β-β-α architecture has the similar structural fold of transcriptional factor Swi5 (Z-score 3.9, r.m.s.d. 2.9 Å, 46 Cα) (Supplementary Figure S1B) (15) and zinc-finger domain of human zinc-finger BED domain containing protein 2 (Z-score, 3.6, r.m.s.d. 4.2 Å, 60 Cα) (Figure 2B, middle panel) (PDB accession: 2DJR). Notably, the structural motif of the zinc-finger domain, including the canonical β-β-α structure and the C-terminal long α helix–kink–helix (HkH) motif, resembles that of IPTase (Figure 2B, right panel) (16). The β-β-α-α(HkH) motif identified in IPTase putatively represents zinc-finger proteins that specifically bind to dsRNA (16).
Figure 2.
SE zinc-finger domain may be involved in RNA binding. (A) Electrostatic potential surface representation of SERRATE core as the same orientation as in Figure 1B. The zinc-finger domain is boxed and highlighted at left panel with ∼45° rotation along y-axis. The SERRATE zinc finger has a highly positive charged surface (colored in blue) for potential nucleic acid binding. (B) Structure-based sequence alignments of SE zinc-finger domain (left panel), C2H2 type zinc-finger domain of human zinc-finger BED domain containing protein 2 (middle panel) and eukaryotic IPTase zinc-finger domain/RNA compelx (right panel). Cysteines and histidines coordinated to zinc are highlighted in yellow and zinc ion is highlighted in red. Residues that form hydrogen bonds with RNA bases via their backbone functional groups in IPTase structure are indicated by asterisks and colored in red. The unique C-terminal HkH motifs within zinc fingers are highlighted in wheat. The views are positioned at the same orientation followed by superimposition.
Role of SE core in SE-HYL1 and SE-DCL1 interactions
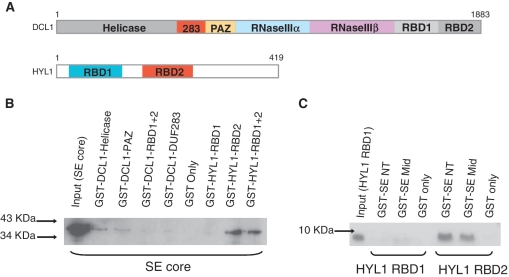
HYL1 and SE are known to bind DCL1 to promote preciseness of pre-miRNA processing (6). However, the domain contacts within the hetero-trimer are yet to be detailed. With the SE core structure in hand, we attempted on mapping of domain interactions between SE, HYL1 and DCL1 by in vitro pull-down assay. DCL1 RNaseIIIa+IIIb domains could not be tested in this assay due to low solubility of this fragment expressed in bacterium. GST-fused DCL1 helicase and PAZ domains showed weak interactions with hexahistidine-tagged SE core, detected by polyclonal anti-SE core antibody raised in rabbit (Figure 3A and B). In addition, both GST-fused HYL1 RBD1+2 and GST-fused HYL1 RBD2 were able to pull-down SE core, whereas GST-fused HYL1 RBD1 failed to pull-down SE core (Figure 3A and B). The observation agrees with the notion proposed by Yang et al. (2010) (17) that HYL1 RBD2, not RBD1, is solely responsible for protein–protein interaction. Next, we asked whether a certain domain within SE core specifically participates in or eludes SE-HYL1 interaction. As expected, both GST-fused SE N-terminal (SE-NT) and SE Middle (SE-Mid) domains were able to pull-down HYL1 RBD2 but not HYL1 RBD1 (Figure 3C). SE C-terminal zinc-finger domain alone could not be expressed in bacterium and so was excluded from this assay.
Figure 3.
Protein network amongst SE, HYL1 and DCL1. (A) Domain architectures of DCL1 (upper panel) and HYL1 (bottom panel). (B) In vitro pull-down assay showing Arabidopsis SE core binds to Arabidopsis DCL1 helicase and PAZ domains as well as Arabidopsis HYL1 RBD2 domain directly. (C) In vitro pull-down assay showing Arabidopsis SE core binds to HYL1 RBD2 via its N-terminal and middle domains.
SE ZF+C is critical to rescue se-1 phenotype
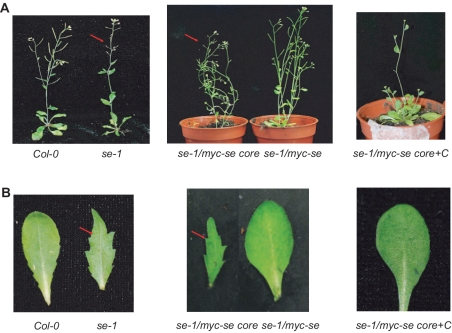
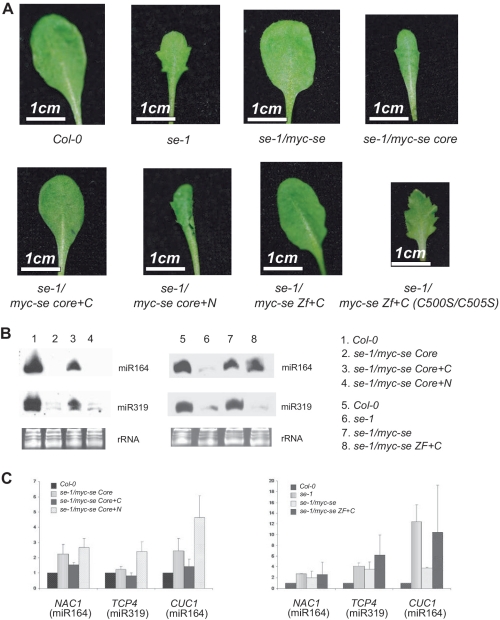
The protein–protein interactions described above were tested within SE core covering only ∼50% of full-length SE sequence. The N- and C-terminal tails outside SE core were predicted to be unstructured and turned out to be non-crystallizable. Nevertheless, these putatively unstructured tails may have important functional impacts for miRNA processing. To investigate this possibility, we generated transgenic lines expressing HA- and myc-tagged full-length SE and SE core under 35S promoter in the se-1 background, respectively. Notably, full-length SE, instead of SE core can considerably rescue the morphological phenotype of se-1 mutant (Figure 4A and B). Typical phenotypes of se-1 mutant, such as leaf serration and abnormal cluster of flowers and siliques (Figure 4A indicated by arrows colored in red), went unobserved by expression of full-length SE protein. By contrast, transgenic Arabidopsis expressing myc-tagged SE core in the se-1 background showed pleiotropic phenotypes characteristic of se-1 mutant line.
Figure 4.
SE C-terminal region is essential to rescue se-1 mutant phenotype. (A and B) Comparison of the morphological phenotype between Col-0, se-1, se-1/35S::myc-se, se-1/35S::myc-se core, se-1/35S::myc-se core+C plants.
Surprisingly, the expression of longer fragment including SE core and C-terminal tail (SE core+C) was sufficient to rescue the miRNA morphology phenotype (Figure 4A and B). Along with the recovery of WT morphology, miRNA levels of se-1 mutant, exampled by miR164 and miR319, as well as their mRNA targets were also restored close to WT expression levels (Figure 5B and C). By contrast, neither SE core nor SE core+N was able to restore the miRNA deficiency morphology (Figure 5B and C). These molecular and morphological observations indicate that SE core+C can largely restore SE deficiency in the se-1 mutant background and its activity is comparable to that of SE.
Figure 5.
Both SE zinc-finger domain and C-terminal region is required to rescue se-1 mutant phenotype. (A) Comparison of the morphological phenotype between Col-0, se-1, se-1/35S::myc-se, se-1/35S::myc-se core, se-1/35S::myc-se core+C, se-1/35S::myc-se core+N, se-1/myc-se Zf+C, se-1/myc-se Zf+C (C500S/C505S) plants. (B and C) Accumulation of small RNAs and target mRNAs (C) in Col-0, se-1, se-1/35S::myc-se, se-1/35S::myc-se core, se-1/35S::myc-se core+C, se-1/35S::myc-se core+N and se-1/35S::myc-se ZF+C plants. Each lane contained 12 µg RNA. rRNAs were used as a loading control.
To narrow down the minimum requirement of SE domains capable of phenotype rescue, we made a truncated fragment consisting only of the zinc-finger domain and the C-terminal tail (SE ZF+C). Remarkably, the expression of SE ZF+C in the se-1 mutant background was able to rescue serration phenotype. The expression of miRNAs was rescued and the expression of their target mRNAs was decreased likewise (Figure 5B and C). Consistent to the notion of the importance of C-terminal ZF domain, the introduction of point mutations at the two cysteine residues involved in Zinc coordination (C500S/C505S double mutant) disrupted the rescue ability and caused reversion of serration phenotype and extremely retarded growth (Figure 5A).
SE core+C binds pre-miRNA in vitro
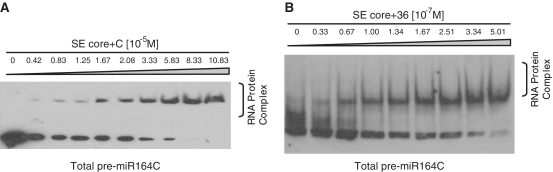
The phenotype rescue experiment indicates that SE core together with some undetermined residues located somewhere in C-terminal tail retain in vivo function. Next, we asked whether the functional role in miRNA processing displayed by SE in vivo is due to the RNA binding ability of SE core+C. To this end, we tested substrate-binding ability of SE core+C by EMSA in vitro, and found that SE core+C can stably associate with biotin-labelled pre-miR164c (Figure 6A). To further narrow down the necessary residues within C-terminal tail that enables SE zinc-finger domain to bind miRNA precursors, we made a series of SE constructs gradually lengthening the tail from SE core toward C-terminus and tested substrate binding by EMSA in vitro. We found out that the fragment comprising SE core plus 36 residues (194–579) is enough to stably associate with internally digoxygenin-labelled pre-miR164c (Figure 6B). These results show that SE is an RNA binding protein and that substrate recognition by SE probably plays a role in phenotype rescue.
Figure 6.
SE core+C binds pre-miR164c in vitro. (A) EMSA results showing that SE core+C (residues 194–720) binds biotin-labelled pre-miR164c. (B) EMSA results showing that SE core+36 amino acid (residues 194–579) binds digoxigenin-labelled pre-miR164c.
DISCUSSION
The biochemical analysis of Arabidopsis SE demonstrated that SE is an RNA binding protein contributed from the non-canonical C2H2 zinc-finger domain and C-terminal tail. The topological similarity between SE C2H2 zinc-finger motif and IPTase HkH zinc-finger domain prompted us to superimpose SE core structure with IPTase/tRNA complex aligned on zinc-finger domain and examine the possible arrangement of dsRNA bound along the SE core structure.
In our computational model, the N-terminal α-helix of the HkH motif packs against the minor groove of the bound dsRNA, whereas the C-terminal helix runs across the major groove (Supplementary Figure S2). Several highly conserved residues (K522, T524 and T528) are positioned to a place near the junction of stem and D-loop. Interestingly, the architecture of tRNA, especially at the junction of stem and D-loop resembles the ssRNA–dsRNA junction within pre-/pri-miRNA, which tempts us to speculate that the recently discovered tRNA-derived small RNAs may adopt a cleavage mechanism similar to that of miRNAs (18).
The RNA recognition model presented here is shared among other dsRNA-specific zinc-finger proteins (16). However, our SE sequence lacks a highly conserved Lys/Arg residue one helical turn right after the second zinc-coordinating histidine residue (16) (Figure 1A), reminding us that SE C2H2 zinc-finger domain is essential but not sufficient for ssRNA–dsRNA junction recognition. Therefore, other critical residues, most likely from the conserved C-terminal tail of SE, are expected to recognize ssRNA–dsRNA junction of pre-miRNA. This hypothesis is consistent with our observation that SE core was not sufficient to rescue the se-1 mutant phenotype in vivo.
Our SE core structure supplemented by rescue experiment in vivo and RNA binding assay in vitro suggest that SE works both in recognition of RNA substrate by ZF domain together with C-terminal tail and in guidance of RNA into the miRNA processing machinery through protein–protein interaction, like a scaffold bridging RNA, HYL1 and DCL1, and guarantee a synergistic stimulating effect on DCL1 catalyzed miRNA processing (6). We speculate that SE homolog, Ars2, probably adopts similar structural principles to recognize miRNA precursors in flies and in mammals.
Having noted that SE ZF+C fragment lacking SE-NT and Mid is capable of phenotype rescue, a question arises of whether SE-NT plus Mid possibly evades a role in DCL1 complex formation, contradicting our afore statement that SE constitutes the ternary complex together with HYL1 mediated by SE-NT and Mid domains. We speculate that although SE ZF+C alone is able to largely rescue the miRNA deficient phenotype in vivo, SE-NT, Mid domains and/or N-terminal tail might be indispensable for yet unidentified protein–protein interactions involved in miRNA processing. This line of speculation was inspired by the observation that rescue of leaf serration with in vivo expression of ZF+C failed to entail recovered accumulation of miR319 and down-regulation of its target (Figure 5B), despite apparently normal leaf morphology and vigor of the plant. More rigorous morphological and genetic analyses would be required to explicate the discrepancy and assess the possibility of SE domain-specific involvement in different miRNA maturation and of having other SE-requiring miRNAs whose deficiency inflicts more significant impacts on leaf morphology than miR319 does. Furthermore, our unexpected discovery of minimal SE-fragment capable of serration rescue further raises intriguing questions of how other parts of SE than ZF+C participate in and facilitate RNA metabolism at the bifurcation of micro- and messenger-RNA processing pathways (19). SE might possess structural features to sort out the substrates among RNAs rich in variety of size and structures, and to allocate binding partners to form either splicing or dicing complexes, for the latter of which our crystallization effort is under way.
ACCESSION NUMBER
Coordinates and the structure factors have been deposited to Protein Data Bank with accession codes 3AX1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Singapore Ministry of Education (T208A3124), Research scholarship from National University of Singapore and intramural research fund from Temasek Life Sciences Laboratory. Funding for open access charge: Intramural fund, Temasek Life Sciences Laboratory.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Saxena at X12C at the National Synchrotron Light Source of the Brookhaven National Laboratory for assistance with data collection and N.-H. Chua for insightful discussion. S.M., H.-Y.C. and Y.A.Y. designed the project; S.M. performed biochemical and genetic work; H.-Y.C. performed crystallographic work; S.M. and Y.A.Y. wrote the article with input from H.-Y.C.
REFERENCES
- 1.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 3.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 5.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl Acad. Sci. USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke JH, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- 8.Prigge MJ, Wagner DR. The arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell. 2001;13:1263–1279. doi: 10.1105/tpc.13.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, et al. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- 10.Gruber JJ, Zatechka DS, Sabin LR, Yong J, Lum JJ, Kong M, Zong WX, Zhang Z, Lau CK, Rawlings J, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabin LR, Zhou R, Gruber JJ, Lukinova N, Bambina S, Berman A, Lau CK, Thompson CB, Cherry S. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 14.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 15.Neuhaus D, Nakaseko Y, Schwabe JW, Klug A. Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of beta-sheet. J. Mol. Biol. 1992;228:637–651. doi: 10.1016/0022-2836(92)90846-c. [DOI] [PubMed] [Google Scholar]
- 16.Andreeva A, Murzin AG. A fortuitous insight into a common mode of RNA recognition by the dsRNA-specific zinc fingers. Proc. Natl Acad. Sci. USA. 2008;105:E128–E129. doi: 10.1073/pnas.0810994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Gene Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Ratsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.