Abstract
The abnormal number of repeats found in triplet repeat diseases arises from ‘repeat instability’, in which the repetitive section of DNA is subject to a change in copy number. Recent studies implicate transcription in a mechanism for repeat instability proposed to involve RNA polymerase II (RNAPII) arrest caused by a CTG slip-out, triggering transcription-coupled repair (TCR), futile cycles of which may lead to repeat expansion or contraction. In the present study, we use defined DNA constructs to directly test whether the structures formed by CAG and CTG repeat slip-outs can cause transcription arrest in vitro. We found that a slip-out of (CAG)20 or (CTG)20 repeats on either strand causes RNAPII arrest in HeLa cell nuclear extracts. Perfect hairpins and loops on either strand also cause RNAPII arrest. These findings are consistent with a transcription-induced repeat instability model in which transcription arrest in mammalian cells may initiate a ‘gratuitous’ TCR event leading to a change in repeat copy number. An understanding of the underlying mechanism of repeat instability could lead to intervention to slow down expansion and delay the onset of many neurodegenerative diseases in which triplet repeat expansion is implicated.
INTRODUCTION
Triplet repeat diseases constitute a class of genetic diseases in which there is an expanded number of repeats in regions of the genome containing repetitive stretches of three nucleotides (1). The abnormal number of repeats found in these diseases arises from a process termed ‘repeat instability’, in which repeats are subject to a change in copy number. Many neurological diseases fall into this category, including a subset, known as polyglutamine diseases, involving an expanded copy number of the trinucleotide CAG in a coding region, leading to proteins with an excessive number of glutamines. There are at least nine polyglutamine diseases, including Huntington's disease (HD), spinobulbar muscular atrophy (Kennedy disease), dentatorubral–pallidoluysian atrophy (Haw River syndrome) and several spinocerebellar ataxias (1–4). Several lines of evidence highlight the importance of repeat number, and consequent repeat expansion, in disease progression. Significantly, in polyglutamine diseases the age of onset correlates inversely with the length of repeats, for which a higher CAG repeat number is indicative of an earlier onset of disease symptoms (1,5). Repeat length is subject to change due to repeat instability, a process that occurs both in germline and somatic tissues (3,6). Somatic instability of CAG repeats is both tissue-specific and age dependent, and has been documented in HD mouse models and HD patients (7–10). Direct analysis of CAG repeat length in HD patients reveals that the highest levels of repeat instability appear in the brain, with the regions, such as the basal ganglia, most affected by HD displaying the highest levels of repeat instability and juvenile onset patients displaying higher levels of repeat instability in these regions as compared to adult onset patients (7). These data are consistent with the idea that the tissue-specific effects observed in HD patients arise from higher repeat instability in affected tissues. Notably, low levels of repeat instability are observed in tissues in which cell turnover is high, such as the blood and intestine, suggesting that somatic instability involves processes other than cell proliferation (7).
Various molecular mechanisms for triplet repeat instability have been proposed, implicating mainly DNA replication, recombination and/or repair [for review see (2,6,11)]. Many of these mechanisms involve the formation of unusual secondary structures within triplet repeat DNA. One of these structures is the hairpin, which can form in palindromic sequences and consists of a single strand folding back on itself, forming Watson/Crick hydrogen bonded bases and adopting an intramolecular B-DNA structure capped by a loop (12). Each of the strands within our current sequence of interest, (CAG)n•(CTG)n, can form mismatched hairpins, whenever the strands separate during DNA replication, recombination or transcription, stabilized by pairing between C and G, and containing either a mismatched A•A or T•T (3). Although both CTG and CAG repeats can form mismatched hairpins, CTG repeat hairpins have been shown to be more stable (13–15). Both the propensity of (CAG)n•(CTG)n to form alternate structures and the probability of repeat expansion increase sharply with the number of repeats (2,3,16,17).
As mentioned above, triplet repeat expansion has been shown to occur in the brain, in which cells are generally not dividing, and therefore a molecular mechanism independent of DNA replication is necessary to account for expansion in these cells. Recent studies provide evidence for a novel expansion/contraction model involving both transcription and DNA repair (18–20). Specifically, these studies provide evidence for a role of transcription-coupled repair (TCR) in repeat instability. TCR, a sub-pathway of nucleotide excision repair (NER), is dedicated to removal of transcription-blocking lesions from the transcribed strand (TS) of active genes. The arrest of RNAPII during transcription due to a DNA lesion is known to trigger TCR and lead to the recruitment of repair proteins (21,22). Studies have shown that upregulation of transcription enhances repeat instability, as assayed by triplet repeat contractions, and that this effect is dependent on TCR proteins (18). The study identified nine proteins involved in the same pathway of transcription-induced instability, including proteins involved in mismatch repair (MMR) (proteins MSH2 and MSH3), TCR (CSB), NER (XPA, ERCC1 and XPG) and proteins that may interact with the stalled RNA polymerase (RNAPII) (TFIIS, BRCA1 and BARD1) (18–20). Proteins involved only in global genome NER, the other sub-pathway of NER, (XPC) and proteins involved in base excision repair (OGG1) were not implicated. From these studies, a transcription-dependent pathway for CAG repeat instability was proposed, involving unusual DNA structures and TCR (18). The first step of the proposed pathway involves the generation of slipped-strand structures upon transcription, formed by (CAG)n•(CTG)n repeats base pairing out of register. The CTG slip-outs are thought to be in a hairpin conformation, while the CAG slip-outs are mainly in a loop conformation, probably due to greater helix disruption of the purine A•A versus the pyrimidine T•T mismatch (3,13). The hairpins are then stabilized by MutSβ, a complex formed by the MMR proteins MSH2 and MSH3. The next translocating RNAPII is then thought to arrest upon encountering the CTG hairpin on the TS, signaling for TCR. DNA repair is initiated and, depending upon where the CTG and CAG slip-outs are relative to each other, can result in expansion or contraction. If a CTG hairpin is opposite a larger CAG loop, then during repair synthesis the larger CAG loop would be copied and expansion would occur. Alternatively, when a CTG hairpin is opposite a strand without a slip-out at that position, contraction would occur (18–20). The arrest of RNAPII by a CTG hairpin is a crucial step in the transcription-dependent repeat-instability model, since it is the event that is thought to trigger TCR. An earlier study with an in vitro synchronized-transcription system had shown that RNAPII could pause within CTG repeats (23). Here, we directly test whether CAG or CTG slip-outs can arrest transcription. We designed DNA constructs containing a (CAG)20 or (CTG)20 slip-out and performed in vitro transcription studies to determine whether RNAPII or T7 phage RNAPII (T7 RNAP) is arrested upon encountering these structures.
MATERIALS AND METHODS
DNA substrates
The general strategy for preparing the DNA substrates is shown in Figure 1A. The promoter fragment, containing the adenovirus major late promoter (RNAPII promoter) and the T7 RNAP promoter, was obtained by restriction digest with EcoRI and BamHI [New England Biolabs (NEB)] of the pWT plasmid (24) which is a derivative of the pUCG7G-TS plasmid (25). The fragment was gel purified as described previously (24), without exposure to either ethidium bromide (EtBr) or UV light. To obtain longer promoter fragments, the plasmid was restricted with EcoRI and BglII and the fragment was purified as described above.
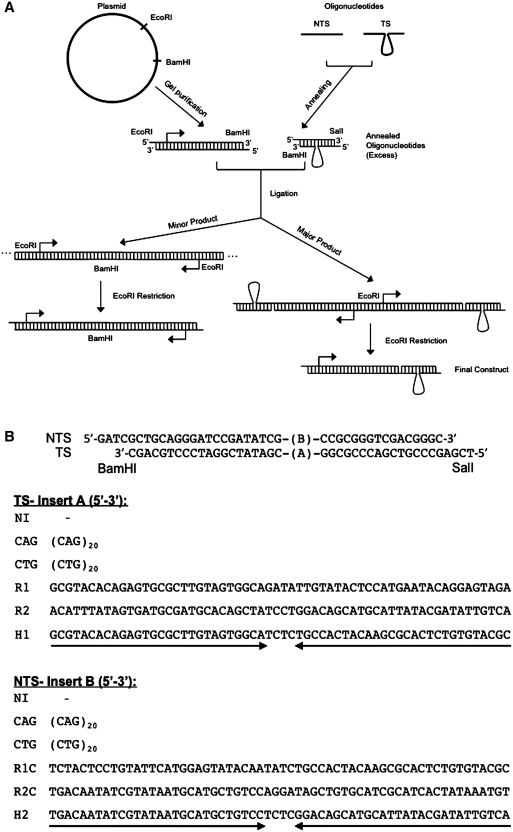
Figure 1.
(A) Preparation of DNA substrates. The plasmid was cut with restriction enzymes EcoRI and BamHI to yield a fragment containing the T7 RNAP and RNAPII promoters. Oligonucleotides were designed such that, when annealed, they would produce a slip-out and contain BamHI and SalI sticky ends. The olgonucleotides were ligated to the promoter fragment through the BamHI site. Oligonucleotides were not 5′ phosphorylated in order to prevent self-ligation and were used in excess to promote ligation of most promoter fragments to an oligonucleotide. A minor fraction, consisting of promoter fragments ligated through the BamHI site still appeared, however, this fraction does not interfere with our analysis because its transcription product is longer than that produced by our construct of interest. Due to lack of 5′ phosphorylation of oligonucleotides, our substrates contain a nick in the NTS, however, the presence of a nick on the NTS had no effect on the observed transcription arrest (Supplementary Figure SI6). Ligation was followed by EcoRI digestion to obtain the final substrates containing one promoter fragment, with T7 RNAP and RNAPII promoters (designated by the arrow), and a slip-out. (B) Sequences used in this study. Oligonucleotides sequences, which will become the TS or the NTS when ligated to a promoter fragment, are shown. Different combinations of oligonucleotides result in different inserts in the middle but have the same flanking sequence (shown on top). The sequence of different inserts is indicated in the 5′ to 3′ direction. NI (‘No insert’) refers to oligonucleotides containing only the flanking sequence. The designation of inserts is as follows: CAG, 20 CAG repeats; CTG, 20 CTG repeats; R1, random sequence; R1C, complementary to R1; R2, random sequence; R2C, complementary to R2; H1, capable of forming a perfect hairpin; H2, capable of forming a perfect hairpin.
Single stranded DNA oligonucleotides with the indicated sequences were purchased from Integrated DNA Technologies (IDT) with PAGE purification. The sequences of these single-stranded oligonucleotides were designed such that the non-TS (NTS) and TS were completely complementary except for the presence or absence of an insert [(CAG)20, (CTG)20, hairpin or random sequence] right in the middle. Annealing of the oligonucleotide strands resulted in 4-nt sticky ends for ligation to the promoter-containing fragment (Figure 1B). Annealing was performed in 10 mM Tris–HCl, 10 mM MgCl2 with 2 µM of each complementary single-stranded oligonucleotide for 10 min at 65°C, followed by 3 h at 37°C.
The annealed oligonucleotides and the promoter fragment were ligated in reactions containing 850 nM of annealed oligonucleotides, 85 ng of pure promoter fragment, 1× ligase buffer (NEB) and T4 Ligase (2000 U) (NEB) in a total volume of 20 µl at 16°C overnight. The reaction was then heated for 30 min at 65°C to inactivate ligase and the constructs were digested with EcoRI to obtain constructs containing one promoter fragment ligated to one oligonucleotide (Figure 1A). The constructs were analyzed by 1.5% agarose gel electrophoresis run for 4 h at 60 V, and visualized by EtBr staining and exposure to UV light (Supplementary Figure SI1).
RNAPII transcription assay
Each reaction contained 2 µl of DNA sample (≈0.8 nM), 8 U of HeLa nuclear extracts (HeLa NE) (HeLaScribe Promega) and 7.5 µl of 1× transcription buffer [20 mM HEPES (pH 7.9 at 25°C), 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol] (Promega) in a total volume of 18 µl. The reaction mixture was incubated at 28°C for 45 min to allow for formation of initiation complexes. After incubation, 40 U of RNasin, 1 µl of NTPs (10 mM ATP, GTP, UTP and 0.4 mM CTP) and 40 µCi of [α-32P] CTP were added and the reaction was incubated at 28°C for 15 min. To each reaction, 1 µl of 1.25 µg/ml heparin was added and the reaction was incubated at 28°C for another 30 min. The reaction was the stopped by addition of stop solution [0.3 M Tris–HCl (pH 7.4), 0.3 M sodium acetate, 0.5% SDS, 2 mM EDTA and 3 µg/ml tRNA]. Nucleic acids were extracted with 50% phenol, 48% chloroform, 2% isoamyl alcohol, precipitated with ethanol and dried using a SpeedVac. The pellets were resuspended in 6 µl of denaturing formamide loading dye and run on a 5% polyacrylamide gel containing 8 M of urea. The gel was run for 1 h at 2000 V, dried and results were visualized by autoradiography and by using a PhosphorImager.
T7 RNAP transcription assay
Reactions were performed in a total volume of 12 µl, containing 2 µl of 5X transcription buffer (Promega), 4 mM DTT, 16 U of RNAsin (Promega), 0.17 mM of each of ATP, GTP and UTP, 0.017 mM of CTP, 10 µCi [α-32P] CTP, 20 U of T7 RNAP (Promega) and ≈1 nM of corresponding DNA substrate. The reactions were incubated at 37°C for 30 min. The reaction was stopped and precipitated as described previously (24). The samples were run in a 5% polyacrylamide gel containing 8 M of urea for an hour and a half at 2000 V. The results were visualized using a PhosphorImager.
T7 RNAP transcription assay in HeLa nuclear extracts
Substrates containing only the T7 RNAP promoter were prepared as described above in ‘DNA substrates’ except that the promoter fragment was obtained by digestion of the pBluescript SK(+) with BamHI and BsmFI. The protocol for RNAPII transcription was followed by the addition of 40 U of T7 RNAP.
RESULTS
CTG and CAG slip-outs arrest RNAPII
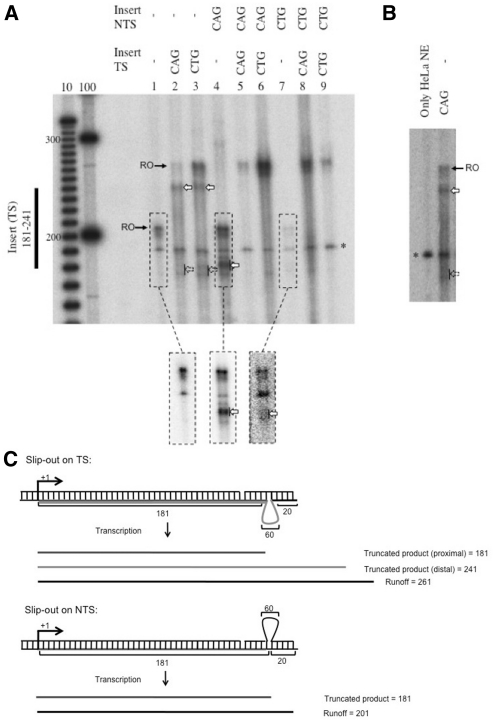
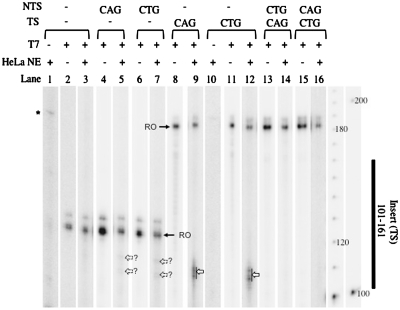
To directly test whether CAG or CTG slip-outs can cause transcription arrest, we prepared linear DNA substrates designed specifically to contain a slip-out. We used DNA oligonucleotides that were completely complementary to each other, except for an insert containing (CAG)20 or (CTG)20 repeats in the middle (Figure 1B). When annealed, these oligonucleotides formed a (CAG)20 or (CTG)20 slip-out. The annealed oligonucleotides were then ligated to a DNA fragment containing a T7 RNAP and an RNAPII promoter (Figure 1A). The CAG and CTG slip-outs formed mismatched hairpins, as evidenced by their mobility in polyacrylamide gels (Supplementary Figure SI4). We used these DNA constructs as substrates to perform an in vitro transcription assay using HeLa nuclear extracts as the source of RNAPII and the required transcription factors. DNA substrates were incubated with HeLa nuclear extracts and radioactively labeled nucleotides to produce radioactively labeled RNA (see ‘Materials and Methods’ section). In the case of constructs with a completely complementary sequence, the predominant band corresponds to the run-off (full-sized) products (Figure 2A, lanes 6 and 8). However, truncated products, which we interpret as resulting from transcription arrest, were found when a (CTG)20 or (CAG)20 slip-out was present either on the transcribed strand (TS) or non-transcribed strand (NTS) (Figure 2A, lanes 2–4, 7). When the slip-out was on the TS, truncated transcription products appeared at ∼180 and 240 nt (Figure 2A, lanes 2 and 3). These product lengths are consistent with the position of the proximal, 181 nt, and distal, 241 nt, ends of the slip-out with respect to the promoter (Figure 2C). Arrest due to a slip-out on the NTS occurred for both CAG and CTG slip-outs (Figure 2A, lanes 4 and 7). When the slip-outs were on the NTS, truncated products were found at around 180 nt, corresponding to the position of the slip-out on the NTS (Figure 2C). Strongest arrest occurred when the CAG slip-out was on the TS; in this case, truncated products comprise ∼85% of total products (Supplementaty Data). When the CAG slip-out was on the NTS, the intensity of truncated products corresponded to ∼40% of total products.
Figure 2.
(A) RNAPII transcription is arrested by a CTG or CAG slip-out on either TS or NTS in HeLa NE. Runoff products (indicated by black arrows and RO) appeared as expected (201 nt for no insert on TS and 261 nt for CAG or CTG insert on TS). Run-off products often contained several closely located bands, probably due to either somewhat increased probability of spontaneous transcription termination near the end of substrates or template independent addition of several nucleotides to the runoff transcript. This heterogeneity occurs in close vicinity of run-off product and does not affect interpretation of our results. Truncated products appeared at ∼240 nt and at ∼180 nt for constructs with a CTG hairpin or a CAG loop on the TS. Truncated products also appeared at ∼180 nt when a CAG or CTG slip-out was on the NTS. Pronounced products are indicated by white-block arrows and less pronounced products are indicated by dashed-block arrows. Dashed boxes show magnification of that area with contrast. Asterisk (*) indicates a 190-nt band that appears in extract even without DNA substrates (see B). (B) Band at 190 nt in HeLa NE is independent of DNA substrates. Transcription reaction was performed in the absence of DNA substrates (lane 1) and the band at 190 nt was still present. (C) Diagram of truncated products that appear when a CTG or CAG slip-out is on either the transcribed or NTS. When a slip-out was on the TS, truncated products correspond to proximal (181 nt) and distal (241 nt) end of the slip-out. When a slip-out was on the NTS, truncated products correspond to the position of the slip-out (181 nt).
A band at ∼190 nt, designated by an asterisk, appeared in every transcription reaction in HeLa nuclear extracts, regardless of the DNA substrate used (Figure 2A). In fact, this band appeared even in the absence of DNA substrates, indicating that it is intrinsic to the HeLa nuclear extracts and unrelated to our DNA constructs (Figure 2B). Previous studies have also found similar bands when using HeLa nuclear and whole-cell extracts. These bands were found to be independent of de novo RNA synthesis and are thought to be produced by end labeling of RNA (particularly rRNA and its breakdown products or tRNA) (26,27). DNA substrates containing the same structures but with longer promoters (new runoff products expected at 246 and 306 nt) were prepared to displace the band from the region of interest in the gels. These substrates produced runoff products of the expected lengths and yielded the same pattern of arrest relative to the runoff as previously observed (Supplementary Figure SI2A). These longer substrates also revealed some truncated products when using substrates containing CAG or CTG on both strands (Supplementary Figure SI2A); these had not been observed in previous experiments because the position overlapped with the 190-nt band (Figure 2A). Additional controls show that the truncated products were not the result of digestion of DNA substrates in the extracts (Supplementary Figure SI3).
Hairpins and loops arrest RNAPII
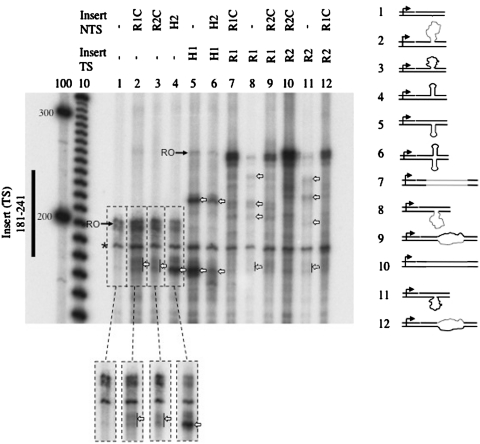
Given the finding that both CTG and CAG slip-outs cause RNAPII arrest, further experiments were performed to explore the nature of this arrest. Is the arrest due to the structures formed by the slip-outs or does it depend both on the structure and its sequence? The structures formed by CTG and CAG slip-outs are either a mismatched hairpin or a loop (15). Therefore, we tested whether either a slip-out forming a hairpin or one forming a loop would also arrest RNAPII transcription. Substrates containing an insert with either a hairpin-forming or a random sequence were prepared in the same way as the substrates containing CAG and CTG inserts (Figure 1). In vitro RNAPII transcription using nuclear extracts indicated that a hairpin or a loop on either strand cause RNAPII arrest (Figure 3). When either a loop or a hairpin was present on the NTS, arrest always occurred at around 170–180 nt (Figure 3, lanes 2–4), which corresponds roughly to the position of the insert in the NTS (Figure 2C), while no arrest was detected in the absence of these structures (Figure 3, lane 1). When the insert was on the TS, RNAPII transcription gave rise to a complicated pattern of truncated products. A hairpin on the TS and a cruciform reproducibly showed well-pronounced arrest. However, the particular pattern of truncated products varied between experiments (compare Figure 3 and Supplementary Figure SI2B), possibly due to variation in the concentrations of DNA binding proteins in extract aliquots (see below).
Figure 3.
RNAPII transcription is arrested by a hairpin or loop on either strand. Hairpin or loop structures present on the TS or NTS cause RNAPII arrest. Runoff (full-length) transcripts (indicated by a black arrow and RO) appeared at 201 nt, as expected, for constructs containing no insert on TS and 261 nt for those with an insert on TS. The presence of a hairpin or loop on the TS gave rise to a complicated pattern of blockage. The presence of a hairpin or loop on the NTS gave rise to truncated products at ∼170–180 nt. White arrows indicate truncated products with intensity comparable to or exceeding that of the run-off and dashed-block arrows indicate less pronounced blockage that is still comparable to the runoff. The designation of inserts is as follows: (−), no insert; R1, random sequence; R1C, complementary to R1; R2, random sequence; R2C, complementary to R2; H1, capable of forming a perfect hairpin; H2, capable of forming a perfect hairpin.
To examine the effect of a loop on transcription, we tested two different inserts with random sequences, R1 and R2 (Figure 1B). For substrates with completely complementary strands, the predominant product corresponds to full-length products, although some minor truncated products appeared, probably due to some weak sequence-specific blockage signal (Figure 3, lanes 7 and 10). However, when either random sequence was looped-out, the observed arrest became much stronger, as compared to the runoff (Figure 3, lanes 8 and 11). Substrates with two non-complementary loops (R1-R2C or R2-R1C) also show sites of arrest but to a much weaker extent (Figure 3, lanes 9 and 12). These results suggest that the pattern of arrest for these random sequences is sequence-dependent but can become exacerbated by the formation of a looped-out structure. However, it is significant that all tested structures, CTG or CAG slip-out, hairpin and loop, show arrest near both the proximal and distal ends of the structure, suggesting that once CTG or CAG repeats form a slip-out, the RNAPII arrest does not depend on the presence of the particular CAG or CTG repetitive sequence.
Distinct arrest of RNAPII and T7 RNAP in HeLa nuclear extracts
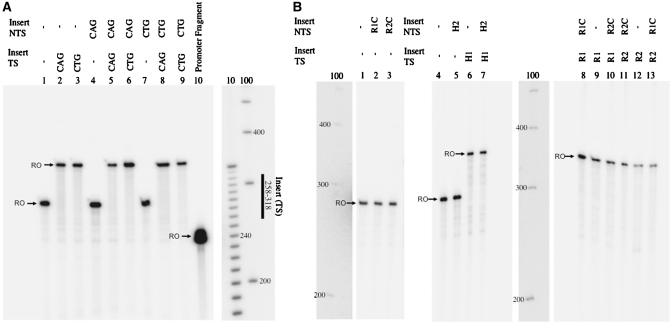
Our results show RNAPII arrests upon encountering a CTG or CAG slip-out on either strand in nuclear extracts. However, T7 RNAP in a purified system does not arrest upon encountering these structures (Figure 4). Several possibilities could account for this difference. First, a purified system was used for T7 RNAP transcription while nuclear extracts were used for RNAPII transcription. Consequently, RNAPII transcription occurs in the presence of other proteins, which could bind the CAG and CTG slip-outs causing RNAPII arrest, while no arrest is caused for T7 RNAP transcription because these proteins are not present. Another possibility is that the two polymerases have distinct properties, causing RNAPII to arrest upon encountering these structures but not T7 RNAP. Finally, it is possible that both of these factors have a role in causing the arrest. To explore these possibilities, we performed a T7 RNAP transcription reaction in the presence of nuclear extracts. If the difference between RNAPII and T7 RNAP behaviors is due to protein binding, then the addition of nuclear extract should cause arrest for T7 RNAP. The experiment was performed using the RNAPII protocol but the substrates only had a T7 RNAP promoter, and thus no RNAPII transcription should occur (see ‘Materials and Methods’ section). As a control, we performed the reaction without the addition of T7 RNAP and, as expected, obtained no transcription products (Figure 5, lanes 1 and 10). For all CAG and CTG slip-out substrates, we performed the reaction in the presence or absence of nuclear extracts and under the same conditions used for RNAPII transcription. All substrates that did not contain a slip-out showed no difference between the transcription products in the presence or absence of nuclear extracts (Figure 5, lanes 2, 3, 13–16). The runoff, as expected, was around 181 or 121 for substrates with or without a slip-out on the TS, respectively. Products a few nucleotides longer than the runoff appeared for all samples and these are most likely related to a known T7 RNAP property of addition of extra nucleotides under non-optimal conditions (28). When the slip-outs are on the NTS, two very faint bands, barely distinguishable from background, appear in the presence of nuclear extracts, between 100 and 110 nt, corresponding roughly to the position at which RNAPII arrest occurred for substrates with the same inserts (Figure 6, lanes 4 versus 5 and 6 versus 7). The intensity of these bands, compared to the runoff, was much lower than that observed for RNAPII arrest, suggesting that even if proteins play some role in causing the difference in polymerase arrest at this position, this role is minor and that other factors, such as difference in polymerase properties, are responsible for the observed differences at this position. T7 RNAP transcription of substrates with a slipped-out CAG or CTG in the TS revealed the appearance of truncated products between 100 and 110 nt in the presence of nuclear extracts (Figure 6, lanes 8 versus 9 and 11 versus 12). This length corresponds to a short distance beyond the position of the proximal end of the slip-out, one of the sites at which RNAPII arrest occurred. However, there is no arrest at the other site of RNAPII arrest (distal end of the slip-out, which would correspond to a length of 161 nt), suggesting that the difference in arrest at the proximal end of the slip-out was mainly due to the presence of proteins, while the difference in arrest at the distal end of the slip-out reflects a difference in polymerase properties. Decreasing the concentration of T7 RNAP 10-fold produced the same amount of arrest when the slip-out was on the TS, indicating that the concentration of T7 RNAP does not affect the amount of transcription arrest found when the reaction is performed in the presence of nuclear extracts (Supplementary Figure SI7).
Figure 4.
T7 RNAP transcription in a purified system is not arrested by a DNA slip-out. (A) T7 RNAP transcription was performed using DNA substrates containing CAG or CTG slip-outs. Transcription product sizes were as expected for full length transcripts (indicated by a black arrow and RO), 278 nt for constructs with no insert in TS and 338 nt for those with either an insert on the TS and no truncated products appeared for any of the tested substrates. (B) T7 RNAP transcription of substrates with a hairpin or a loop produced full-length transcripts (338 nt or 278 nt) and no truncated products.
Figure 5.
Addition of nuclear extracts causes partial T7 RNAP transcription arrest by a CAG or CTG slip-out. DNA substrates contained only the T7 RNAP promoter and the RNAPII protocol for transcription was followed with addition of T7 RNAP. Full-length products (indicated by a black arrow and RO) appeared, as expected at 181 or 121 nt. Truncated products appeared when the slip-out was on the TS at ∼100–110 nt (lanes 9 and 12). For substrates with a CAG or CTG slip-out on the non-TS, two very faint bands appear at ∼100–110 nt in the presence of nuclear extracts (lanes 5 and 7, indicated by dashed arrow). Overall T7 RNAP transcription was weaker in the presence of nuclear extracts; therefore each lane is shown separately to have comparable exposures. Lanes 1 and 10 show a control in which reactions were performed without the addition of T7 RNAP and as expected, no transcription products appear. The 190-nt band, which was observed on all previous experiments using nuclear extracts, is not readily seen here because of lower exposure of the gel, which is required to not overexpose the run-off products, since the yield is much higher for T7 RNAP than for RNAPII. However, the band is seen in lane 1 (control where no transcription is taking place) as it is shown with a higher exposure.
Figure 6.
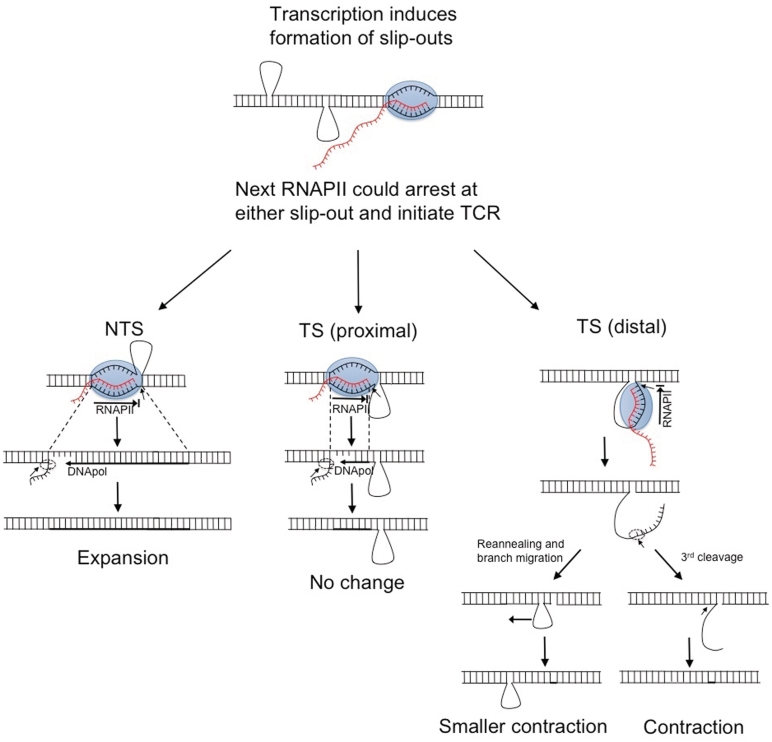
Model for transcription-dependent repeat instability. In genes involved in polyglutamine diseases CTG repeats are present on the TS and CAG repeats are on the NTS. After the passing of RNAPII has induced the formation of slipped-strand structures, the next RNAPII can arrest at either a CAG slip-out on the NTS or a CTG slip-out on the TS (at either the proximal or distal position). If arrest at a CAG slip-out on the NTS initiates TCR and repair proteins cut the TS, the CAG slip-out would get copied during repair patch formation leading to expansion. TCR due to arrest at the proximal end of the CTG slip-out on the TS could lead to no change in repeat number if incisions are made before the slip-out. TCR due to arrest at the distal end of the CTG slip-out, which is bigger than the transcription bubble, on the TS could lead to either complete contraction of the slip-out or a smaller contraction. If, after the TCR has excised the DNA fragment, the remaining slip-out re-anneals and branch migrates it would escape further contraction. However, complete contraction could occur if an additional cleavage event occurs, removing the remaining part of the slip-out.
DISCUSSION
In this study, we have shown that RNAPII is arrested upon encountering a (CAG)20, (CTG)20, hairpin or loop slip-out on either strand in HeLa nuclear extracts. Previous studies have shown that transcription can be arrested by DNA sequences capable of forming unusual DNA structures as well as by lesions in the DNA (24,29–32). DNA sequences that arrest transcription include those capable of forming Z-DNA (left-handed helix), H-DNA (triplex DNA), G-quadruplexes and extra-stable R-loops (24,31–33). The mechanisms by which unusual DNA structures arrest transcription are not completely understood and most likely vary, depending on which structure is causing the arrest. There are several possible mechanisms that may account for the transcription arrest observed in the presence of a DNA a slip-out. One possibility is that it occurs due to lack of upstream re-annealing of DNA strands after the passage of RNAPII. It is thought that template re-annealing can promote elongation by displacing the RNA from DNA onto an RNA binding site on the polymerase, stabilizing the elongation complex (34,35). The presence of a slip-out on one strand, such as a loop or hairpin, may therefore arrest transcription because after passing through the slip-out the two DNA strands cannot re-anneal and displace the RNA. However, if this were the case we would expect to see the same arrest for two non-complementary loops, but we see only weak arrest for these structures. Therefore, it is unlikely that the inability to re-anneal the DNA strands after passage of the polymerase plays a major role in producing the arrest observed in this study. Another possibility is that arrest is caused by these structures due to steric constraints placed on the polymerase. If this were the case, it would explain why the blockage seen with one slip-out is much stronger than that seen with two non-complementary loops. Finally, it is quite possible that protein binding to the slipped-out structures plays a role in causing transcriptional arrest. In fact, the finding that truncated products appear when T7 RNAP transcription is performed in the presence of HeLa nuclear extracts suggests that there is a role for proteins in causing the arrest. However, the T7 RNAP arrest was significantly weaker in some cases than that seen with RNAPII and it is not observed for all RNAPII arrest sites, suggesting that differences in polymerase properties also play a role. One possibility is that the MMR protein complex, MutSβ, which has been implicated in both transcription-induced repeat instability and somatic instability, plays a role in causing transcription arrest (18,36,37). However, it is also possible that MutSβ only plays a role in instability by assisting in the formation of the slip-outs or by stabilizing them so that they exist for a longer time, rather than directly causing transcription arrest, and that other proteins are responsible for the transcription arrest. Regardless of which proteins are involved, the observed RNAPII arrest due to slip-outs is likely due to a combination of protein factors and steric constraints that impede passage of the polymerase.
Our observation that slip-outs cause RNAPII transcription arrest has potential implications for genetic instability. Arrest caused by these structures in vivo could lead to mutagenesis by eliciting ‘gratuitous’ TCR (21,22,38). The finding that (CTG)20 and (CAG)20 slip-outs cause arrest is consistent with a transcription-induced repeat instability model in which expansion or contraction might arise from gratuitous TCR initiated by RNAPII arrest at a CAG or CTG slip-out. Future studies are needed to directly demonstrate if TCR can be induced by the transcription arrest caused by the CAG or CTG slip-outs. The current transcription-induced repeat instability model proposes that RNAPII arrests upon encountering a CTG hairpin on the TS, initiating TCR and leading to expansion or contraction depending upon where the slip-outs are localized relative to each other (18). Our results allow us to elaborate this model based on the observations that: (i) RNAPII arrest is caused by a CAG slip-out on the NTS and (ii) RNAPII arrest due to a CTG slip-out on the TS can occur either at the proximal or distal end of the slip-out. The current model suggests that expansion occurs when a large CAG loop on the NTS is opposite a smaller CTG hairpin on the TS; however, slip-outs opposite each other would have a high probability of collapse into a double-stranded structure and, thus, should exist only very transiently. The finding that RNAPII arrests on a CAG slip-out on the NTS provides a plausible scenario for expansion without requiring slip-outs to be opposite each other. This scenario involves arrest at a CAG slip-out on the NTS initiating TCR, leading to excision of the TS, followed by repair patch formation in which the CAG slip-out would be copied, leading to expansion (Figure 6). This model assumes that (i) RNAPII arrest due to a slip-out on the NTS initiates TCR and (ii) TCR initiation leads to excision of part of the TS. The first assumption is a reasonable one, given that even though the arrest is caused by an obstacle on the non-transcribed rather than the TS, the signal for initiation of TCR is the same: RNAPII arrest. However, whether TCR is initiated by arrest due to an obstacle on the NTS and whether this leads to cutting of the TS needs to be determined experimentally. From the biological role of TCR, it is likely that the TS is usually cut, given that most lesions that can arrest transcription are present on the TS. In fact, all DNA lesions studied so far cause a significant block to transcription only when present on the TS (21,22,38). The only known instances in which a block to transcription is due to an impediment on the NTS are unusual structures, specifically G-quadruplexes (31) and slip-outs, as shown in our study. Repair factors recruited by RNAPII arrest may be loaded based on the orientation of the polymerase, leading always to excision of the TS. Although speculative, that scenario could account for expansion observed in human diseases. However, further studies into the mechanism of TCR when the obstacle is on the NTS are required to elucidate its potential involvement in repeat instability.
The finding that RNAPII arrests both at the proximal and distal end of a CTG slip-out on the TS allows us to incorporate this into the model. The outcome of TCR due to arrest at a slip-out on the TS would depend upon where the strand is cut. Normally, TCR initiation leads to recruitment of NER proteins, including XPG and the XPF-ERCC1 complex (21,22,38). It is thought that before these proteins can act, RNAPII must be displaced and once NER proteins gain access to the DNA, the XPF–ERCC1 complex cuts the TS 5′ to the transcription bubble. This is followed by initiation of repair patch formation by DNA polymerase. Finally, XPG cuts 3′ to the transcription bubble and ligase closes the remaining nick (21,22,38). If RNAPII arrests at the proximal end of the slip-out, then it is possible that TCR would not lead to a change in repeat number (Figure 6). This would occur, however, only if the XPF–ERCC1 complex cuts before the beginning of the slip-out. Further studies are necessary to determine if TCR is initiated by arrest caused by these structures and if so, exactly how far XPF–ERCC1 excision occurs. If RNAPII arrests at the distal end of the slip-out and initiates TCR, this could lead to either contraction of the entire slip-out or to a smaller contraction (Figure 6). After the second NER cleavage (XPG cleavage 3′ to transcription bubble), the single-stranded DNA could be subject to additional cleavage by a nuclease that removes single stranded flaps, leading to contraction of the entire slip-out. Alternatively, the single-stranded DNA might re-anneal and branch migrate, therefore escaping further contraction (Figure 6). These different scenarios may help to explain the repeat expansion bias observed in human triplet repeat diseases, given that arrest at a CAG slip-out on the NTS would always lead to expansion while arrest at a CTG slip-out on the TS would lead to complete contraction only when an additional cleavage event occurs.
In this study, slipped-out (CAG)20 and (CTG)20 structures were pre-formed and shown to cause transcription arrest. For transcription-induced repeat instability to occur, these structures would have to form in vivo and the extent of instability would depend on both the probability of formation of slipped-out structures and on the probability that they cause arrest and initiate TCR. CAG and CTG slip-outs have been shown to occur in vitro and their propensity to form increases with increasing repeat length (2,15,16). Recent studies have provided evidence for the formation of hairpin structures within CTG•CAG repeats during replication and transcription in vivo (39,40). A study implicating the formation of R-loops in CTG•CAG transcription-induced repeat instability, shows that single-stranded DNA exists on the non-transcribed (CAG) strand in human cells, in a pattern consistent with hairpin formation on the non-transcribed strand during transcription (40,41). R-loops, which consist of stretches of RNA–DNA hybrids, can form in GC-rich DNA regions because of the high stability of rGdC base pairing. R-loops can form in disease-associated triplet repeats, including CTG•CAG, and were recently shown to increase CTG•CAG repeat instability (40–42). The formation of stable R-loops during transcription leads to a longer exposure of the NTS in its unpaired state, thus increasing the probability of the formation of a secondary structure in that strand. Furthermore, after the R-loop is ‘removed’ and the DNA strands are re-annealed, the secondary structure already present in the NTS may enhance the formation of slipped-strand structures. As discussed above, these structures could then potentially lead to RNAPII arrest, TCR and instability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute; National Institutes of Health (grant CA077712 to P.C.H.); UAR Major Grant (to V.S. at Stanford University). Funding for open access charge: National Institutes of Health (grant CA077712) from the National Cancer Institute to the laboratory of P.C.H. at Stanford University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anne Pipathsouk for her help with experiments, Graciela Spivak for showing us the protocol for RNAPII transcription, Sergei Mirkin, Silvia Tornaletti and all the members of the Hanawalt lab for helpful discussions.
REFERENCES
- 1. Wells, R.D. and Ashizawa, T. (2006) Genetic Instabilities and Neurological Diseases, 2nd edn. Elsevier Inc, San Diego, CA.
- 2.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 3.Sinden RR, Potaman VN, Oussatcheva EA, Pearson CE, Lyubchenko YL, Shlyakhtenko LS. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci. 2002;27:53–65. doi: 10.1007/BF02703683. [DOI] [PubMed] [Google Scholar]
- 4.Usdin K, Grabczyk E. DNA repeat expansions and human disease. Cell Mol. Life Sci. 2000;57:914–931. doi: 10.1007/PL00000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, Folstein S, Ross C, Franz M, Abbott M, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat. Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 6.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 7.Telenius H, Kremer B, Goldberg YP, Theilmann J, Andrew SE, Zeisler J, Adam S, Greenberg C, Ives EJ, Clarke LA, et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat. Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 8.Kennedy L, Evans E, Chen CM, Craven L, Detloff PJ, Ennis M, Shelbourne PF. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 9.Ishiguro H, Yamada K, Sawada H, Nishii K, Ichino N, Sawada M, Kurosawa Y, Matsushita N, Kobayashi K, Goto J, et al. Age-dependent and tissue-specific CAG repeat instability occurs in mouse knock-in for a mutant Huntington's disease gene. J. Neurosci. Res. 2001;65:289–297. doi: 10.1002/jnr.1153. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy L, Shelbourne PF. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington's disease? Hum. Mol. Genet. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- 11.Wells RD, Dere R, Hebert ML, Napierala M, Son LS. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res. 2005;33:3785–3798. doi: 10.1093/nar/gki697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghavan SC, Lieber MR. DNA structure and human diseases. Front Biosci. 2007;12:4402–4408. doi: 10.2741/2397. [DOI] [PubMed] [Google Scholar]
- 13.Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petruska J, Hartenstine MJ, Goodman MF. Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J. Biol. Chem. 1998;273:5204–5210. doi: 10.1074/jbc.273.9.5204. [DOI] [PubMed] [Google Scholar]
- 15.Pearson CE, Tam M, Wang YH, Montgomery SE, Dar AC, Cleary JD, Nichol K. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearson CE, Sinden RR. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 17.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin Struct. Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol. Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 20.Lin Y, Hubert L, Jr, Wilson JH. Transcription destabilizes triplet repeats. Mol. Carcinog. 2009;48:350–361. doi: 10.1002/mc.20488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanawalt PC. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 22.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 23.Parsons MA, Sinden RR, Izban MG. Transcriptional properties of RNA polymerase II within triplet repeat-containing DNA from the human myotonic dystrophy and fragile X loci. J. Biol. Chem. 1998;273:26998–27008. doi: 10.1074/jbc.273.41.26998. [DOI] [PubMed] [Google Scholar]
- 24.Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 25.Tornaletti S, Patrick SM, Turchi JJ, Hanawalt PC. Behavior of T7 RNA polymerase and mammalian RNA polymerase II at site-specific cisplatin adducts in the template DNA. J. Biol. Chem. 2003;278:35791–35797. doi: 10.1074/jbc.M305394200. [DOI] [PubMed] [Google Scholar]
- 26.Manley JL, Fire A, Samuels M, Sharp PA. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- 27.Pugh BF. Preparation of HeLa nuclear extracts. Methods Mol. Biol. 1995;37:349–357. doi: 10.1385/0-89603-288-4:349. [DOI] [PubMed] [Google Scholar]
- 28.Nacheva GA, Berzal-Herranz A. Preventing nondesired RNA-primed RNA extension catalyzed by T7 RNA polymerase. Eur. J. Biochem. 2003;270:1458–1465. doi: 10.1046/j.1432-1033.2003.03510.x. [DOI] [PubMed] [Google Scholar]
- 29.Tornaletti S. DNA repair in mammalian cells: Transcription-coupled DNA repair: directing your effort where it's most needed. Cell Mol. Life Sci. 2009;66:1010–1020. doi: 10.1007/s00018-009-8738-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tornaletti S. Transcription arrest at DNA damage sites. Mutat. Res. 2005;577:131–145. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ditlevson JV, Tornaletti S, Belotserkovskii BP, Teijeiro V, Wang G, Vasquez KM, Hanawalt PC. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 2008;36:3163–3170. doi: 10.1093/nar/gkn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc. Natl Acad. Sci. USA. 2010;107:12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopal V, Brieba LG, Guajardo R, McAllister WT, Sousa R. Characterization of structural features important for T7 RNAP elongation complex stability reveals competing complex conformations and a role for the non-template strand in RNA displacement. J. Mol. Biol. 1999;290:411–431. doi: 10.1006/jmbi.1999.2836. [DOI] [PubMed] [Google Scholar]
- 35.Andrecka J, Treutlein B, Arcusa MA, Muschielok A, Lewis R, Cheung AC, Cramer P, Michaelis J. Nano positioning system reveals the course of upstream and nontemplate DNA within the RNA polymerase II elongation complex. Nucleic Acids Res. 2009;37:5803–5809. doi: 10.1093/nar/gkp601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manley K, Shirley TL, Flaherty L, Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 37.Sinden RR. Neurodegenerative diseases. Origins of instability. Nature. 2001;411:757–758. doi: 10.1038/35081234. [DOI] [PubMed] [Google Scholar]
- 38.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu G, Chen X, Bissler JJ, Sinden RR, Leffak M. Replication-dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010;6:652–659. doi: 10.1038/nchembio.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y, Dent SY, Wilson JH, Wells RD, Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl Acad. Sci. USA. 2010;107:692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIvor EI, Polak U, Napierala M. New insights into repeat instability: role of RNA.DNA hybrids. RNA Biol. 2010;7:551–558. doi: 10.4161/rna.7.5.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy K, Tam M, Bowater RP, Barber M, Tomlinson M, Nichol Edamura K, Wang YH, Pearson CE. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2010;39:1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.