Abstract
RNA degradation is among the most fundamental processes that occur in living cells. The continuous decay of RNA molecules is associated not only with nucleotide turnover, but also with transcript maturation and quality control. The efficiency of RNA decay is ensured by a broad spectrum of both specific and non-specific ribonucleases. Some of these ribonucleases participate mainly in processing primary transcripts and in RNA quality control. Others preferentially digest mature, functional RNAs to yield a variety of molecules that together constitute the RNA degradome. Recently, it has become increasingly clear that the composition of the cellular RNA degradome can be modulated by numerous endogenous and exogenous factors (e.g. by stress). In addition, instead of being hydrolyzed to single nucleotides, some intermediates of RNA degradation can accumulate and function as signalling molecules or participate in mechanisms that control gene expression. Thus, RNA degradation appears to be not only a process that contributes to the maintenance of cellular homeostasis but also an underestimated source of regulatory molecules.
INTRODUCTION
In higher eukaryotes, the majority of genomic DNA is transcribed, but only a small portion of the resultant RNA encodes proteins (1,2). The non-protein-coding fraction of the transcriptome can be divided into two general classes: housekeeping RNA and regulatory RNA (3,4). The former comprises constitutively expressed RNAs (of which rRNA and tRNA are the most abundant species) that are indispensable for fundamental cellular processes. The latter class includes a diverse spectrum of RNAs that are present temporarily and control gene expression in response to a variety of stimuli. The non-protein-coding RNAs (npcRNAs) have justifiably been described as ‘the architects of eukaryotic complexity’ because their number increases with evolutionary advancement (5). RNA-based mechanisms that regulate gene expression appeared in bacteria and Archaea (6,7), but far more complex regulatory strategies, involving npcRNA, were developed in eukaryotes. Regulatory RNAs (often called riboregulators) can affect almost all stages of eukaryotic gene expression. These RNAs can shape genome structure, influence mRNA stability and repress translation (5,8,9). Thus, there is an increasing amount of evidence that riboregulators participate in a broad spectrum of biological processes.
Transcription and the processing of the resultant transcripts have been recognized as the two main stages of the biogenesis of most long (>40-nt) and short npcRNAs. After transcription, long-npcRNA precursors most frequently undergo mRNA-like maturation that involves capping, polyadenylation and sometimes splicing (4). Primary transcripts representing short npcRNA precursors can also be capped and polyadenylated. In addition, to form functional RNA, the precursors need to be subjected to a series of cleavages by specific ribonucleases. Precursors of the regulatory RNAs generally lack other functionality (they are only substrates for regulatory RNA production). Exceptions to the rule are some precursors of small interfering RNAs (siRNAs), for example, viral RNA that operate as genomic RNA.
Accumulating evidence indicates that, in addition to primary transcripts, mature functional RNA can also be a source of short npcRNAs. In 2005, a specific tRNA cleavage in Tetrahymena thermophila was discovered as a response to amino acid deprivation (10). In this case, degradation targeted mature deacylated tRNA and resulted in an accumulation of so-called tRNA halves. The observed mechanism was proposed to be an adaptation to starvation (10). Accordingly, tRNA halves appeared to be markers of an early starvation response. Several reports have demonstrated that an endonucleolytic cleavage of tRNA is a widespread phenomenon in eukaryotes as phylogenetically distant as fungi, plants and mammals (11–14). A new perspective in research on tRNA-related npcRNAs has been opened by a recent finding that molecules excised from pre-tRNA are capable of modulating RNA-silencing pathways (14). What is more, similar molecules derived from other RNA species [tRNA, rRNA, small nucleolar RNA (snoRNA)] were also identified (11,15–18). Some of these molecules were shown to influence gene expression in a miRNA-like fashion (17). These findings demonstrated that mature, functional RNAs can be a source of riboregulators more often than one would expect. The data collected suggest that these new regulatory molecules are stable intermediates of RNA degradation, one of the fundamental processes that continuously occur in cells. Accordingly, one can hypothesize that in addition to being the key element of nucleotide turnover, RNA maturation and quality control, RNA degradation also plays an important role in the biogenesis of functional npcRNAs.
This article presents recent progress in RNA degradome research. First, we briefly describe basic RNA degradation pathways. Then we attempt to systematize what is currently known about stable intermediates of RNA degradation, focusing on their origin, their classification and their proven or putative functions.
RNA DEGRADATION PATHWAYS
RNA decay is one of the key processes that shape cellular transcriptomes. For a long time, RNA degradation was considered to be a series of random events. Recently, it has become increasingly clear that it is a well-ordered, strictly controlled and reproducible process, inseparably connected with all three of the main stages of RNA metabolism: (i) maturation of primary transcripts; (ii) quality control; and (iii) RNA turnover. Because RNA degradation pathways have been comprehensively described in earlier reviews (19–24), here we delineate only issues most relevant to the current survey.
RNA maturation
Primary transcripts are rarely functionally ready to fulfill their biological roles without any additional modifications. Almost all eukaryotic, and many prokaryotic, RNAs must undergo numerous transformations, including cleavage, to achieve their mature form. Three eukaryotic rRNAs and all prokaryotic rRNAs are excised from single primary transcripts in a sequence of cleavages and trimmings that results in a progressive release of functional molecules and in the degradation of discarded fragments. In Escherichia coli, these cleavages are performed by RNases III, E, G and T (20), whereas in Bacillus subtilis this occurs through RNase J1, RNase M5 (25) and mini RNase III (26). In eukaryotes, pre-rRNA maturation is much more complex and involves numerous small RNAs and proteins, among them several endonucleases (RNase III and MRP) and exonucleases (Rrp44, Rat1, Xrn1, Rex1 and Rex2) (27).
The processing of pre-tRNA also requires nucleolytic cleavages. The mature 5′ terminus of prokaryotic and eukaryotic tRNA is generated by RNase P. In eukaryotes, the 3′ terminus is targeted by tRNase Z, which removes a 3′ trailer prior to CCA sequence synthesis (28). In bacteria, tRNA 3′-ends containing a genome-encoded CCA sequence are processed differently from those in which a CCA sequence is added post-transcriptionally (20,22). In addition to terminus formation, some tRNA precursors are spliced (28,29).
Precursors of eukaryotic mRNA are also extensively cleaved before they are transformed into the mature molecules. The splicing machinery removes introns, which are subsequently degraded or processed to snoRNAs or miRNAs. Additionally, the 3′ terminus of pre-mRNA is cleaved prior to polyadenylation (19). Furthermore, it has been shown that many human pre-mRNAs contain several potential 3′-end cleavage sites, providing alternatives that are used during different processes, such as in development. It has been suggested that there is some correlation between the patterns of 3′-end cleavage and alternative splicing (30). Most bacterial mRNAs do not require processing. Nevertheless, some polycistronic mRNA precursors are subjected to splicing or site-specific digestion (23).
Nucleases also participate in the biogenesis of non-coding regulatory RNAs. Among short non-coding regulatory RNAs, the most intensively studied are miRNAs and siRNAs. The former are generally produced in a multi-step process catalyzed by Drosha and Dicer. The latter are generated by Dicer alone (31). Long non-coding regulatory RNAs are usually subjected to pre-mRNA-like processing. However, they can also undergo an alternative maturation in which RNase P cleaves the transcript downstream of a genomically encoded poly(A) tract. This cleavage simultaneously generates the 3′-end of the mature long npcRNA and the 5′-end of a small tRNA-like molecule (32). As a result, two or more non-coding RNAs of different function and subcellular localization can be produced from a single locus.
RNA quality control
RNA degradation also plays a pivotal role in controlling the quality of RNA molecules. Defective RNA molecules, which are often mature but dysfunctional (for example, truncated, incorrectly synthesized or modified molecules), need to be rapidly degraded. Otherwise they can interfere with many cellular processes, especially gene expression. Our knowledge of the mechanisms controlling RNA quality in prokaryotes is very limited. Recently, it has been shown that two ribonucleases, RNase R and polynucleotide phosphorylase (PNPase), participate in this process (20). In eukaryotes, the main component of the RNA quality control system is the exosome accompanied by its activating complexes, for example, TRAMP and Ski. However, other exonucleases can also take part in the degradation of aberrant RNAs (24). In addition, there are highly specialized pathways that eliminate only defined classes of defective RNAs, for example, the non-functional rRNA decay (NRD) (33), rapid tRNA decay (28), nonsense-mediated decay (NMD) (34), no-stop decay (NSD) (35) and no-go decay (NGD) (36) pathways. The last three pathways remove mRNA molecules that have premature stop codons, lack stop codons or form structural barriers for ribosomes, respectively.
RNA turnover
There are several lines of evidence that particular classes of RNA do not contribute equally to nucleoide turnover. Depending on their function, RNAs are stably maintained in the cell (e.g. rRNA and tRNA) or exist for only a short time (e.g. mRNA or small regulatory RNAs). At present, mRNA turnover seems to be best characterized. Numerous cis- and trans-acting factors affecting mRNA stability have been identified, for example, 3′- and 5′-end modifications, which make RNA less susceptible to degradation. Eukaryotic mRNA is capped and polyadenylated, whereas prokaryotic mRNA often contains a triphosphate group at the 5′-end and a stem-loop structure at the 3′-end (37). In bacteria, the first endonucleolytic cleavage of mRNA is usually mediated by RNase E. This enzyme has been identified as an important component of the degradosome (38,39). Nonetheless, there are numerous other endo- and exonucleases that participate in prokaryotic mRNA decay (20,38–40). In eukaryotes, a cap structure and a poly(A) tail are usually removed prior to degradation. Afterwards, mRNA is sequentially digested by the 3′–5′ exonucleases of the exosome complex (41–43) and by Xrn1, the main eukaryotic 5′–3′ exoribonuclease (44).
Under physiological conditions, the degradation rate of housekeeping npcRNAs (especially rRNA and tRNA) is relatively low. However, stress conditions can trigger their decay both in prokaryotes and eukaryotes (11,21,45). Interestingly, the degradation of these RNAs does not necessarily lead to their rapid decomposition into mononucleotides. Instead, stable intermediates of RNA degradation can accumulate in cells (11,13,15,18,46,47). In bacteria, cleavage of housekeeping npcRNAs is most likely performed by the degradosome or its components (20). In addition, upon cell membrane damage, periplasmic RNase I can contribute to stable RNA degradation. The corresponding mechanisms and enzymes involved in the degradation of eukaryotic housekeeping npcRNAs are less defined. An example of an enzyme that has been found to degrade rRNA during apoptosis in higher eukaryotes is RNase L (48).
STABLE INTERMEDIATES OF RNA DEGRADATION
Despite the high efficiency of the RNA degradation machinery, sometimes RNA molecules are not instantly digested into single nucleotides. Instead, relatively stable degradation intermediates are generated. Why these fragments do not immediately undergo further digestion remains to be elucidated. Hypothetically, the products released by initial cleavages become transiently resistant to nucleases. This temporal stability of degradation intermediates may suggest that they are functional.
During the past few years, many studies have focused on such partially degraded RNA molecules. Accordingly, a large number of degradation products derived from several classes of RNA have been identified. Fragments of mRNA have been surveyed to identify targets for miRNAs and siRNAs. To this end, molecules that lack the cap structure but are polyadenylated, and that originate from the small RNA-directed cleavage of mRNA, were selectively captured and further analyzed. The collection of these mRNA-derived fragments was referred to as the RNA degradome (49–56). Products of miRNA-directed RNA cleavage were found in many organisms, including plants and mammals (56,57). Global analysis of the mammalian RNA degradome revealed that fragments generated by miRNA-independent endonucleolytic cleavage are also widespread in cells (56,58–60). It has been postulated that some of these molecules are the products of post-splicing RNA cleavage and subsequent secondary capping. According to this mechanism, 3′-UTR-associated RNAs (uaRNAs) are most likely formed. They have been shown to undergo developmental and tissue/cell-specific regulation (58,61).
Nevertheless, it has been shown that in addition to mRNA fragments, derivatives of other RNA species also accumulate in cells. Several recent studies have demonstrated the existence of a very interesting pool of small RNAs that correspond primarily to fragments of tRNA (10–15,18,46,47,62–66), rRNA (11,13,15,18,46,47), as well as snRNA and snoRNA (11,13,15,18,46). These products of incomplete RNA degradation are present in a wide range of organisms, including: (i) the bacterium Streptomyces coelicolor (47); (ii) the fungi Aspergillus fumigatus (13) and Saccharomyces cerevisiae (11); (iii) the protozoans Tetrahymena thermophila (10), Trypanosoma cruzi (66) and Giardia lamblia (46); (iv) the fruit fly Drosophila (67); and (v) the plants Arabidopsis thaliana (11,63) and Cucurbita maxima (15), as well as mammalian cell lines (11,12,14,18,62,64,68,69). These observations provide a surprising link to findings made three decades earlier, showing that tRNA breakdown products accumulate in the urine of patients with different cancers (70,71).
Although the exploration of the products of incomplete RNA degradation has only just begun, some characteristic features of stable intermediates have already been depicted. Molecules derived from tRNA are among the most frequently analyzed and are relatively well described. The intermediates of tRNA degradation are usually 3′- or 5′-halves generated by a cleavage at the anticodon loop. In A. fumigatus, the majority of 5′-halves contain the anticodon sequence at their 3′-ends. Accordingly, most of the 3′-halves start right after the anticodon (13). In HepG2, a liver carcinoma cell line, the tRNA cleavage occurs at the anticodon loop or within the T loop region (18). However, in G. lamblia, tRNAs are cut at the anticodon left arm (46); in pumpkin, they are cut at the D loop (15). In the case of the latter, the fragments generated are >60-nt long. Ultra-high-throughput sequencing of small RNA fractions from human cell lines (12,62) and Arabidopsis phosphate-starved roots (63) has revealed that tRNA fragments shorter than halves (∼17–26-nt) can also accumulate. These fragments come from both ends of mature tRNA molecules. An additional class of the tRNA-related fragments identified includes 3′ trailers removed from pre-tRNAs during their maturation (12,14). Intriguingly, the short tRNA derivatives (∼17–26-nt-long), together with molecules derived from the processing of 3′-end of tRNA precursors, constitute the second most abundant class of small RNAs, the most abundant being miRNAs (12).
None of the tRNA fragments identified so far contained 5′ leader, intron or 3′ trailer sequences of pre-tRNA. This implies that tRNA halves are generated from fully mature molecules rather than from their precursors (10–12,46,47,62,66,69). In Tetrahymena, the majority of 3′ tRNA fragments lack the terminal CCA sequence (10), indicating that the cleavage most likely involves deacylated and thus unprotected tRNA molecules. In contrast, in other organisms, most of the cloned 3′ tRNA fragments contain the CCA sequence (11,12,15,46,47,69). This observation supports the opinion that they are cleaved from fully mature molecules. The phenomenon of tRNA cleavage affects all tRNA isoacceptors (10,46,47), although in bacteria, most degradation intermediates are derived from tRNAs carrying the most frequently used anti-codons (47). Moreover, no correlation has been found between the gene copy number of a given tRNA and the abundance of its cleavage products (10).
Fragments of rRNA have also been identified, but their characteristics are still obscure. In Aspergillus, rRNA fragments are derived from the cytoplasmic and mitochondrial pools of all rRNA molecules (13). In yeast, most rRNA degradation intermediates represent the 3′-end of 25 S rRNA, and they only rarely are derived from 5 S and 18 S rRNAs (11). The length of rRNA fragments ranges from 17 to 53 nt (11). As suggested by Elbarbary et al., rRNA fragments found in various human cell lines can be closely related to Piwi interacting RNAs (piRNAs) (72). Although rRNA derivatives usually constitute a significant fraction of all RNA degradation intermediates, their contribution to the RNA degradome is significantly smaller than the contribution of rRNA to total cellular RNA (>80%). This strongly suggests that rRNA fragments are not derived from random rRNA cleavage or non-specific digestion.
Fragments of snRNAs and snoRNAs form a small and relatively poorly characterized group of stable degradation intermediates. It has been shown that snRNA-derived molecules detected in human cell lines originate from the 3′ portions of U1, U4 and U5, and most of them are 24–31-nt long (18). Most (90%) of the snoRNA degradation products identified came from a 34-nt-long fragment located in the 3′-end of a single C/D box type molecule (18). Recent reports revealed that in animals (mouse, chicken and fruit fly) and humans, snoRNA-derived fragments originate from the 3′-end of H/ACA snoRNA (mainly 20–24-nt-long molecules) and from the 5′-end of C/D snoRNA (17–19 and over 27-nt-long molecules) (16). A more detailed analysis showed that the locations of cleavage sites and, consequently, the lengths of the degradation products are evolutionarily conserved for the majority of snoRNAs from Arabidopsis, fission yeast and animals (16). It has also been found that many highly abundant fragments are derived from snoRNAs, the genes of which are weakly expressed (16). This finding suggests that the stable intermediates of snoRNA degradation, similar to the rRNA fragments described above, cannot be considered products of random RNA cleavage.
RNA DEGRADOME—DEFINITIONS AND CLASSIFICATIONS
The two recent decades brought unparalleled progress in our understanding of complex gene expression networks, revealing the previously hidden role of RNA in these pathways. It has been noted that the existing definitions are barely sufficient to describe the whole repertoire of currently known RNA species. Therefore, novel RNA classes have been designated. The expanding spectrum of the terms can sometimes be puzzling because they often emerge rapidly and are not widely assimilated. Among such newly distinguished types are sno-derived RNA (16), tRFs (tRNA-derived RNA fragments) (12) and tsRNA (tRNA-derived small RNA) (14). It seems that the last two names were assigned to the same class of molecules. The latest findings in RNA field challenged previously established definitions, even relatively new ones such as the one of miRNA (73). In addition, the standard classification of RNA species (rRNA, tRNA and so on) has been found to be inconvenient, as it often does not reflect the whole range of functions that these molecules perform (18).
In view of these latest reports, incomplete RNA degradation that yields stable intermediates emerges as a common and, at least in some cases, an evolutionarily conserved phenomenon. Unfortunately, the current classification of stable intermediates of RNA degradation seems inconsistent. On the one hand, several specific terms have been introduced to describe the products of tRNA or snoRNA degradation. On the other hand, a very broad term, ‘RNA degradome’, is frequently used to describe only the pool of mRNA fragments that are the products of RNA silencing pathways (49–55). We believe that it would be more reasonable to apply the term RNA degradome to the whole set of RNA degradation products that accumulates in a given cell or organism. Accordingly, narrower terms might be used with reference to specific classes of degradants, for example, mRNA degradome and tRNA degradome. In our opinion, such nomenclature would adequately reflect the complexity of this significant fraction of cellular RNA.
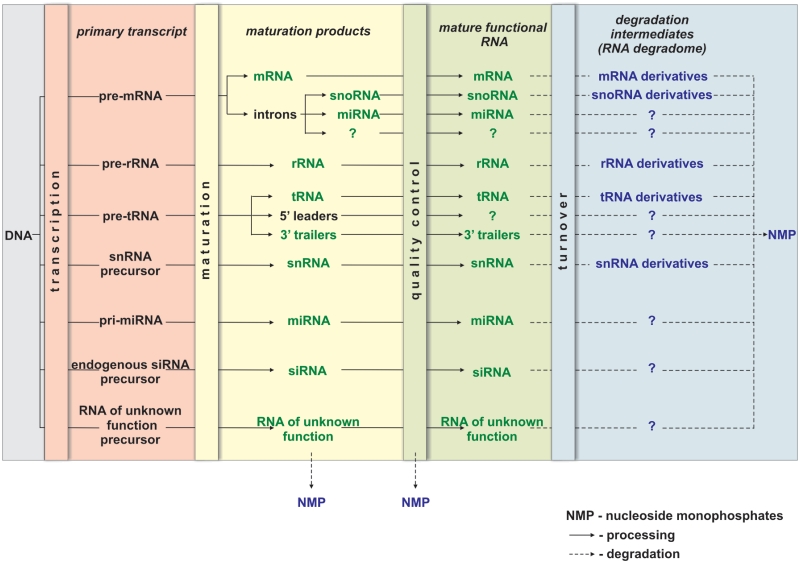
Considering the data presented above, we suggest that the RNA degradome should be defined as the collection of all molecules that result from the incomplete degradation of mature, functional RNAs (Figure 1). Such a definition of the RNA degradome excludes products formed during RNA maturation and RNA quality control.
Figure 1.
The RNA life cycle—from transcription to degradation. The primary transcript is processed to yield mature RNA. Processing (solid lines) usually involves several steps, including precursor cleavage. Surveillance systems provide quality control for all steps of RNA maturation. During maturation, certain RNA fragments are discarded and ultimately degraded (dashed lines) to nucleoside monophosphates (NMPs). Aberrant, dysfunctional molecules are also rapidly digested to nucleoside monophosphates (some enzymes, however, can generate alternative products, e.g. nucleoside diphosphates). Different classes of mature functional RNA display various levels of stability, but all functional RNA molecules eventually undergo turnover. Turnover-associated degradation can be very rapid and can yield no stable intermediates. Alternatively, stable intermediates of RNA degradation can form and accumulate in cells. It seems that at least some of these intermediates can operate as regulatory or signaling molecules. The scheme contains the names of previously described classes of RNA, and question marks indicate potential, undiscovered classes. Components of the RNA degradome are also indicated as defined in the text. Immature RNA is represented in black, mature RNA in green, degradation products in blue.
The RNA degradome could potentially also be defined as the set of all molecules that are generated in any process involving RNA cleavage. However, in this case, all RNA molecules, except primary transcripts, would be classified as degradants, as RNA maturation usually consists of sequential RNA cleavages. Alternatively, the definition of the RNA degradome could include the so-called by-products of RNA maturation, which are excised from primary transcripts. However, as our knowledge of the complex RNA landscape increases, it is becoming more and more difficult to indicate explicitly which molecules are indeed the by-products. At present, it is clear that a single primary transcript can give rise to different mature molecules representing various classes of RNA. For instance, introns that had previously been considered by-products of mRNA maturation have been found to be precursors of snoRNA and miRNA (74,75). Similarly, a 3′ trailer excised from pre-tRNA by RNase Z was observed to act as a regulator capable of modulating RNA-silencing pathways (14). These examples illustrate that primary transcripts are multi-functional molecules that can yield a wide variety of products. Our proposed definition of the RNA degradome also excludes cleavage products generated during RNA quality control. RNA surveillance systems are mechanisms that eliminate defective RNA molecules (e.g. truncated molecules or improperly synthesized or matured molecules). Aberrant molecules are typically very short-lived and do not contribute significantly to the pool of cellular RNAs (19).
It appears that all stable degradation intermediates identified to date are derived from mature RNAs. One should, however, always keep in mind that unambiguous classification of a particular RNA molecule is only possible when its biogenesis has been fully characterized. As long as the biogenesis pathway remains unknown, degradants devoid of any precursor-derived sequences could be assumed to originate from mature RNAs.
ENZYMES IMPLICATED IN THE PRODUCTION OF STABLE INTERMEDIATES OF RNA DEGRADATION
Although molecular mechanisms underlying the origin of stable intermediates of tRNA, rRNA, snRNA and snoRNA degradation are not fully understood, several enzymes are postulated to be involved in their biogenesis. Interestingly, these enzymes are different from those that take part in typical RNA turnover.
Nucleases implicated in the fragmentation of tRNA have been the most thoroughly studied so far. The first reports describing specific cleavage at the anticodon loop came from research on E. coli PrrC endonuclease, which cleaves tRNA in response to bacteriophage infection (76). In addition to PrrC, there are other enzymes, called colicins, that specifically cleave tRNAs. Colicins are produced by certain E. coli strains and inhibit the growth of other bacteria. Some of these ribonucleases are very specific and could potentially participate in the release of stable intermediates of RNA degradation. Colicin D catalyzes the cleavage of four isoaccepting tRNAs for Arg. Site-specific hydrolysis occurs within anticodon loops between the 38th and 39th nucleotides (77). Colicin E5 cleaves tRNAs for Tyr, His, Asn and Asp, which contain a modified base (queuine) at the wobble position of the anticodon. This ribonuclease hydrolyzes tRNA at the 3′ side of the modified nucleotide (78). Other colicins can target rRNA molecules. Colicin E3 cleaves 16 S rRNA between nucleotides 49 and 50, counting from the 3′-end, causing ribosome inactivation. However, it does not appear that colicins are responsible for the production of all stable RNA degradants, primarily because the phenomenon observed is not restricted to colicin-sensitive molecules. The tRNA and rRNA molecules that are not substrates for colicins have also been identified as a source of stable intermediates of RNA degradation. Recently, some degradation hot-spots within human tRNA have been mapped. It was observed that tRNAs are cleaved preferentially between conserved U–U nucleotides located within the T loop (18). Moreover, in prostate cancer cell lines, fragments derived from the 5′ portion of tRNA were found to be preferentially cleaved after A, whereas both ends of fragments released from the 3′ portion of tRNA were created by cleaving between A and U nucleotides (12).
It has recently been shown that tRNA is cleaved in yeast by Rny1p, a member of the RNase T2 family (79). In humans, tRNA is cleaved by angiogenin (68). Endonuclease Rny1 is released from the vacuole into the cytosol during oxidative stress. Independently of its catalytic activity, the enzyme inhibits cell growth and promotes cell death. Mutant yeast strains lacking Rny1 protein (rny1Δ) are unable to produce tRNA fragments during oxidative stress and other stress conditions, or during entry to stationary phase. The production of tRNA fragments in these mutant strains can be rescued by providing rny1 on an expression plasmid. In contrast, expression of the human ortholog of endonuclease Rny1, RNASET2, in yeast rny1Δ mutants causes the production of tRNA fragments to be only partially recovered. It has also been shown that the silencing of the RNASET2 gene in human cells does not have a significant influence on the accumulation of tRNA degradants. Although Rny1 can cleave a wide spectrum of RNAs, including rRNAs, it seems that other factors are involved in the production of rRNA degradants (79).
Angiogenin, a ribonuclease required for tRNA cleavage in humans, is a small protein that binds to the surface receptors of endothelial cells and, after internalization, promotes blood vessel growth and cell division. This multi-functional protein is activated during oxidative stress, heat shock or ultraviolet irradiation, resulting in an accumulation of tRNA-derived small RNAs and translation arrest. Angiogenin knockdown in mammalian cells impairs these processes, whereas knockdown of the angiogenin inhibitor, RNH1, leads to an increased production of tRNA fragments and enhances the inhibition of protein synthesis (68).
A pool of highly abundant small RNA fragments derived from mature tRNA in HeLa cells seems to have a different origin from tRNA fragments released by angiogenin cleavage. It has been shown that RNAi machinery can be involved in the biogenesis of these fragments. There is evidence that the processing of the human tRNA-Gln fragment in vivo depends on Dicer and that Dicer cleaves tRNA in vitro (62). In mouse embryonic stem cells, a small tRNA fragment is excised by Dicer from the tRNA that can fold into a pre-miRNA-like secondary structure (64). Other studies have shown that Dicer is also required for biogenesis of a small RNA class that originates from snoRNAs in human cells (17), fruit flies and mice (16). This evidence for Dicer participation in the biogenesis of tRNA and snoRNA fragments leads one to assume that these molecules can in fact take part in RNA silencing.
Although several candidates contributing to tRNA and snoRNA cleavage have been identified, ribonucleases involved in the production of specific rRNA or snRNA fragments remain mostly unidentified. Therefore, the origin of stable intermediates of RNA degradation requires further elucidation.
POTENTIAL FUNCTIONS OF RNA DEGRADANTS
Although stable intermediates of RNA degradation commonly occur in a number of organisms, their biological role remains unclear. There are several lines of evidence that they may be regulators of gene expression (12,14,15,68,72,80) or signaling molecules (15). So far, some putative molecular mechanisms underlying RNA degradants’ activities have been postulated for tRNA derivatives.
It has been shown that endogenous 5′-halves of tRNAs, as well as some rRNA fragments, can serve as guide molecules for tRNase Z (72). Human cytosolic tRNase Z participates in the maturation of the 3′-end of tRNAs, but it can also function as a ribonucleoprotein complex composed of an endonuclease and a small guide RNA (sgRNA). The role of sgRNA is to interact with the complementary sequence of the target RNA. Owing to these interactions, a pre-tRNA-like or a hook-like structure is formed. This motif is then recognized by a nuclease that cleaves the target RNA in a site-specific manner. Recently, it has been shown that some RNA degradants can act as sgRNAs. For example, there are 432 predicted human mRNAs whose expression could potentially be regulated by the complex of tRNase Z with the 5′-half of tRNA-Glu. There is also experimental evidence that protein phosphatase 1 F and cytoplasmic dynein heavy chain mRNAs are targets of tRNase Z guided by the 5′-half of tRNA-Glu or by the 28 S rRNA fragment, respectively (72).
The potential connections between stable RNA degradants and cellular silencing networks have been intensively investigated. However, the results obtained are contradictory. Whereas some reports have indicated that tRNA fragments do not efficiently associate with human Argonaute (Ago) proteins (62), others have showed that tRNA fragments are present in a pool of small RNAs purified from complexes with Piwi and Ago proteins (67,81). Therefore, it cannot be excluded that RNA degradants can act in a manner similar to the canonical miRNAs and piRNAs. In addition, a recent study demonstrated that Dicer-dependent tRNA fragments associate with human Ago proteins and that this class of tRNA-derived small RNAs can participate in gene silencing in trans (14).
The snoRNA-derived molecules also display extensive similarity to small regulatory RNAs, especially to miRNAs (16,17). It has been shown that the level of snoRNA-derived degradants correlates with the amount of RNAi pathway components present in the cell. What is more, these molecules associate with Ago proteins in Arabidopsis, fission yeast (16) and humans (17) and are capable of guiding the silencing of target gene expression, as demonstrated by a luciferase assay (17).
A recent report shows that tRNA halves, as well as the fragments of rRNA and snRNA, can diffuse into a vascular extract of pumpkins and can spread in plants (15). Long-distance transport of degradation products may indicate that these molecules take part in signaling mechanisms, delivering information about the metabolic state of the source tissue to distant organs. In addition, RNA extracted from phloem can inhibit translation in vitro by a currently unidentified mechanism. Interestingly, this inhibition is abolished when the RNA sample is denatured, which implies that the secondary and tertiary structures of these molecules determine their biological function (15).
It has been shown that tRNA 5′-halves are also involved in the stress response in mammalian cells, where they participate in the formation of stress granules (65). Stress granules are RNA–protein complexes containing untranslated mRNA, translation initiation components, and other proteins affecting mRNA function. Stress granules have been proposed to affect mRNA translation and stability and have been linked to apoptosis and nuclear processes (82). A similar mechanism was discovered in Trypanosoma cruzi, in which tRNA-derived molecules are recruited to cytoplasmic granules. Interestingly, northern blot analysis has revealed that fragments derived from 5′ and 3′ halves of tRNA do not co-localize and are associated with different cytoplasmic granules (66).
As mentioned before, the functions of RNA degradants can depend on their secondary and tertiary structure (15). It has also been shown that 5′ but not 3′ tRNA halves affect protein synthesis (68). It has been suggested that the smaller 5′ halves (∼30 nt) bind protein cofactors more efficiently than the bigger 3′ halves (∼40 nt). It seems that the presence of phosphate groups on the 5′ and 3′ termini of these tRNA fragments can also determine their potential function. The 5′-halves released by angiogenin, like many other small regulatory RNAs, are likely to be 5′ monophosphorylated and their 3′ terminus contains 2′, 3′ cyclic phosphate. In contrast, both termini of the 3′ halves most likely lack phosphate groups (68).
Our knowledge of the biological function of mRNA derivatives is very limited. Nevertheless, it has been postulated that uaRNA molecules can fulfill different roles than mRNAs, with which they are normally associated (61). This assumption is supported by the observation of discordant expression of uaRNA and its corresponding coding sequence (CDS) in mouse embryos. It was shown that uaRNA can be absent from cells in which its CDS is present, and vice versa. Both molecules can also differ in their subcellular localization. Earlier reports have shown that 3′-UTR sequences of many genes can control cell proliferation and differentiation in trans in the absence of an assigned coding region. For example, the expression of the 3′-UTR of oskar gene in Drosophila melanogaster is sufficient to rescue an oogenesis defect in oskar-null mutants (83). The observation that the oskar 3′-UTR exists as an independent entity in vivo strongly supports the idea that uaRNAs can operate as trans-acting non-coding regulators. However, alternative functions of uaRNAs have also been under consideration. It has been suggested that they may be involved in sequestration of important factors like miRNAs or might serve as scaffolds for regulatory complexes (61).
PERSPECTIVES
The complex landscape of cellular RNAs has recently been enriched with novel classes of molecules being the products of mature RNA degradation. Some of these molecules have been shown to control gene expression or act as signalling molecules. Consequently, RNA decay has emerged as a process yielding regulatory molecules. Despite differences between RNA degradation in prokaryotes and eukaryotes, these mechanisms have common features, implying their ancient origin and evolutionary conservation (24). In this context, an intriguing question arises: whether the extant small regulatory RNAs (e.g. miRNAs and endogenous siRNAs) could have evolved from stable degradation intermediates. It seems likely that primordial cells contained a large number of nonspecific RNA degradants competing for binding with other biomolecules. Some of these could have achieved a higher level of specialization, a higher selectivity and a higher efficacy by forming ribonucleoprotein complexes.
A number of issues regarding the composition and functions of the RNA degradome remain to be addressed. One issue is whether the involvement of degradants in regulatory processes is a widespread phenomenon. It will also be important to decipher the molecular mechanisms of the action of degradants. The current experimental findings promote the hypothesis that the functions of these degradants go beyond merely guiding molecules for the RNA silencing machinery.
Another unanswered question concerns the biogenesis of RNA degradants. The available data suggest that stable intermediates of mature RNA degradation are generated by enzymes other than those involved in a regular turnover pathway. Nevertheless, the repertoire of nucleases engaged in each of these processes has hardly been explored. Therefore, it remains to be established whether these two pathways overlap or whether they developed as independent systems. In addition, the mechanisms regulating the production of RNA degradants need elucidation. Clearly, these mechanisms must be finely tuned to provide a balance between mature functional RNAs and the stable products of their degradation.
At present, there are no reports comparing the composition of cellular RNA degradomes under various conditions. Considering the earlier analyses of the cellular transcriptome or its fractions, one can hypothesize that the profile of the RNA degradome can fluctuate in response to environmental or internal stimuli. This implies that the profiling of the RNA degradome will give new insight into cellular processes and will presumably allow new biomarkers to be identified.
FUNDING
Funding for open access charge: Polish Government through a grant from the Ministry of Science and Higher Education (PBZ-MNiI-2/1/2005 to M.F.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 2.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szymanski M, Barciszewski J. Regulatory RNAs in mammals. Handb. Exp. Pharmacol. 2006;173:45–72. doi: 10.1007/3-540-27262-3_3. [DOI] [PubMed] [Google Scholar]
- 4.Rymarquis LA, Kastenmayer JP, Huttenhofer AG, Green PJ. Diamonds in the rough: mRNA-like non-coding RNAs. Trends Plant Sci. 2008;13:329–334. doi: 10.1016/j.tplants.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennis PP, Omer A. Small non-coding RNAs in Archaea. Curr. Opin. Microbiol. 2005;8:685–694. doi: 10.1016/j.mib.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 10.Lee SR, Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 11.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jochl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Huttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Sun L, Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 20.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deutscher MP. Degradation of stable RNA in bacteria. J. Biol. Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 22.Condon C. Maturation and degradation of RNA in bacteria. Curr. Opin. Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 24.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 25.Condon C, Pellegrini O, Mathy N, Benard L, Redko Y, Oussenko IA, Deikus G, Bechhofer DH. Assay of Bacillus subtilis ribonucleases in vitro. Methods Enzymol. 2008;447:277–308. doi: 10.1016/S0076-6879(08)02215-5. [DOI] [PubMed] [Google Scholar]
- 26.Olmedo G, Guzman P. Mini-III, a fourth class of RNase III catalyses maturation of the Bacillus subtilis 23S ribosomal RNA. Mol. Microbiol. 2008;68:1073–1076. doi: 10.1111/j.1365-2958.2008.06203.x. [DOI] [PubMed] [Google Scholar]
- 27.Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhold-Hurek B, Shub DA. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- 30.Wilusz JE, Spector DL. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–266. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol. Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 35.Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 36.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 37.Schoenberg DR. The end defines the means in bacterial mRNA decay. Nat. Chem. Biol. 2007;3:535–536. doi: 10.1038/nchembio0907-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain C. Degradation of mRNA in Escherichia coli. IUBMB Life. 2002;54:315–321. doi: 10.1080/15216540216036. [DOI] [PubMed] [Google Scholar]
- 39.Kushner SR. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson KL, Dunman PM. Messenger RNA Turnover Processes in Escherichia coli, Bacillus subtilis, and Emerging Studies in Staphylococcus aureus. Int. J. Microbiol. 2009;2009:525491. doi: 10.1155/2009/525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 42.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat. Struct. Mol. Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu CL, Stevens A. Yeast cells lacking 5′–>3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mroczek S, Kufel J. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res. 2008;36:2874–2888. doi: 10.1093/nar/gkm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Luo J, Zhou H, Liao JY, Ma LM, Chen YQ, Qu LH. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang SL, Quirk D, Zhou A. RNase L: its biological roles and regulation. IUBMB Life. 2006;58:508–514. doi: 10.1080/15216540600838232. [DOI] [PubMed] [Google Scholar]
- 49.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meng Y, Gou L, Chen D, Wu P, Chen M. High-throughput degradome sequencing can be used to gain insights into microRNA precursor metabolism. J. Exp. Bot. 2010;61:3833–3837. doi: 10.1093/jxb/erq209. [DOI] [PubMed] [Google Scholar]
- 51.Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010;62:960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhou M, Gu L, Li P, Song X, Wei L, Chen Z, Cao X. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica) Front Biol. 2010;5:67–90. [Google Scholar]
- 53.Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15:2112–2121. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- 56.Bracken CP, Szubert JM, Mercer TR, Dinger ME, Thomson DW, Mattick JS, Michael MZ, Goodall GJ. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr110. doi: 10.1093/nar/gkr110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karginov FV, Cheloufi S, Chong MM, Stark A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mercer TR, Dinger ME, Bracken CP, Kolle G, Szubert JM, Korbie DJ, Askarian-Amiri ME, Gardiner BB, Goodall GJ, Grimmond SM, et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20:1639–1650. doi: 10.1101/gr.112128.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 60.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 61.Mercer TR, Wilhelm D, Dinger ME, Solda G, Korbie DJ, Glazov EA, Truong V, Schwenke M, Simons C, Matthaei KI, et al. Expression of distinct RNAs from 3' untranslated regions. Nucleic Acids Res. 2011;39:2393–2403. doi: 10.1093/nar/gkq1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsieh LC, Lin SI, Kuo HF, Chiou TJ. Abundance of tRNA-derived small RNAs in phosphate-starved Arabidopsis roots. Plant Signal Behav. 2010;5:537–539. doi: 10.4161/psb.11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Ronalte-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2010;171:64–73. doi: 10.1016/j.molbiopara.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 67.Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 68.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 70.Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977;37:3362–3366. [PubMed] [Google Scholar]
- 71.Speer J, Gehrke CW, Kuo KC, Waalkes TP, Borek E. tRNA breakdown products as markers for cancer. Cancer. 1979;44:2120–2123. doi: 10.1002/1097-0142(197912)44:6<2120::aid-cncr2820440623>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 72.Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. Modulation of gene expression by human cytosolic tRNase Z(L) through 5′-half-tRNA. PLoS One. 2009;4:e5908. doi: 10.1371/journal.pone.0005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pederson T. Regulatory RNAs derived from transfer RNA? RNA. 2010;16:1865–1869. doi: 10.1261/rna.2266510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ying SY, Lin SL. Intron-derived microRNAs-fine tuning of gene functions. Gene. 2004;342:25–28. doi: 10.1016/j.gene.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 75.Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum. Mol. Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- 76.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc. Natl Acad. Sci. USA. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- 79.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Zhou H. tRNAs as regulators in gene expression. Sci. China C Life Sci. 2009;52:245–252. doi: 10.1007/s11427-009-0039-y. [DOI] [PubMed] [Google Scholar]
- 81.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 82.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jenny A, Hachet O, Zavorszky P, Cyrklaff A, Weston MD, Johnston DS, Erdelyi M, Ephrussi A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]