Abstract
The human DEK gene is frequently overexpressed and sometimes amplified in human cancer. Consistent with oncogenic functions, Dek knockout mice are partially resistant to chemically induced papilloma formation. Additionally, DEK knockdown in vitro sensitizes cancer cells to DNA damaging agents and induces cell death via p53-dependent and -independent mechanisms. Here we report that DEK is important for DNA double-strand break repair. DEK depletion in human cancer cell lines and xenografts was sufficient to induce a DNA damage response as assessed by detection of γH2AX and FANCD2. Phosphorylation of H2AX was accompanied by contrasting activation and suppression, respectively, of the ATM and DNA-PK pathways. Similar DNA damage responses were observed in primary Dek knockout mouse embryonic fibroblasts (MEFs), along with increased levels of DNA damage and exaggerated induction of senescence in response to genotoxic stress. Importantly, Dek knockout MEFs exhibited distinct defects in non-homologous end joining (NHEJ) when compared to their wild-type counterparts. Taken together, the data demonstrate new molecular links between DEK and DNA damage response signaling pathways, and suggest that DEK contributes to DNA repair.
INTRODUCTION
The human Dek oncogene was first discovered as a fusion with the gene encoding the CAN nucleoporin protein, NUP214, in a subset of acute myeloid leukemia (AML) patients (1). Apart from its original association with cancer as a fusion protein, upregulated DEK expression has also been linked to various solid human tumors, as well as AML types that do not exhibit the above translocation (2–10). We previously reported that DEK overexpression promotes the transformation of human keratinocytes, and that Dek knockout mice are partially resistant to chemically induced papilloma formation (11,39). The overexpression of DEK inhibits cell death, and its knockdown results in apoptosis in HeLa cells and primary keratinocytes, in part through the stabilization and transcriptional activation of p53 (12,40). Other reports similarly are consistent with a role for DEK in cellular survival (13,14). Together, these findings support a role for DEK as an oncogene. The underlying molecular mechanisms by which it functions, however, remain poorly understood.
DEK is a highly abundant, evolutionarily conserved and ubiquitous nuclear protein that can be regulated at the level of transcription and post-translational modifications (15–21). Evidence suggests that it may function as a nuclear architectural protein. Similar to the classical chromatin architectural high mobility group (HMG) proteins, DEK contains regions enriched in acidic amino acids (22,23) and preferentially binds to supercoiled and cruciform DNA (24). Perhaps related to such a role, the literature supports distinct intracellular functions for DEK in DNA replication (25), positive and negative regulation of gene transcription (26–30), histone acetylation (31,32), mRNA splicing (20,33) and nucleosome assembly (34). Several reports have already suggested that DEK may modulate genome stability. First, a C-terminal fragment of DEK could partially rescue Ataxia–Telangiectasia (A–T) fibroblast mutagen sensitivity, high spontaneous recombination rates, and radio-resistant DNA synthesis (35). Second, DEK knockdown sensitized HeLa cells to genotoxic stress (18) and resulted in chemosensitivity to doxorubicin in melanoma cells (14). Accordingly, we hypothesized that DEK expression is important for DNA damage sensing and/or repair.
Sustaining DNA integrity is a continuous cellular challenge in the face of endogenous and exogenous DNA damage insults. Sensor kinases detect damage and signal the appropriate cellular responses. Proper repair of a lesion is followed by the resumption of normal cellular proliferation and continued growth, whereas incomplete repair results in prolonged cell-cycle arrest and ultimately, cell death. The protein kinase ataxia telangiectasia mutated (ATM) undergoes autophosphorylation on Ser1981 in response to DNA double-strand breaks (DSBs), and then phosphorylates and activates downstream targets including structural maintenance of chromosomes 1 (SMC1) (36,37). The DNA-dependent protein kinase (DNA-PK), which is composed of the Ku70/80 heterodimer and the DNA-PK catalytic subunit (DNA-PKcs), responds to DSBs by the initial recruitment of Ku70/80, binding and autophosphorylation of DNA-PKcs, and subsequent recruitment of the DNA repair machinery. The most extensively studied types of DSB DNA repair are homologous recombination (HR) and non-homologous end joining (NHEJ). HR utilizes the ATM pathway proteins and homologous DNA sequences as a template for DNA repair, while NHEJ fuses broken ends of DNA with little regard for homology (38).
Here we demonstrate that the loss of DEK expression, either by knock-down in human cells or by genetic deletion in murine cells, is sufficient to activate the DNA damage response and to exaggerate phenotypic responses to exogenous stress. DEK loss results in stimulated ATM activity, attenuated Ku70/80 recruitment, and repressed DNA-PK activity, in correlation with a decrease in repair by NHEJ. Together, these data suggest that DEK expression modulates ATM and DNA-PK signaling and the efficiency of DNA DSB repair.
MATERIALS AND METHODS
Cell culture
U2OS human osteosarcoma and HeLa cervical cancer cell lines (ATCC, Manassas, VA, USA) were maintained in Dulbecco's modified Eagle medium (DMEM) (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) and antibiotics. The SAOS-2 human osteosarcoma cell line (ATCC) was maintained in McCoy's 5a modified medium (ATCC) with 15% FBS and antibiotics. Dek wild-type, heterozygous and knockout mice [described in (39)] were used to generate MEFs. MEFs from Day 13.5 embryos were explanted and maintained in high glucose DMEM containing 10% FBS, 2 mM l-glutamine, 100 μM MEM non-essential amino acids, 0.055 mM β-mercaptoethanol and 10 μg/ml gentamycin. Immortalized MEFs were obtained by 3T3 serial passaging. Cells were treated with 1 mM hydroxyurea, 1 μM camptothecin and 4 μM or 25 μM etoposide for indicated periods of time (all from Sigma) or with 10 Gy γ-irradiation (IR) and collected post-treatment at the indicated times.
Plasmids and viral constructs
Ad-GFP and AdDEKsh adenoviruses were described previously (40). Retroviral R780 and R780-DEK expression vectors were described previously, with the exception that vector particles were Eco pseudotyped (11).
Adenoviral and retroviral infections
For adenoviral infections, cells were infected as previously published, using 10 infectious units per cell (40). For retroviral infections, cells were incubated with virus for 4 h in medium containing 2 μg/ml polybrene, then washed and overlaid with fresh medium. Retrovirally infected cells were sorted for GFP and expanded as a polyclonal population.
Western blot analyses
Whole cell lysates were harvested with RIPA buffer containing phosphatase inhibitors and analyzed as described previously (40). For detection of DNA damage response proteins, cells were lysed in 50 mM Tris–HCl, pH 7.4, 250 mM NaCl and 5 mM EGTA, supplemented with protease and phosphatase inhibitors (BD Biosciences, San Jose, CA, USA). Primary antibodies: DEK (1:1000; BD Biosciences, San Diego, CA, USA), γH2AX (1:1000; Upstate, Charlottesville, VA, USA); p53ser15 (1:1000; Santa Cruz Biotechnologies, Santa Cruz, CA, USA), DNA-PKcs (1:500; Santa Cruz), phospho-ATM (Ser1981) (1:1000; R&D, Minneapolis, MN, USA), ATM (1:1000; Cell Signaling, Boston, MA, USA), phospho-SMC1 (Ser957) (1:1000; Cell Signaling), SMC1 (1:1000; Abcam, Cambridge, MA, USA), phospho-DNA-PKcs (Ser2056) (1:500; Abcam); and actin (1:20,000; a gift from James Lessard, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, USA). Membranes were exposed to enhanced chemiluminescence reagents (Perkin Elmer, Boston, MA, USA) and bands detected by chemiluminescence. Band intensities were quantified using ImageJ software, and first normalized to actin in each case, and then expressed as fold change for AdDEKsh relative to Ad-GFP control sample. FANCD2 monoubiquitination was expressed as the intensity of the monoubiquitinated (L, long) relative to the non-ubiquitinated (S, short) form of the protein.
Xenograft injections
For DEK depletion in xenograft tumors, 1 × 106 U2OS cells were injected into each flank of 4- to 8-week-old female athymic nude mice (Harlan Laboratories, Indianapolis, IN, USA). Tumors were allowed to grow to 150–250 mm3, and 108 infectious units of Ad-GFP or AdDEKsh were injected into tumors on the left and right flank, respectively (39). Tumors were virally injected on Days 0, 3 and 6, and harvested 24 h later. Tumor tissue was fixed in 4% paraformaldehyde, washed with PBS, and gradually dehydrated in 50% and 70% ethanol for paraffin embedding.
Immunofluorescence microscopy
Cells were plated onto 100 μg/ml poly-d-lysine hydrobromide (Sigma) coated coverslips, infected with Ad-GFP or AdDEKsh adenoviruses, and fixed with 2% paraformaldehyde. Coverslips were incubated in 0.2% Triton X-100 for 3 min, blocked with 5% normal goat serum, and incubated with primary antibody for 1 h at 37°C. Antibody dilutions were as follows: DEK (1:20; BD Biosciences); γH2AX (1:1000; Upstate); FANCD2 (1:1000; Novus, Littleton, CO, USA); and SC35 (1:2000; Sigma). Coverslips were incubated in secondary anti-mouse or anti-rabbit rhodamine- or FITC-conjugated antibody (1:200; Jackson Immunoresearch, West Grove, PA, USA) for 30 min at 37°C, and mounted onto glass slides with DAPI Vector Vectashield mounting media (Vector Laboratories, Burlingame, CA, USA). For immunofluorescence analysis of Ku70/80 dimer mobilization, we utilized heterodimer-specific anti-Ku70/80 antibody (clone 162) (1:300; Abcam), following established methodologies (41). Subsequent detection was with secondary anti-mouse rhodamine-conjugated antibody (1:200, Jackson Immunoresearch). For analysis of tumor sections, eight micron sections from paraffin embedded tissue were deparaffinized and sequentially rehydrated. Sections were boiled in 10 mM sodium citrate buffer for 20 min, washed in PBS, blocked for 1 h, and incubated in GFP primary antibody (1:200; Abcam) for 1 h. Following washes, sections were incubated in anti-rabbit FITC-conjugated secondary antibody for 30 min at room temperature, and mounted using Vector Vectashield mounting medium. Images in Figure 1 were taken using ×100 magnification on a Zeiss fluorescence microscope (Zeiss, Thornwood, NY, USA) with an Axiovision camera (Lucas Microscope Service, Skokie, IL) driven by Axiovision software. Images in Figure 3 were taken using ×100 magnification on a Zeiss LSM 510 laser scanning confocal microscope using LSM510 and Zen software.
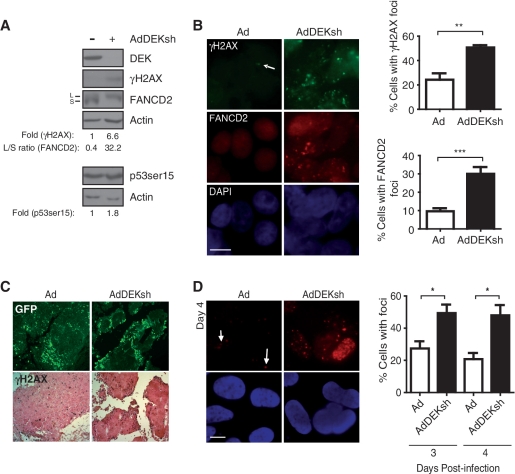
Figure 1.
DEK depletion increases DNA damage markers in vitro and in vivo. (A) U2OS cells were infected with Ad-GFP or AdDEKsh adenovirus and harvested 3 days post-infection, then analyzed by western blot analysis for DEK, γH2AX, FancD2 and p53ser15, and quantified as described in the ‘Materials and Methods’ section. (B) U2OS cells were infected as in (A) and focus formation was visualized by immunofluorescence on Day 3 post-infection using either γH2AX or FANCD2 primary antibodies together with FITC- or rhodamine-conjugated secondary antibodies. At least five fields and 200 cells, from three independent experiments, were counted for quantification of the data, which is presented in the graphs on the right. Columns, mean values of 200 cells per experiment; bars, SEM; magnification = 100×; scale bar = 10 μm. (**P = 0.01, ***P = 0.0001) (C) Xenografts generated in nude mice were injected with 108 infectious units Ad-GFP or AdDEKsh in the left and right flanks, respectively, three times at 3 day intervals. Tumors were removed, fixed, and two consecutive sections in each case were subjected to immunofluorescent detection of GFP and immunohistochemical detection of γH2AX. (D) SAOS-2 cells were infected as in (A) and probed with γH2AX primary and rhodamine-conjugated secondary antibody, and counterstained with DAPI on Day 4 post-infection. The graphs represent the percentage of cells positive for foci quantitated on Days 3 and 4 post-infection. Columns, mean values of 200 cells per experiment; bars, SEM; magnification = 100×; scale bar = 10 μm. (*P < 0.05).
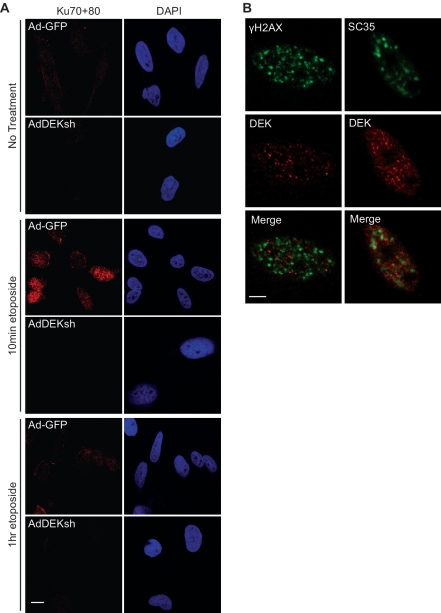
Figure 3.
DEK depletion impairs Ku70/80 heterodimer mobilization. (A) HeLa cells were infected with Ad-GFP or AdDEKsh, treated with 25 μm etoposide for 10 min and 1 h 3 days post-infection, and treated with extraction buffer for 2 min prior to fixation in paraformaldehyde. Cells were analyzed by immunofluorescence using a Ku70/80 heterodimer specific antibody (red). (B) HeLa cells were treated with 25 μM etoposide for 3 h and analyzed for DEK (red) and γH2AX or SC35 (green) localization. Magnification = 100×; scale bars = 10 μm.
Immunohistochemistry
Sections were prepared as described above. For analysis of γH2AX, samples were blocked in M.O.M mouse IgG (Vector Laboratories) and incubated in γH2AX primary antibody (1:1000; Upstate) for 1 h. Following washes, sections were incubated with biotinylated secondary antibody for 30 min. Staining was developed in 3,3′ diaminobenzidine solution (Vector Laboratories) for 2–4 min and counterstained with Nuclear fast Red (Biomeda, Foster City, CA, USA). Sections were dehydrated in ethanol followed by xylene, and mounted with Permount (Fisher Scientific, Suwanee, GA, USA).
Alkaline comet assays
HeLa cells were infected with Ad-GFP and AdDEKsh adenoviruses, and MEFs were treated with 1 mM hydroxyurea for 24 h. Detection of both single and double strand DNA breaks was determined using the Trevigen CometAssay Kit (Trevigen, Gaithersburg, MD, USA) per the manufacturer's instructions. Images were captured using ×20 magnification on a Zeiss fluorescence microscope with an Axiovision camera driven by Axiovision software. Images were saved as bitmap files and tail moments calculated using TriTek CometScore™ Freeware v1.5.
DNA-PK kinase assays
HeLa cells were infected with Ad-GFP or AdDEKsh, and nuclear extracts prepared Day 3 post-infection as previously described (42). Nuclear extracts were run through 1 ml DEAE Sepharose columns (GE Healthcare, Piscataway, NJ, USA) to remove endogenous DNA using an AKTApurifier (GE Healthcare). The nuclear extracts were then used for kinase assays to determine DNA-PK activity according to manufacturer's protocol (Promega, Madison, WI, USA). DNA-PK enzyme activity (in pmol ATP/min/μg of protein) was determined using the following equation: [(cpmreaction with activator − cpmreaction without activator) × reaction volume]/[(volume reaction in μl on membrane) × (timemin) × (μg protein in reaction) × (specific activity of [γ-32P] ATP).
BrdU incorporation and cell-cycle analysis by flow cytometry
Primary MEFs grown to 70–80% confluency in 10 cm plates were pulsed with 10 μM BrdU for 45 min before collection by trypsinization, prepared using the BD Pharmingen APC BrdU Flow Kit per the manufacturer's instructions and as previously published (40), and analyzed on a BD FacsCalibur (BD Biosciences).
Drug sensitivity assay
One hundred thousand primary wild-type and Dek knockout MEFs were plated in six-well plates in medium only or medium containing 4 μM etoposide, which was replenished every 24 h. Attached cells were trypsinized and counted at 4 h for plating efficiency and at 24, 72 and 120 h post-plating for cell numbers. Additionally, cells were fixed and stained for SA-β-galactosidase to determine levels of senescence at 120 h, as described in (43).
DSB repair assays
An established strand break rejoining assay using the plasmid pEGFP-Pem1-Ad2 was used to determine levels of NHEJ (44). pEGFP-Pem1-Ad2 contains two I-SceI restriction enzyme sites, which when digested, generates non-compatible DNA ends. Repair by NHEJ results in the expression of EGFP, which can be quantitated by flow cytometry relative to a DsRed transfection efficiency control vector. pEGFP-Pem1-Ad2 is incapable of expressing EGFP in the absence of repair by NHEJ. An amount of 6 μg I-SceI-linearized pEGFP-Pem1-Ad2 plasmid and 4 μg pCMV-DsRed-Express were co-transfected into 1 × 106 immortalized wild-type and Dek knockout MEFs using Amaxa Nucleofector (Lonza, Allendale, NJ, USA). pEGFP-Pem1, which does not contain the Ad2 exon and expresses EGFP, was used as a control for normalization of linearized plasmids. Seventy-two hours post-transfection, cells were trypsinized, fixed with 2% paraformaldehyde, resuspended in 3% FBS in PBS and 50 000 cells per sample were analyzed by flow cytometry using a BD LSRII instrument (BD Biosciences) and FACSDiva software. Data was analyzed by taking the ratio of total EGFP positive cells to total DsRed positive cells and normalizing linearized vector samples to pEGFP-Pem1 controls. A second established assay for DSB repair via NHEJ was performed according to a previously published protocol (45). The construct harbors a 5-bp microhomology on both sides of the lesion and assesses DSB end processing and re-ligation by microhomology-mediated NHEJ. Briefly, wild-type and Dek knockout MEFs were transfected with DNA plasmid mix (4 μg of DNA/well, six-well plate), containing a repair plasmid designed to measure NHEJ and I-SceI expressing vector (to induce DNA breaks), using Fugene HD (Roche) at 2 μl/μg DNA. On the following day, cells were processed and analyzed by flow cytometric analysis using a BD FacsCalibur. DSB repair frequency was determined by calculating the percentage of EGFP positive cells divided by the percentage of transfection efficiency each. The statistical significance of differences was determined using two-tailed Wilcoxon matched pairs test.
RESULTS
DEK depletion activates endogenous DNA damage response pathways
DNA damage was previously shown to occur in HeLa cells in response to DEK knockdown (18). HeLa cells harbor human papillomavirus (HPV) sequences and express the viral oncoproteins E6 and E7, as well as wild-type p53 which is kept inactive by E6. Since both oncoproteins modulate DNA damage pathways, we chose to use HPV-negative, p53 proficient U2OS cells to rule out the possibility that DNA damage in response to DEK knockdown is due to HPV. Adenoviral DEK short hairpin RNA (AdDEKsh) delivery resulted in efficient DEK knockdown and showed an increase in phosphorylation of H2AX (γH2AX), as well as an increase in mono-ubiquitinated FANCD2 by western blot analysis (Figure 1A). These data were further corroborated by the appearance of γH2AX, ubiquitinated FANCD2 and Rad51 foci in DEK depleted cells (Figure 1B and data not shown), with an ∼2- to 3-fold increase in the number of cells containing γH2AX and FancD2 foci compared with control infected cells. A slight increase in the phosphorylation of p53 at ser15 was also detected (Figure 1A). We previously observed that DEK depletion leads to p53 protein stabilization (40). Due to the modest increase of ser15 phosphorylation observed here, we do not believe that this modification can fully account for p53 stabilization. However, a role for other p53 residues that are phosphorylated in response to DNA damage such as ser20 and ser37 is possible. Alternatively, phosphorylation-independent mechanisms may contribute to p53 accumulation. To determine whether DNA damage occurred in response to DEK depletion in vivo, we performed human U2OS tumor xenograft studies in athymic nude mice. Tumors injected with GFP-tagged AdDEKsh virus exhibited increased levels of γH2AX compared to those injected with Ad-GFP virus (Figure 1C), suggesting that DEK depletion is sufficient for the induction of DNA damage markers in vitro and in vivo.
One prominent feature of apoptotic cells is DNA fragmentation, and consequently, the appearance of DSBs. To exclude the possibility that the appearance of γH2AX foci in response to DEK depletion was a consequence, rather than a cause, of apoptosis, we infected SAOS-2 osteosarcoma cells with AdDEKsh adenovirus to knock down DEK expression. SAOS-2 cells are genetically deleted for p53, and do not undergo apoptosis in response to DEK depletion (40). Similar to the results obtained in U2OS cells, we observed a significant accumulation of γH2AX foci in DEK-depleted SAOS-2 cells as compared to controls (Figure 1D). Together, these data indicate that the appearance of DNA damage foci occurs in the absence of p53 or apoptosis, thus suggestive of a scenario whereby DEK depletion first results in DNA damage and, subsequently, in p53-dependent cell death.
DEK modulates ATM and DNA-PK activity
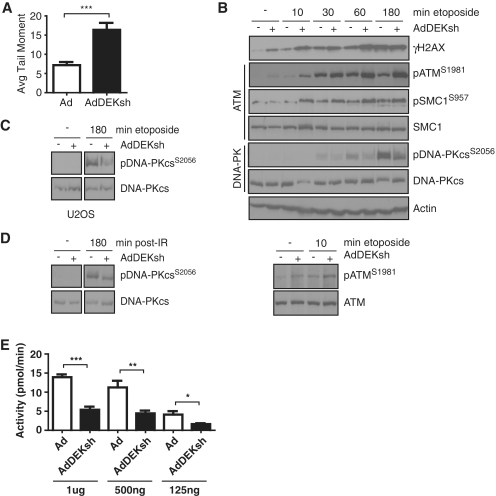
Because DEK depletion resulted in the phosphorylation of H2AX and p53, and the ubiquitination of FANCD2, we next examined the effect of DEK depletion on DNA damage response pathways in HeLa cells. Like U2OS and SAOS-2 cells, HeLa cells exhibited increased DNA damage upon DEK depletion as observed by increased levels of DNA strand breaks (Figure 2A). U2OS and SAOS-2 cells trended towards similar increases (Supplementary Figure S1). Furthermore, DEK-deficient HeLa cells exhibited increased γH2AX levels as detected by western blot analysis (Figure 2B). The phosphorylation of H2AX in response to DSBs is mediated by the sensor kinases ATM and DNA-PK. Kinase activation results in the phosphorylation of a cascade of target proteins and the recruitment of DNA damage response modulators. To begin to elucidate whether DEK regulates these kinase-driven pathways, HeLa cells were infected with Ad-GFP or AdDEKsh, and treated with etoposide for the time points indicated to induce DSBs, and analyzed by western blot analysis. As shown in Figure 2B, DEK-depleted HeLa cells displayed increased ATM pathway activation as determined by increased levels of autophosphorylation of ATM at serine 1981 in both untreated and etoposide-treated cells (top and bottom panel). Activation of the ATM pathway was further supported by increased modest phosphorylation of the ATM target SMC1 in etoposide treated DEK-deficient cells, when compared to controls (Figure 2B). Based on the induction of DNA strand breaks in DEK-depleted human cells (Figure 2A), it is likely that ATM is indirectly activated by DEK knockdown via accumulation of DNA damage. Since significant ATR kinase activation was not observed in several experiments (data not shown), these data implicate ATM kinase activity in the observed increased H2AX phosphorylation in DEK-deficient human cells. Interestingly, in contrast to the observed activation of ATM signaling, DNA-PKcs activity was decreased, as determined by reduced levels of autophosphorylation at Ser2056 following etoposide treatment (Figure 2B). Decreased DNA-PKcs autophosphorylation in response to DEK knockdown was also observed in γ-irradiated HeLa cells (Figure 2D), in U2OS cells (Figure 2C) and in Caski cervical cancer cells (data not shown). Overall, these data suggest that DEK loss results in an increase in ATM kinase activity and signaling, and a decrease in DNA-PK activity in response to DSBs.
Figure 2.
DEK knockdown leads to increased ATM and decreased DNA-PK signaling. (A) HeLa cells were infected with Ad-GFP and AdDEKsh adenoviruses. Three days post-infection, cells were collected, suspended in agarose gel, lysed and subjected to electrophoresis for comet assay analysis. Cells were then stained with SYBR green and assessed for DNA damage by determining the tail moment. Columns, mean value of tail moment from three independent experiments; bars, SEM. (***P < 0.0001) (B) HeLa cells were infected as in (A), and treated with 25 μM etoposide for the time points indicated 3 days post-infection. Whole cell lysates were subjected to western blot analysis using antibodies specific for γH2AX, pATMSer1981, total ATM, pSMC1Ser957, total SMC1, pDNA-PKcsSer2056 and total DNA-PKcs. (C) U2OS cells were infected as in (A) and treated with 25 μm etoposide for 3 h at 3 days post-infection and analyzed for pDNA-PKcsSer2056 and total DNA-PKcs by western blot analysis. (D) HeLa cells were infected as in (A) and treated with 10 Gy IR 3 days post-infection, allowed to recover for 3 h, and whole cell lysates analyzed by western blot analysis. Western blots depicted in (C) and (D) are from different parts of the same immunoblot, respectively. (E) HeLa cells were infected with Ad-GFP or AdDEKsh, and nuclear extracts collected 3 days post-infection. Endogenous DNA was removed from the nuclear extracts by passage through a DEAE sepharose column, and then kinase assays were performed using a biotinylated DNA-PK specific p53 target substrate, and activity determined using a scintillation counter. Columns, mean value of activity of 3 independent experiments; bars, SEM. (*P = 0.05, **P = 0.02, ***P = 0.001).
To determine whether decreased DNA-PKcs phosphorylation in DEK-deficient cells translated into reduced DNA-PK activity, kinase activities in HeLa cell nuclear lysates were compared between cells infected with AdDEKsh and those infected with control adenovirus. A fragment of p53 that is specifically phosphorylated by DNA-PK was utilized as a substrate. Consistent with reduced DNA-PKcs autophosphorylation (Figure 2B–D), DNA-PK kinase activity was compromised in DEK-depleted cells, as compared to controls (Figure 2E). Recruitment of the Ku70/80 heterodimer to DSBs is essential for the formation of the DNA-PK complex and subsequent DNA-PKcs autophosphorylation. We therefore determined whether DEK depletion affected the mobilization of Ku70/80 in etoposide treated cells. Using an established immunofluorescence approach, based upon cell permeabilization prior to fixation, and confocal microscopy, DEK-deficient cells exhibited impaired Ku70/80 mobilization when compared to controls (Figure 3A). Defects in Ku70/80 recruitment to strand breaks could account for the observed repression of DNA-PK kinase activity and autophosphorylation in DEK-deficient cells.
As the ATM and DNA-PK pathways respond to DSBs, we asked whether DEK itself could relocalize to DNA damage foci in etoposide treated HeLa cells. DEK foci were not damage inducible (data not shown). Additionally, DEK did not colocalize with γH2AX following treatment with etoposide as shown by confocal microscopy (Figure 3B, left panels). DEK has been reported to localize to sites of RNA processing at interchromatin granule clusters (ICGs) in response to treatment with deacetylase inhibitors (16). When treated with etoposide, DEK did not colocalize with the splicing factor SC35, which is concentrated at ICGs (Figure 3B, right panels). Collectively, these data indicate that while DEK affects specific DNA damage signaling responses, it does not appear to be physically relocated to sites of DNA damage.
DEK expression promotes resistance to exogenous stress
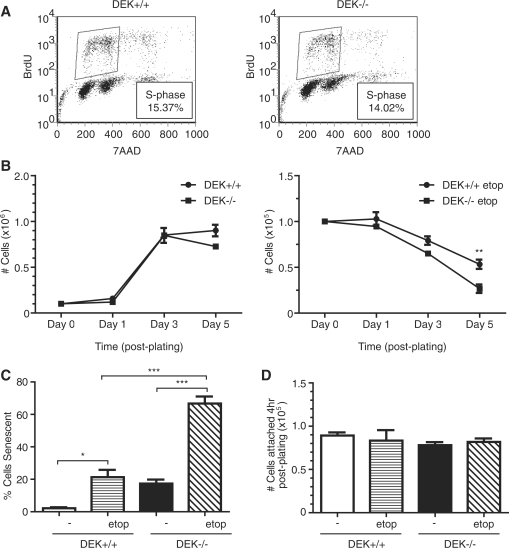
Upregulated ATM and suppressed DNA-PK signaling upon DEK depletion in cancer cell lines indicated that DEK may regulate DNA damage response and repair pathways. Since adenoviral DEK shRNA delivery could potentially influence DNA damage responses, and since cancer cells have evolved aberrant responses to damage, we examined primary MEFs generated from Dek proficient and deficient mice (39). Dek knockout mice were viable, indicating that DEK is not a necessary requirement for embryonic development and survival. To examine murine DNA damage phenotypes in response to genotoxic agents, we characterized primary MEFs from Dek knockout, heterozygous or wild-type mice for the detection of markers of DNA damage. Importantly, BrdU incorporation assays demonstrated similar cell-cycle distributions regardless of DEK presence (Figure 4A). However, following exposure to low doses of etoposide, Dek knockout MEFs exhibited decreased cellular growth compared to controls (Figure 4B), accompanied by an increase in the number of cells undergoing premature senescence (Figure 4C). Untreated wild-type and knockout cells grew at similar rates except for a small decrease of DEK-deficient over wild-type cells on Day 5 when the cells were completely confluent. To rule out differential plating efficiency as a possible contributor for growth differences, we plated equal cell numbers and quantified cellular attachment at 4 h post-plating. No differences in cellular attachment were noted between wild-type and Dek knockout cells in the presence or absence of etoposide (Figure 5D). Clonogenic cellular growth assays, either in the presence or absence of etoposide, also revealed substantial defects in Dek knockout MEFs, as compared to wild-type cells (Supplementary Figure S2). We conclude that Dek knockout, as compared to wild-type, MEFs are growth impaired when subjected to genotoxic or other stress conditions.
Figure 4.
Dek knockout MEFs are hyper-sensitive to genotoxic stress. (A) Asynchronous populations of primary wild-type and Dek knockout MEFs were analyzed for BrdU incorporation via flow cytometry. The percentage of cells in S-phase is indicated. (B) One hundred thousand wild-type or Dek knockout MEFs were plated in six-well plates in media only (left graph) or media containing 4 μM etoposide (right graph) and counted on Days 1, 3 and 5 post-plating for cell viability. Media +/− etoposide was replenished every 24 h. Line graphs, mean value of total number of cells in three independent experiments; bars, SEM. (**P = 0.0168) (C) Cells were plated and treated as in (B), and fixed 5 days post-plating for SA-β-galactosidase assays to determine levels of senescence. Columns, mean value of 200 cells counted in three independent experiments; bars, SEM. (*P < 0.05, ***P < 0.001) (D) Wild-type and Dek knockout cells were plated and treated as in (B). At 4 h post-plating, cells were trypsinized and counted to determine plating efficiency. Columns, mean value of attached cells from three independent experiments; bars, SEM.
Figure 5.
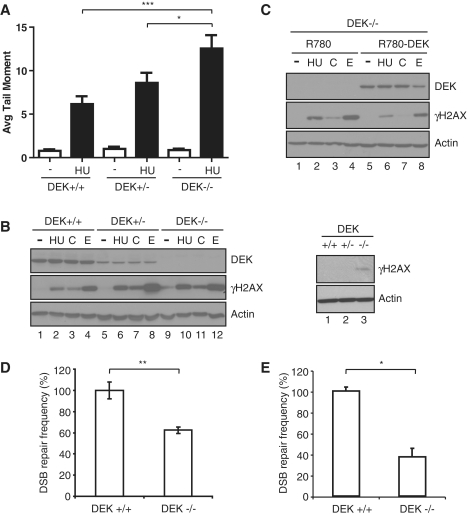
Dek-deficient MEFs exhibit increased levels of DNA strand breaks and decreased repair by NHEJ. (A) Primary MEFs were isolated from Dek wild-type, heterozygous, and knockout MEFs and treated with 1 mM HU for 24 h. Cells were then collected, suspended in agarose gel, lysed and subjected to electrophoresis. Cells were stained with SYBR green and assessed for DNA damage by determining the tail moment. Columns, mean value of the tail moment from three independent experiments; bars, SEM. (*P < 0.05, ***P < 0.001). (B) Primary MEFs were subjected to hydroxyurea (HU), camptothecin (C) and etoposide (E) treatment for 3 h, and protein lysates harvested and analyzed by western blot analysis for DEK, γH2AX and actin. To confirm the slight baseline increase of γH2AX in the knockout MEFs (compare lanes 1, 5 and 9), untreated samples were rerun separately (right panels). (C) Dek knockout MEFs were retrovirally infected with an empty vector (R780) or a DEK-expressing vector (R780-DEK). Following flow cytometric sorting, the cells were treated and analyzed as in (B). (D) Wild-type and Dek knockout MEFs were co-transfected with pEGFP-Pem1-Ad2 linearized with I-SceI along with DsRed-Express to control for transfection efficiency. The cells were harvested after 3 days and analyzed by flow cytometric analysis. DSB repair frequency was calculated from the percentage of EGFP positive cells divided by the percentage of transfection efficiency. DSB repair frequency of Dek wild-type MEFs were set to 100% each (absolute mean value: 80 × 10−2). Columns, mean value of four independent experiments; bars, SEM. (**P = 0.01) (E) Wild-type and Dek knockout MEFs were transfected with a repair reporter plasmid to detect NHEJ along with an I-SceI expressing vector. One day post-transfection, cells were collected and analyzed for EGFP expression by flow cytometry to determine repair efficiency. DSB repair frequency was calculated from the percentage of EGFP positive cells divided by the percentage of transfection efficiency in each case. DSB repair frequency of Dek wild-type MEFs were set to 100% each (absolute mean value: 30.0 × 10−3). Columns, mean values of 10 measurements per cell type and construct; bars, SEM (*P < 0.05).
DEK knockout MEFs are hyper-sensitive to DNA damaging agents and deficient in NHEJ
To determine whether MEF stress responses were linked to the presence of DNA strand breaks, Dek knockout, heterozygote and wild-type cells were treated with hydroxyurea (HU) for 24 h and subjected to alkaline comet assay for the direct detection of DNA strand breaks. Increased frequencies of DNA strand breaks were observed in Dek heterozygous MEFs, and were further increased in Dek knockout cells (Figure 5A). The cells were also subjected to the genotoxic agents hydroxyurea, camptothecin and etoposide, and analyzed for γH2AX levels by western blot analysis. Consistent with the knockdown results in human cancer cell lines, Dek knockout MEFs exhibited increased baseline levels of γH2AX compared to the wild-type and heterozygous controls (Figure 5B, left panel, lanes 1, 5 and 9; run separately in the right panel). Elevated γH2AX levels were also observed in heterozygous and knockout MEFs compared to wild-type cells that were treated with DNA damaging agents (Figure 5B). The slightly higher levels of γH2AX in etoposide treated heterozygous compared to homozygous knockout cells were not consistently observed. These data indicated that loss of DEK function was sufficient to induce an endogenous DNA damage response, and that just a 50% reduction in DEK expression could stimulate cellular DNA damage responses to exogenous stress. The murine and human DEK proteins share 89% sequence identity (Supplementary Figure S3). In order to determine whether retroviral expression of the human DEK protein can functionally complement Dek-deficient murine cells, we transduced immortalized Dek knockout MEFs with a retroviral human DEK expression vector (R780-DEK) as previously reported for keratinocytes (11). The resulting control R780 and R780-DEK cell lines were treated with hydroxyurea, camptothecin and etoposide, and were subjected to DEK and γH2AX specific western blot analysis (Figure 5C). DEK expression in Dek knockout MEFs resulted in a decrease in the phosphorylation of H2AX in response to each DNA damaging agent (compare lanes 2 with 6, 3 with 7, and 4 with 8). This indicated that the human DEK protein can rescue Dek-deficient murine cells from exaggerated cellular responses to DNA damage, extending previous findings by Kappes et al. from HeLa to murine cells.
Based on the observed defects in DNA-PK activity in DEK deficient human cells (Figure 2), and similar observations in MEFs (Supplementary Figure S4), we determined whether Dek knockout MEFs are deficient for NHEJ. To this end, we utilized a plasmid-based DNA repair assay (44). The indicator plasmid pEGFP-Pem1-Ad2 was linearized with I-SceI to generate non-compatible DNA ends whose processing and re-ligation by NHEJ results in intracellular EGFP expression. Dek knockout and wild-type cells were co-transfected with linearized template along with a DsRed plasmid, and repair by NHEJ was quantitated by flow cytometric detection of EGFP relative to DsRed. Consistent with decreased DNA-PKcs phosphorylation and kinase activity in DEK-deficient cells, DNA repair by NHEJ in the knockout MEFs was significantly reduced in four independent experiments when compared to wild-type MEFs (Figure 5D). A second NHEJ reporter assay was utilized that measures the role of DEK in response to intracellular I-SceI cleavage and microhomology-mediated NHEJ. Dek wild-type and knockout MEFs were co-transfected with the reporter together with an expression vector coding for the I-SceI endonuclease. Successful repair was detected by EGFP expression using flow cytometric analysis. Dek knockout compared to wild-type cells again exhibited a significant decrease in repair by NHEJ (Figure 5E). Together these data suggest that DEK expression promotes DNA-PK activity and DNA damage repair by NHEJ.
DISCUSSION
Our previous studies of DEK as an apoptosis inhibitor have demonstrated that transient DEK depletion leads to the stabilization of p53 and cell death in HeLa and primary cells. Unable to detect a direct interaction between DEK and p53, we examined the possibility that p53 regulation by DEK might be indirect via DNA damage responses. Indeed, DEK-deficient human and mouse cells exhibited increased levels of DNA damage, as assessed by the presence of nuclear DNA repair foci, by western blot analysis, and by comet assays. This response was independent of p53 status or apoptosis, occurring in p53 negative SAOS-2 cells which are resistant to apoptosis induced by DEK knockdown (40). DEK depletion was previously shown to delay the kinetics of exogenously-induced DNA damage repair, thus explaining sensitization of those same cells to DNA damaging agents. Also, DEK expression conferred chemoresistance to melanocytes in a manner which was independent of p53 (14). Our data support this scenario and extend it to murine cells in which DNA damage was exacerbated in DEK-deficient cancer cells and primary MEFs. The finding that elevated γH2AX levels in Dek knockout MEFs can be rescued by the expression of human DEK protein suggests that responsible DEK activities are evolutionarily conserved between mice and humans. Finally, the induction of γH2AX foci and apoptosis was not limited to cultured cells, systems which are inherently prone to DNA damage and cellular stress conditions (40,46). DEK depletion in a xenograft tumor model upregulated the number of γH2AX positive cells in the tumors in vivo, suggesting that DNA damage induction may underlie previous observations of cell death in these tumors (39). The finding that DEK knockdown is sufficient for γH2AX induction differs from previously published data (18). Kappes et al. showed elevated γH2AX levels following lentiviral DEK knockdown in the presence, but not in the absence, of genotoxic stress. This difference may be due to the extent of DEK knockdown which is more pronounced with adenoviral as compared to lentiviral shRNA delivery in our hands. Alternatively, small differences in culture conditions and resulting endogenous DNA damage levels could be at play. Given that apoptosis in response to DEK depletion was greatly attenuated in proliferating primary keratinocytes versus cancer cells (21,40,47,48), and was not observed in differentiated keratinocytes (11), our data supports the possibility that DEK might be a target for cancer treatment, perhaps in combination with conventional chemotherapy treatments. The fact that DEK loss induced DNA strand breaks in primary MEFs, however, should also raise concerns about long term DEK inhibition which could conceivably result in sustained DNA strand breaks, oncogenic mutations and secondary tumorigenesis.
In addition to sensitizing cells to exogenous genotoxic agents, the loss of DEK was sufficient for causing DNA strand break accumulation, ATM activity and γH2AX induction in cancer cells. Together with the established role of DEK as a chromatin topology modifier, and the finding that DEK does not itself re-localize to sites of DNA damage, it is unlikely that DEK knockdown activates the ATM response directly, but rather indirectly via DNA repair dysfunction and subsequent DNA damage accumulation. Of note, we have neither observed nor fully excluded a role for ATR in response to DEK loss, thus leaving open a possible role for DEK in DNA replication associated DNA damage response pathways. With regards to DNA repair defects, DEK loss distinctly decreased the level of Ku70/80 heterodimer recruitment, DNA-PKcs autophosphorylation, and DNA-PK kinase activity. Given that Ku70/80 recruitment is a prerequisite for DNA-PK assembly, detailed studies of the underlying molecular mechanism by which DEK loss impairs Ku70/80 recruitment are now critical. Moreover, DNA-PKcs autophosphorylation at Ser2056 is required for the full activity of DNA-PK (49,50), and may further impair DNA-PK function in DEK-deficient cells. Because DEK regulated DNA damage response pathways, but did not itself localize to DNA lesions, we consider it likely that the observed effects occur via specific DEK interactions with chromatin. It is important to note, however, that we cannot rule out a rapid or transient interaction of DEK with DSBs. In many cases, DNA repair requires chromatin unpacking prior to repair and chromatin restoration after repair. Indeed, DEK has been linked to chromatin topology and functions through preferential binding to structured DNA and histones and as a histone chaperone (24,25,34). Mechanisms by which these DEK-chromatin interactions might be linked to its role in DNA damage signaling and repair remains to be determined.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Cancer Institute (T32 CA59268 to G.M.K., T.M.W., and R.J.M.); the National Cancer Institute Public Health Service (CA102357 and CA116316 to S.I.W.); the National Institutes of Health (R01 GM072915 to J.M.W., R01 DK061458 to J.J.B., R01 ES012695, R01 ES016625 to P.J.S., R01 HL085587 to P.R.A.); the Center for Environmental Genetics (P30-ES006096 to G.F.B.); and the Deutsche Forschungsgemeinschaft (DFG Klinische Forschergruppe 167: “Apoptoseregulation und ihre Störungen bei Krankheiten” to B.G. and L.W.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Abdullah Ali and Ruhikanta Meetei for technical assistance with the AKTApurifier, Birgit Ehmer for technical assistance with confocal microscopy, and James Lessard for the monoclonal actin antiserum. The authors thank Elizabeth Hoskins, Teresa Morris and Fan Zhang for their technical assistance, and the Hematology/Oncology flow cytometry core at the Cincinnati Children's Hospital Research Foundation. The authors thank Lisa Privette Vinnedge for critical evaluation of this manuscript, and are grateful to Gerard Grosveld for providing the Dek knockout mice and for continued scientific advice.
REFERENCES
- 1.von Lindern M, Breems D, van Baal S, Adriaansen H, Grosveld G. Characterization of the translocation breakpoint sequences of two DEK-CAN fusion genes present in t(6;9) acute myeloid leukemia and a SET-CAN fusion gene found in a case of acute undifferentiated leukemia. Genes Chromosomes Cancer. 1992;5:227–234. doi: 10.1002/gcc.2870050309. [DOI] [PubMed] [Google Scholar]
- 2.Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, Muller H. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5:1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 3.Casas S, Nagy B, Elonen E, Aventin A, Larramendy ML, Sierra J, Ruutu T, Knuutila S. Aberrant expression of HOXA9, DEK, CBL and CSF1R in acute myeloid leukemia. Leuk. Lymphoma. 2003;44:1935–1941. doi: 10.1080/1042819031000119299. [DOI] [PubMed] [Google Scholar]
- 4.Evans AJ, Gallie BL, Jewett MA, Pond GR, Vandezande K, Underwood J, Fradet Y, Lim G, Marrano P, Zielenska M, et al. Defining a 0.5-mb region of genomic gain on chromosome 6p22 in bladder cancer by quantitative-multiplex polymerase chain reaction. Am. J. Pathol. 2004;164:285–293. doi: 10.1016/S0002-9440(10)63118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita M, et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–4996. [PubMed] [Google Scholar]
- 6.Kroes RA, Jastrow A, McLone MG, Yamamoto H, Colley P, Kersey DS, Yong VW, Mkrdichian E, Cerullo L, Leestma J, et al. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156:191–198. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- 7.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Carbayo M, Socci ND, Lozano JJ, Li W, Charytonowicz E, Belbin TJ, Prystowsky MB, Ortiz AR, Childs G, Cordon-Cardo C. Gene discovery in bladder cancer progression using cDNA microarrays. Am. J. Pathol. 2003;163:505–516. doi: 10.1016/S0002-9440(10)63679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savli H, Sirma S, Nagy B, Aktan M, Dincol G, Salcioglu Z, Sarper N, Ozbek U. Real-time PCR analysis of af4 and dek genes expression in acute promyelocytic leukemia t (15;17) patients. Exp. Mol. Med. 2004;36:279–282. doi: 10.1038/emm.2004.38. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Hoffmann MJ, Hartmann FH, Schulz WA. Amplification and overexpression of the ID4 gene at 6p22.3 in bladder cancer. Mol. Cancer. 2005;4:16. doi: 10.1186/1476-4598-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wise-Draper TM, Morreale RJ, Morris TA, Mintz-Cole RA, Hoskins EE, Balsitis SJ, Husseinzadeh N, Witte DP, Wikenheiser-Brokamp KA, Lambert PF, et al. DEK proto-oncogene expression interferes with the normal epithelial differentiation program. Am. J. Pathol. 2009;174:71–81. doi: 10.2353/ajpath.2009.080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Munger K, Wells SI. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J. Virol. 2005;79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johung K, Goodwin EC, DiMaio D. Human papillomavirus E7 repression in cervical carcinoma cells initiates a transcriptional cascade driven by the retinoblastoma family, resulting in senescence. J. Virol. 2007;81:2102–2116. doi: 10.1128/JVI.02348-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz DM, Soengas MS. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein–an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 16.Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, Cole PA, Markovitz DM. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J. Biol. Chem. 2005;280:31760–31767. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 17.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol. Cell. Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Burkle A, Markovitz DM, Ferrando-May E. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol. Cell. Biol. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol. Cell. Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 21.Tabbert A, Kappes F, Knippers R, Kellermann J, Lottspeich F, Ferrando-May E. Hypophosphorylation of the architectural chromatin protein DEK in death-receptor-induced apoptosis revealed by the isotope coded protein label proteomic platform. Proteomics. 2006;6:5758–5772. doi: 10.1002/pmic.200600197. [DOI] [PubMed] [Google Scholar]
- 22.Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- 23.Thomas JO, Travers AA. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 24.Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14:1308–1312. [PMC free article] [PubMed] [Google Scholar]
- 26.Campillos M, Garcia MA, Valdivieso F, Vazquez J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31:1571–1575. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu GK, Grosveld G, Markovitz DM. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc. Natl Acad. Sci. USA. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat. Struct. Mol. Biol. 2007;14:548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 29.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 30.Sammons M, Wan SS, Vogel NL, Mientjes EJ, Grosveld G, Ashburner BP. Negative regulation of the RelA/p65 transactivation function by the product of the DEK proto-oncogene. J. Biol. Chem. 2006;281:26802–26812. doi: 10.1074/jbc.M600915200. [DOI] [PubMed] [Google Scholar]
- 31.Hu HG, Scholten I, Gruss C, Knippers R. The distribution of the DEK protein in mammalian chromatin. Biochem. Biophys. Res. Commun. 2007;358:1008–1014. doi: 10.1016/j.bbrc.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 32.Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, Lee KS, Woo KM, Baek JH, Choo JK, Seo SB. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580:3217–3222. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- 33.McGarvey T, Rosonina E, McCracken S, Li Q, Arnaout R, Mientjes E, Nickerson JA, Awrey D, Greenblatt J, Grosveld G, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell. Biol. 2000;150:309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suwatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, et al. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 2010;24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyn MS, Lu-Kuo JM, Herzing LB. Expression cloning of multiple human cDNAs that complement the phenotypic defects of ataxia-telangiectasia group D fibroblasts. Am. J. Hum. Genet. 1993;53:1206–1216. [PMC free article] [PubMed] [Google Scholar]
- 36.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 37.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 38.McVey M, Lee SE. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA, Cripe TP, Grosveld GC, Wells SI. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol. Cell. Biol. 2006;26:7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J. Biol. Chem. 2005;280:7060–7069. doi: 10.1074/jbc.M410746200. [DOI] [PubMed] [Google Scholar]
- 42.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc. Natl Acad. Sci. USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bohringer M, Wiesmuller L. Fluorescence-based quantification of pathway-specific DNA double-strand break repair activities: a powerful method for the analysis of genome destabilizing mechanisms. Subcell. Biochem. 2010;50:297–306. doi: 10.1007/978-90-481-3471-7_15. [DOI] [PubMed] [Google Scholar]
- 46.Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell. Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim DW, Chae JI, Kim JY, Pak JH, Koo DB, Bahk YY, Seo SB. Proteomic analysis of apoptosis related proteins regulated by proto-oncogene protein DEK. J. Cell. Biochem. 2009;106:1048–1059. doi: 10.1002/jcb.22083. [DOI] [PubMed] [Google Scholar]
- 48.Secchiero P, Voltan R, di Iasio MG, Melloni E, Tiribelli M, Zauli G. The oncogene DEK promotes leukemic cell survival and is downregulated by both Nutlin-3 and chlorambucil in B-chronic lymphocytic leukemic cells. Clin. Cancer Res. 2010;16:1824–1833. doi: 10.1158/1078-0432.CCR-09-3031. [DOI] [PubMed] [Google Scholar]
- 49.Cui X, Yu Y, Gupta S, Cho Y-M, Lees-Miller SP, Meek K. Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 2005;25:10842–10852. doi: 10.1128/MCB.25.24.10842-10852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.