Abstract
Although many proteins are known to function in microRNA (miRNA)-based translational repression, we lack a comprehensive understanding of temporal relationships between the mRNA, miRNA and their constituent proteins. To understand the dynamics of miRNA and protein interactions, we created a synthetic inducible miRNA system in mammalian cells. By visualizing single mRNAs and observing their co-localization with proteins over time, we produced a temporal association map of miRNA-associated factors. Argonaute2, Dcp1a, hedls and Rck co-localize with miRNA-regulated mRNA after 24 h of miRNA induction, and RNAi knockdown of any one of these proteins affected the co-localization of any of the other proteins with miRNA-regulated mRNA, demonstrating that these proteins could interact with each other in a complex. We identified Argonaute2 and hedls as proteins that co-localize and interact with miRNA-regulated mRNA, indicating that processing body components are involved in long-term storage of miRNA-regulated mRNA.
INTRODUCTION
microRNA (miRNA)-directed translational repression, a process in which miRNA serves as a guide for the RNA-induced silencing complex (RISC) to bind complementary target mRNA and arrest translation, regulates many genes including those involved in development (1) and heterochromatin formation (2). Defects in miRNA regulation result in a wide range of disorders including leukemia, lymphoma and fragile X syndrome (3–5). Despite much research over the last decade, questions remain about the fate of mRNA following miRNA-directed translational repression.
Processing bodies (P bodies) are known as sites of mRNA degradation for nonsense or AU-rich element-mediated decay (6,7). Similarly, it was thought that Argonaute proteins in the RNA-induced silencing complex (RISC) direct mRNA to P bodies during translational repression for future degradation (8). However, recent evidence suggests that Argonaute-directed repression and mRNA degradation occur in the absence of P bodies, indicating that miRNA-regulated mRNA localization to P bodies is not necessary for mRNA degradation (9). Additionally, not all translationally repressed mRNA is degraded; up to 50% of repressed mRNA can be left intact in P bodies (10) and re-enter translation (11,12). These observations suggest that P bodies can store repressed mRNA for rapid translational reactivation.
Stress granules, which form under adverse conditions, are also implicated in general translational repression. Stress granules not only share some of the same proteins as P bodies, but also posses unique translation initiation proteins such eIF3, eIF4G and PABP (13), and contain abortive translation initiation complexes (14). Evidence indicates that miRNA-directed translational repression could occur by inhibiting translation initiation (15), thus RISC could interact with translation initiation components in stress granules. Stress granules also lack mRNA decapping enzymes (6,13); they therefore do not degrade mRNA and could store mRNA for future translation. Many P body and stress granules components have already been identified, but we do not know how they contribute to degradation or storage of miRNA-regulated mRNA.
To understand how RISC, P body and stress granule components interact temporally with miRNA-regulated mRNA during and after translational repression, we used single molecule fluorescent in situ hybridization (smFISH) in combination with immunofluorescent staining (IF) of endogenous proteins. We found that Argonaute2, Dcp1a, hedls and Rck co-localize with miRNA-regulated mRNA after 24 h of miRNA induction, suggesting that these proteins could be involved in mRNA storage. We found that RNAi knockdown of any one of these proteins affected the co-localization of other proteins with miRNA-regulated mRNA, indicating that these proteins could interact with each other in a complex. We further demonstrated that Arognaute2 and hedls interact with the miRNA-regulated mRNA, indicating that P body components are involved in miRNA-regulated mRNA storage after miRNA-based mRNA degradation.
MATERIALS AND METHODS
Transgene construction
JDS33 was constructed using Biobrick construction methods (16) and previously characterized Biobrick parts. It consists of a Tet-On Doxycycline promoter fused to luc-1601 miRNA (17), two copies of YFP and a nuclear localization sequence inserted into a modified pcDNA3.1+/Puro vector (Invitrogen) with a deleted CMV promoter. The transcribed RNA would therefore be an intronic miRNA spliced out during mRNA maturation and two linked copies of YFP transcript attached to RNA encoding a nuclear localization signal. JDS48 and other luciferase reporter target plasmids were constructed by inserting four to five copies of target sequences to luc-1601 miRNA that are either of mismatched (CXCR4), imperfect (miRNA, JDS48) or perfect (siRNA) complementarity into the MCS of pmirGLO vector (Promega), which expresses both firefly and renilla luciferase.
Cell culture and cell line construction
All cell lines originated from U2OS cells and were grown in McCoy’s 5 A media (Invitrogen) supplemented with 10% tetracycline-screened FBS (HyClone) and 1% penicillin/streptomycin (Invitrogen). All transfections were conducted with Lipofectamine 2000 and OptiMEM media using the standard manufacturer’s protocol (Invitrogen). JDS33 plasmid was transfected into U2OS Tet-On cell line (ClonTech), randomly integrated and single clones isolated using puromycin (Sigma) selection for low basal YFP levels and high YFP levels upon induction. JDS48 was then co-transfected with pCMV/Zeo (Invitrogen) into the integrated JDS33-expressing cell line, randomly integrated and single clones isolated using zeocin (Invitrogen) selection for high-luciferase activity to produce JDS33/48.
Single molecule FISH—immunofluorescent co-staining
smFISH-IF co-staining protocol was modified from a protocol used by Raj et al. (18) 65 probes against the luc2 gene in the pmirGLO vector were designed using the program available online at http://www.singlemoleculefish.com. Probes were ordered with 3′-amino modifications (BioSearch Technologies), coupled to Cy-5 fluorophore (GE Healthcare) and HPLC purified. JDS33/48 cells were plated for 24 h in multi-well 1.5 thickness glass bottom culture plates (MatTek). To JDS33/48 cells, 1 µg/ml doxycycline was added for the indicated time periods. mRNA transcription was inhibited by adding 50 µM dichlorobenzimidizole riboside (DRB) 5 h prior to fixing cells. At the indicated times, JDS33/48 cells were fixed with formaldehyde, washed with PBS and stored overnight in 70% ethanol. The next day they were washed with buffer (2× SSC, 10% foramide, RNase-free water) and then incubated overnight at 28°C in hybridization buffer (10% dextran sulfate, 2 mM vanadyl-ribonucleoside complex, 0.02% RNAse-free BSA, 1 mg/ml Escherichia coli tRNA, 2× SSC, 10% foramide, RNase-free water) with Cy-5 labeled probes and primary antibody against the protein of interest. We did not perform a blocking step prior to primary antibody incubation to prevent introduction of RNases. All incubations were conducted in the dark. Primary antibody sources and working concentrations are detailed on Supplementary Table S1. The next day cells were aspirated, washed, and incubated for one hour at room temperature in hybridization buffer with species-specific Cy-3 secondary antibodies (Jackson ImmunoResearch) at 1/200 dilution. They were then washed, incubated for 30 min at room temperature in wash buffer containing 5 ng/ml DAPI, and after a final wash mounted in freshly prepared oxygen depleted mounting media.
Data acquisition and analysis
Live cell time-lapse experiments were conducted in a 5% CO2 incubation chamber set at 37°C on a Nikon TE2000E motorized inverted microscope with Perfect Focus System, Prior Proscan II motorized stage and shutters and Prior LumenPro light source. Images were acquired each hour with a ×20 objective with a 500-ms exposure time from a Hamamatsu ORCA-ER cooled CCD camera with Metamorph image acquisition software (Molecular Devices). smFISH-IF fixed cell images were taken on a Nikon TE2000E inverted fluorescence microscope with Nikon motorized stage, excitation and emission wheels and X-Cite 120Q Fluorescence Illumination system (Lumen Dynamics). Images were acquired with a ×100 objective with 100, 200 and 1500 ms exposure times for DAPI, Cy-3 and Cy-5, respectively, from a Hamamatsu ORCA-ER cooled CCD camera with Metamorph image acquisition software (Molecular Devices). All images were analyzed using ImageJ software. For the live cell time-lapse images, base threshold was set at 3× the average intensity of the entire image at t = 0 h. smFISH-IF images were analyzed by local background subtraction and thresholding, with data from the nucleus excluded by DAPI staining. Co-localization was scored using the AND function on ImageJ. Heat map numbers were generated by calculating the ratio of co-localized foci normalized to the total number of protein foci multiplied by 100. This value is essentially the percentage of co-localized foci normalized to the total amount of protein. Number of cytoplasmic mRNA transcripts per cell was approximated by totaling the number of mRNA foci outside of nuclei then dividing by the total number of complete nuclei in each image. Typical images had ∼3–5 nucleii. Luciferase activity assays were conducted using the Dual-Glo Luciferase Assay System (Promega). mRNA transcription was inhibited by adding 50 µM DRB 5 h prior to taking data points. Firefly luciferase activity levels were normalized to renilla luciferase activity levels to control for differences in transcriptional and translational efficiency.
RNA immunoprecipitation and qPCR
Magnetic Dynabeads Protein G and A (Invitrogen) were washed four times in 500 µl RNA immunoprecipitation (RIP) buffer (50 mM Tris, pH 7.0, 0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, 10% glycerol) using a magnetic rack. Antibodies were precoupled to beads in a solution comprised of 445 µl RIP buffer, 10 µl antibody, 50 ul 10 mg/ml BSA and 5 µl yeast tRNA. This solution was rocked for 2 h at 4°C and afterwards washed three times in 1 ml RIP buffer. Cells were washed with DEPC-PBS, scraped off, spun down, resuspended in 1 ml of freshly made cell lysis buffer [RIP buffer, 1:40 RNaseOUT (Invitrogen), 1:100 PMSF (17.5 mg/ml in methanol), 1:2200 RNasin (Promega)] and rocked for 10 min at 4°C. Cell lysate was then spun down to remove debris and the supernatant was added to the magnetic beads and rocked for 4 h at 4°C. Afterwards the beads were then washed 6× in 1 ml RIP buffer with freshly added 1:200 PMSF (17.5 mg/ml) and 1:100 1 M DTT. The beads were then resuspended in 200 µl elution buffer [RNA storage solution (Ambion), 1% SDS, 1 mM EDTA, 1:100 1 M DTT] per sample. They were eluted for 10 min at 65°C, and quickly vortexed every 3–4 min. RNA was then acid/phenol/chloroform extracted from the elution, separated with Heavy Phase Lock Gel tubes (Fisher) and ethanol precipitated overnight. The RNA was reverse transcribed with SuperscriptIII (Invitrogen) according to the standard protocol and qPCR was performed using SYBR Green (Applied Biosystems). Primers for the qPCR of firefly luciferase were 5′-gccatgaagcgctacgccctgg-3′ and 5′-tcttgctcacgaatacgacggtgg-3′. Primers for renilla luciferase were 5′-tcagtggtgggctcgctgca-3′ and 5′-ctttggaaggttcagcagctcg-3 ′
Western blot
First NuPage LDS sample buffer (Invitrogen) was added to each sample, and they were then heated at 100°C for 10 min and air cooled. Samples were run on a NuPage 4–12% Bis–Tris gel (Invitrogen) with 1× NuPage MES buffer (Invitrogen) with Precision Plus Protein Dual Color Standard (Biorad) at 120 V for 90 min. Proteins were transferred to PVDF membranes (Millipore) in NuPage transfer buffer (Invitrogen) using a semidry transfer apparatus at 150 mA for 1 h. Membranes were blocked with 5% non-fat dry milk in PBS + 0.1% Tween (blocking buffer) for 1 h, then exposed to the primary antibody (Argonaute2—1:200, Dcp2—1:500, Dcp1a—1:2000, hedls—1:1000, Rck—1:3000) overnight at 4°C. Membranes were washed with three times with PBS + 0.1% Tween and exposed to 1:5000 dilution species-specific HRP antibody in blocking buffer for 1 h. They were then washed three times with PBS + 0.1% Tween and 1 ml each of Western Lightning Plus-ECL Luminol and Oxidixing Reagents were added for 1 min. Membranes were exposed to Kodak Scientific Imaging Film and developed in a darkroom.
mRNA lifetime calculation
JDS33/48 cells were plated for 24 h in multi-well 1.5 thickness glass bottom culture plates (MatTek). At 0, 1, 2, 3 or 4 h prior to cell fixation, 50 µM dichlorobenzimidizole riboside (DRB) was added and smFISH staining was conducted (18). Approximately 50 cells were imaged for each time point, and number of cytoplasmic mRNA per cell was approximated by totaling the number of mRNA foci outside of nuclei in pictures then dividing by the total number of complete nuclei in each picture. The median of each hour was taken and used to fit a linear mRNA decay equation. The average mRNA lifetime was then extrapolated from that equation.
siRNA transfection
Standard siGENOME SMARTpool siRNAs for Argonaute2 (eIF2C2), Dcp1a, hedls (EDC4) and Rck (DDX6) were ordered from Dharmacon. siRNA transfection was conducted using HiPerFect Transfection Reagent and the standard manufacturer’s protocol (Qiagen). All siRNAs used in RNAi studies were transfected 54 h before fixation. Induction with doxycycline occurred 24, 12, 6 and 0 h prior to fixation, respectively.
RESULTS AND DISCUSSION
Designing miRNA-based silencing of a target reporter gene
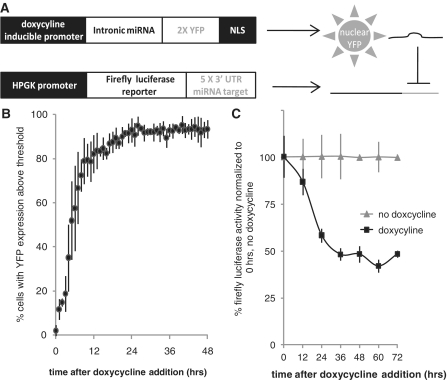
In order to conduct a temporal study of RISC, P body and stress granule component co-localization with miRNA-regulated mRNA, we constructed a synthetic miRNA expression system capable of inducible silencing of a target reporter mRNA. In human U2OS cells, we stably expressed an intronic miRNA (luc-1601 miRNA) (17) coupled to two copies of the gene encoding the yellow fluorescent protein (YFP) with a nuclear localization signal (NLS) under a doxycycline-inducible promoter. Nuclear-localized YFP expression serves as an indirect indicator of miRNA expression. We then stably expressed a firefly luciferase gene with five copies of a target sequence in the 3′ untranslated region (UTR) that is imperfectly complementary to the miRNA under a constitutive human phosphoglycerate kinase (HPGK) promoter (Figure 1A). We call this cell line JDS33/48. In the absence of doxycycline inducer, YFP and miRNA are not expressed and JDS33/48 cells generate a strong firefly luciferase reporter signal. When doxycycline is added, miRNA and YFP production is induced resulting in a substantial increase in nuclear YFP signal as well as a 50–60% decrease in firefly luciferase activity (Figure 1B and C) and a 10-fold decrease in firefly luciferase mRNA levels after 48 h (Supplementary Figure S1A). Dichlorobenzimidizole riboside (DRB), an RNA Polymerase II inhibitor, was added to cells during miRNA induction for 5 h prior to assaying to inhibit transcription of new mRNAs. Over 80% of JDS33/48 cells have significant YFP expression after 12 h while luciferase activity does not substantially decrease until 24–36 h after doxycycline induction. This delay reflects the lag time between miRNA transcription and maturation, miRNA-based silencing of the target mRNA and degradation of the luciferase protein.
Figure 1.
Doxycycline induction of miRNA correlates with increased nuclear YFP and reduced luciferase reporter levels. (A) Schematic of constructs stably integrated in U2OS cells. Doxycycline induction produces intronic miRNA targeting the 3′-UTR of a luciferase reporter mRNA. Nuclear YFP is also expressed as an indirect readout of miRNA production. (B) The majority of cells produce significant levels of nuclear YFP by 12 h of doxycycline induction. Threshold is 3× mean background levels of YFP expression before doxycyline induction (0 h). N ∼ 300 cells, three replicates per data point. (C) Doxycycline induction results in reduced luciferase activity. N = 3 replicates per data point. Error bars = standard deviation.
To confirm that the miRNA specifically targets the luciferase reporter and that the decrease in luciferase activity is due to miRNA-specific targeting, we conducted transient co-transfections of the doxycycline-inducible luc-1601 miRNA-YFP construct with luciferase reporters containing different targets in their 3′-UTR. After co-transfection we induced miRNA expression and measured luciferase activity levels after 48 h (Supplementary Figure S1B). No silencing was observed for firefly luciferase reporters lacking target sequences or containing mismatched CXCR4 target sequences in the 3′-UTR, demonstrating that luciferase reporter activity is not dampened by nuclear YFP expression and that non-specific target sequences are not recognized by the luc-1601 miRNA. Target sequences with imperfect (miRNA) or perfect (siRNA) complementarity display significant silencing after 48 h, indicating that the luc-1601 miRNA does specifically target mRNAs with complementary target sequences. Therefore, we have successfully designed and validated an inducible miRNA expression system.
Co-localization of proteins with miRNA-regulated mRNA
To determine the temporal association of RISC, P body and stress granule components with miRNA-regulated mRNA, we used smFISH (18) to localize firefly luciferase mRNA combined with IF of various P body and stress granule components to examine JDS33/48 cells fixed at various time points after doxycycline induction of miRNA. Choosing timepoints based on YFP expression and luciferase reporter silencing assays (Figure 1), we fixed cells at 0, 6, 12 and 24 h after doxycycline induction in order to capture the widest range of temporal association. DRB was added to cells 5 h prior to fixation, following the doxycyline-mediated miRNA induction to terminate reporter mRNA transcription. We used smFISH to measure mRNA numbers 0–4 h after transcription block with DRB to produce an mRNA decay curve and measured average mRNA lifetime to be 14.07 ± 1.76 h (Supplementary Figure S1C). Therefore each time point represents mRNA transcripts that are at least 5 h old at the sampling time.
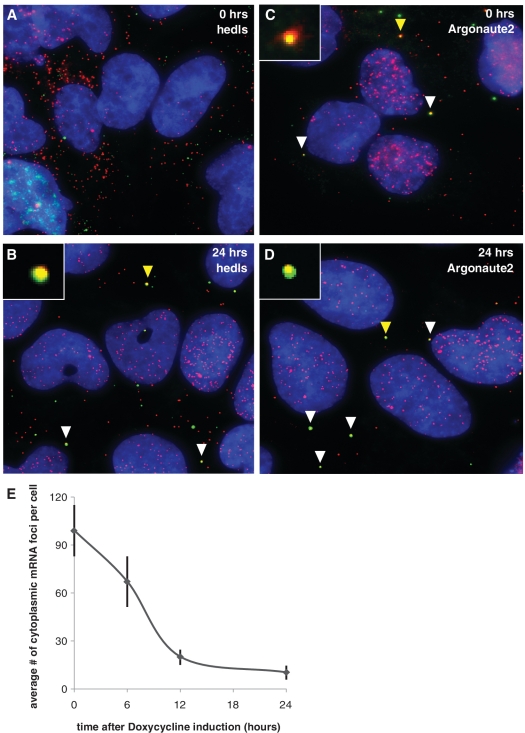
As a proof of concept experiment, we conducted smFISH-IF co-staining of mRNA with antibodies against Argonaute2 and hedls. smFISH-IF co-staining followed by wide-field microscopy imaging with background subtraction produced images showing punctate spots of mRNA probe and protein antibody. We found no co-localization of target mRNA with hedls prior to miRNA induction (Figure 2A), while Argonaute2 initially displayed significant levels of mRNA co-localization (Figure 2C). Upon induction of miRNA, co-localization of Argonaute2 with mRNA remained high while hedls co-localization steadily increased such that 24 h after miRNA induction there were high levels of mRNA co-localization for both proteins (Figure 2B and D).
Figure 2.
Single molecule FISH combined with immunofluorescence shows mRNA co-localization with RISC and P body proteins when miRNA is expressed. (A–D) smFISH-IF co-staining of protein antibody (green, Cy-3) and luciferase reporter mRNA (red, Cy-5) at various times after doxycycline induction of mRNA. Blue = DAPI. White arrows point to examples of co-localization. Yellow arrows point to co-localization shown at 5× size in the inset of the same picture. (A) Hedls, 0 h. (B) Hedls, 24 h. (C) Argonaute, 0 h. (D) Argonaute2, 24 h. (E) Luciferase reporter mRNA levels are reduced when miRNA is expressed. Average number of cytoplasmic mRNA foci per cell 0, 6, 12 and 24 h after doxycycline induction of miRNA. N = 52 cells for 0 h, 48 cells for 6 h, 51 cells for 12 h and 50 cells for 24 h. Error bars = 95% confidence interval for means.
In addition to identifying intracellular mRNA localization, smFISH also allows us to approximate cytoplasmic mRNA levels per cell as each spot of minimal intensity is equivalent to one mRNA (18). An average of cytoplasmic mRNA transcripts per cell shows a substantial decrease in mRNA levels after miRNA induction, with mRNA levels remaining low and relatively unchanged after 24 h of miRNA induction (Figure 2E). This visual demonstration of mRNA molecules decreasing when miRNA targeting the mRNA is expressed is in agreement with ribosome profiling data (19), which shows a large decrease in endogenous target mRNA levels upon miRNA expression. Additionally, our results indicate that mRNA degradation happens within the first 24 h of miRNA expression in mammalian cells, with a lag time for a correlated decrease in protein activity based on the protein half-life.
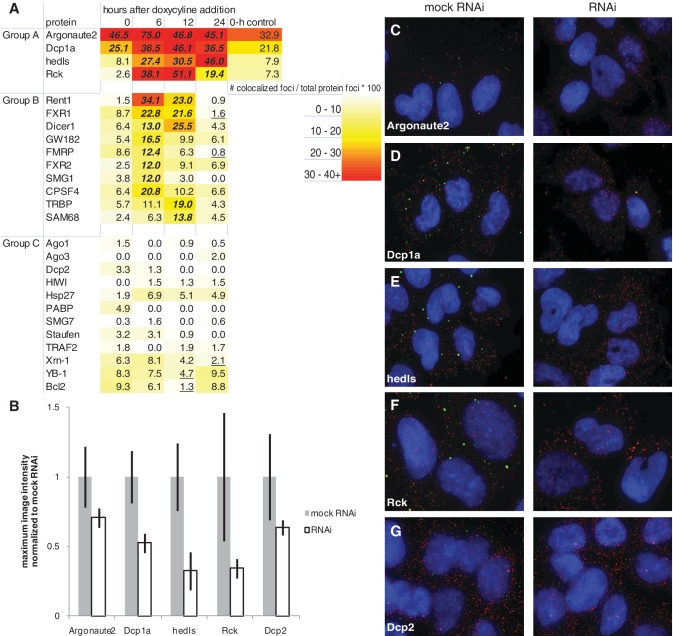
We conducted a targeted temporal association screen of RISC, P body and stress granule components in order to determine which proteins co-localize with mRNA targeted by miRNA at specific times. A heat map summarizing the results of the experiments shows that there are patterns of temporal protein co-localization with miRNA-regulated mRNA that can be organized into three groups (Figure 3A). We generated the numbers on the heat map by calculating the ratio of co-localized foci normalized to the total number of protein foci and multiplying by 100. This value is the percentage of co-localized foci normalized to the total amount of protein.
Figure 3.
P body and RISC proteins temporally co-localize with miRNA-regulated mRNA. (A) Heat map of protein temporal association with miRNA-regulated mRNA. Data points are taken 0, 6, 12 and 24 h after doxycyline induction of miRNA. Number of co-localized foci is divided by number of total protein foci to normalize for variable protein levels and get percentage of protein interacting with the target mRNA. Group A = proteins with high scores when miRNA is expressed. Group B = proteins with high scores in the first 6–12 h, presumably during active RNAi degradation of mRNA. Group C = proteins with low scores overall. Zero-hour control is the association score in a cell line with only the luciferase reporter to control for miRNA leakiness. Each data point is an aggregate of five images with ∼ 20 cells total. A statistical analysis was conducted comparing differences between population proportions of the 0-h time point to each other time point for each protein, except for Argonaute2 and Dcp1a which were compared to hedls at 0 h. Bold/italicized numbers indicate results significantly >0-h time point and underlined numbers indicate results significantly <0-h time point (P < 0.05). (B) The average maximum image intensity for each antibody either mock transfected (mock RNAi) or transfected with siRNA targeting the protein (RNAi). Data for each protein was normalized to mock RNAi. Error bars = standard deviation. N = 15. (C–G) Images of JDS33/48 cells either mock transfected (left) or transfected with siRNA (right) targeting (C) Argonaute2, (D) Dcp1a, (E) hedls, (F) Rck or (G) Dcp2. Cells were fixed 48 h after transfection and smFISH-IF co-staining of protein, the appropriate antibody (green, Cy-3) and luciferase reporter mRNA (red, Cy-5) was conducted. Blue = DAPI.
Group A proteins, comprised of Argonaute2, Dcp1a, hedls and Rck, co-localize with mRNA throughout miRNA induction. The consistent co-localization indicates that they could be involved in miRNA-based mRNA storage as they are still co-localized with mRNA after 24 h of miRNA induction. Argonaute2 is a well-known RISC component that is also associated with P bodies (20). Dcp1a, hedls and Rck are P body components involved in mRNA decapping and degradation. Argonaute2 and Dcp1a both have high levels of protein interaction with the target mRNA prior to miRNA induction, suggesting that there may be leakiness in the inducible promoter. To control for miRNA leakiness, we conducted smFISH-IF experiments for group A proteins in a cell line containing only the luciferase reporter (Figure 3A, 0-h control). Statistical analysis comparing the differences between population proportions for each of the group A proteins revealed no statistical difference (P > 0.05) between the 0-h controls and the 0-h timepoints, demonstrating that some Argonaute2 and Dcp1a interaction with the reporter mRNA occurs without the presence of miRNA.
Group B proteins, such as Rent1, Dicer1, FMR1, GW182 and TRBP, co-localize with the target mRNA 6–12 h after miRNA expression, indicating that they are associated with miRNA-targeted mRNA degradation as the rapid decrease in reporter mRNA levels occurs at these times (Figure 2E). That the majority of proteins in Group B are RISC components, such as FXR1, FXR2, FMR1, GW182 and TRBP, further supports the idea that miRNA-targeted mRNA degradation occurs at this time.
Group C proteins do not significantly co-localize with the target mRNA at any time in our screen. However, proteins in Group C cannot be defined as lacking any association with miRNA-regulated mRNA and may simply not associate for enough time to be observed in our assay. For example, it is interesting to note that predominantly stress granule proteins were not found in Group A proteins. Evidence from other groups shows that individual mRNAs interact for only a few minutes with stress granules during stress-induced translational arrest (21), implying that individual mRNA transcripts do not associate with stress granules long enough to be detectable in our screen.
Hedls and Rck have both been shown to interact with Dcp2 and Dcp1a, with hedls promoting the complex formation between Dcp2 and Dcp1a as well as enhancing Dcp2 decapping activity (22). However, we do not detect Dcp2 co-localizing with targeted mRNA at any time in our experiments. To validate the protein–mRNA co-localization seen in our screen (Figure 3A), we transfected pooled siRNAs corresponding to each of the Group A proteins and Dcp2 into the JDS33/48 cell lines. If the protein staining is specific then RNAi knockdown of each individual protein would lead to a reduction in antibody staining. Indeed, we found antibody staining intensity for each protein dropped when compared to a mock RNAi control (Figure 3B–G), validating both the efficacy of RNAi against each protein and the co-localization of Group A proteins with miRNA-regulated mRNA seen in the screen (Figure 3A). This indicates that Dcp1a, hedls and Rck are co-localizing with mRNA independent of Dcp2 activity, implying a non-decapping function for these proteins without Dcp2 present. The modularity of P body components has been noted before. In Drosophila S2 cells EDC3, a P body component involved in mRNA decapping (22), and TraI, a P body component involved in translational control of maternal mRNA (23), do not interact with each other and are mutually exclusive because they bind to the same binding sites on Dcp1 and Me31B (24). However, EDC3 interacts with both Dcp1 and Dcp2 while TraI only interacts with Dcp1, demonstrating that Dcp1 and Dcp2 can be in separate protein complexes with unique functions (24). Other alternative explanations are that the supply of Dcp2 protein is limited such that most of it cannot co-localize with the synthetic mRNA or that any Dcp2 interaction with the target mRNA is too brief to detect since any active Dcp2 could lead to mRNA decapping and degradation.
Finding functionality in the temporal association of proteins with miRNA-regulated mRNA
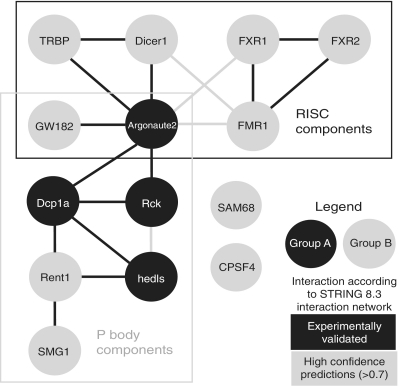
We speculate that proteins with the same temporal association could interact with each other or be part of the same complex. To see if the protein in the groups we characterized (Figure 3A) are known or predicted to interact with each other, we constructed an interaction network of Groups A and B proteins based on the known and predicted interactions of the online STRING 8.3 Interaction Network (Figure 4) (25). Group A proteins cluster in the center of the network, while Group B are separated into two distinct groups. Additionally, RISC and P body proteins cluster in their own sub-networks, with RISC and P body components united by Argonaute2 which is both a RISC and P body component (20). Our association data correlates well with this interaction network.
Figure 4.
STRING 8.3 Interaction Network of Groups A and B proteins.
The interaction network can infer which proteins will interact with each other in an integrated protein complex. For example, based on the interaction network we predict that Group A proteins, which are all known components of P bodies, should interact with each other. If this is true, then modulating one of the Group A P body proteins should affect the other Group A P body proteins. Additionally, we can infer from our predicted interaction network that Group B proteins should be in two different complexes, one RISC-based and the other P body-based.
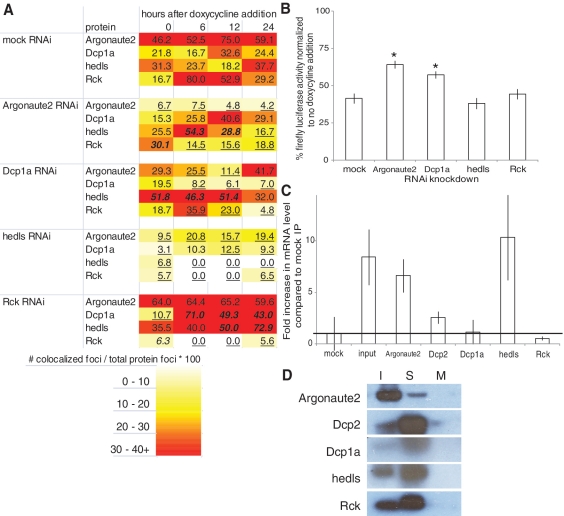
To validate our inferences from the interaction network (Figure 4) for Group A proteins, we transfected pooled siRNAs corresponding to each of the Group A proteins into the JDS33/48 cell lines, induced miRNA expression, fixed cells at the previously determined timepoints and conducted smFISH-IF co-staining of Group A proteins. If Group A proteins interact with each other in a protein complex, then RNAi knockdown of one Group A protein would have an effect on how other Group A proteins co-localize with the target mRNA. Additionally, if a protein functions in miRNA-regulated mRNA degradation of the target mRNA, then knockdown of that protein should result in less efficient silencing and increased luciferase protein levels. A heat map of Group A protein interactions with the target mRNA after RNAi knockdown shows that knockdown of any of the Group A proteins modulates the co-localization of other Group A proteins with miRNA-regulated mRNA (Figure 5A). This data supports the idea that Argonaute2, Dcp1a, hedls and Rck interact with each other when co-localizing with miRNA-regulated mRNA.
Figure 5.
RNAi and RNA immunoprecipitation studies of Group A proteins. (A) Heat map of Group A protein interaction with miRNA-regulated mRNA after siRNA transfection. Number of co-localized foci is divided by number of total protein foci to normalize for variable protein levels and get percentage of protein interacting with the target mRNA. Each data point is an aggregate of five images with ∼20 cells total. A statistical analysis was conducted comparing differences between population proportions of the mock RNAi control to each RNAi knockdown. Bold/italicized numbers indicate results significantly greater than the mock RNAi control and underlined numbers indicate results significantly less than the mock RNAi control (P < 0.05). (B) RNAi silencing of luciferase reporter activity is inhibited when Argonaute2 and Dcp1a are targeted by siRNAs when compared to a mock knockdown. N = 3 replicates per data point. *P < 0.001 compared to mock IP, Student’s t-test. (C) Quantitative RT–PCR of targeted luciferase mRNA immunoprecipitated with antibodies to the indicted proteins. Cell extracts were obtained after cells were induced to express miRNA for 24 h. Input is RNA extracted directly from cell extracts. The mRNA levels were normalized to mock immunoprecipitation (mock = 1). The line is the mRNA level of the mock immunoprecipitation. N = 2 replicates per data point. Error bars = standard deviation. (D) Western blot of the RNA IP shows that the protein is immunoprecipitated by the antibody. The sample (S) for each RNA IP was run in between the input (I) and a mock IP control (M). Antibodies used are noted to the left of the images.
Hedls knockdown reduces all of the other proteins’ association with miRNA-regulated mRNA suggesting that it could be required for protein interactions with the target mRNA. However, hedls does not have any known RNA binding domains, so it is likely that it acts as a scaffold protein to facilitate protein interaction and other proteins with RNA binding domains directly interact with the target mRNA. Interestingly, while hedls knockdown decreases the co-localization of other proteins with target mRNA, Rck knockdown increases co-localization of Dcp1a and hedls with target mRNA, indicating that hedls and Rck could be antagonistic regulators of P body association with miRNA-regulated mRNA.
Protein co-localization with miRNA-regulated mRNA could signify a direct role for the protein in miRNA-based translational repression. We therefore conducted RNAi knockdowns of each Group A protein and asked if reducing expression levels affects miRNA silencing of luciferase reporter activity (Figure 5B). Argonaute2 is known to be functionally involved in miRNA-based mRNA degradation (26), so its knockdown serves as a control. We found that knockdown of Argonaute2 and Dcp1a increased reporter activity, indicating that they do have an impact on RNAi efficacy. Meanwhile, knockdown of hedls and Rck did not affect reporter activity, demonstrating that although they co-localize with miRNA-targeted mRNA, they are not actively involved in degradation of miRNA-regulated mRNA.
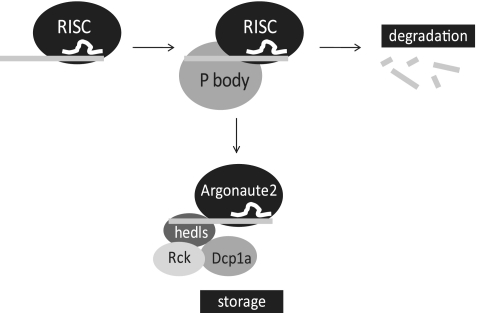
Although hedls and Rck are not actively involved in degradation, they could be important for storage of undegraded miRNA-regulated mRNA. The observation that Group A proteins co-localize with the miRNA-regulated mRNA at the 24-h time point makes them candidates for proteins involved in storage of miRNA-regulated mRNA. Alternatively, some Group A proteins may have no role in storage but are co-localized as a consequence of interactions with proteins that do have a storage role, much like hedls and Rck are physically associated with proteins that are involved in active RNAi. To determine which Group A proteins interact with miRNA-regulated mRNA after RNAi-based mRNA degradation, we conducted an RNA immunoprecipitation (RIP) followed by quantitative RT–PCR of JDS33/48 cells induced to express miRNA for 48 h (Figure 5C and D). If any of the proteins interact with miRNA-regulated mRNA at this time, RIP would result in enrichment for the target mRNA. We found that Argonaute2 and hedls bound the target mRNA compared to a mock RIP, indicating they interact with miRNA-regulated mRNA. In contrast, Dcp1a and Rck mRNA levels were indistinguishable from the mock RIP, indicating that although these proteins co-localize with miRNA-regulated mRNA, they do not interact with the mRNA. Although Dcp1a and Rck do not interact with the target mRNA, they do co-localize with it (Figure 3), and Dcp1a, Rck and hedls are known to interact with Dcp2 as part of a large complex (Figure 4) (27). However, Dcp2 does not co-localize with the target mRNA (Figure 3), and RIP with Dcp2 antibody resulted in low target mRNA levels despite high Dcp2 protein levels detected on the Western blot (Figure 5C and D). Taken together, this signifies that Dcp1a and Rck could be co-localizing with the target mRNA due to their interactions with hedls or other unidentified protein(s). The observation that Dcp2 is not involved in this interaction implies that while normally Dcp1a, Rck and hedls do recruit Dcp2 for decapping, when Dcp2 is not present then they may have a role in storage of miRNA-regulated mRNA (Figure 6).
Figure 6.
Model for protein interaction with miRNA-regulated mRNA. After miRNA (white) is guided by RISC to its target mRNA sequence (light gray), P body proteins are recruited through protein–protein interactions, probably via Argonaute2. While the majority of mRNAs are degraded within the first 12 h after miRNA induction, mRNAs that are stored are bound in a RNA–protein complex that includes Argonaute2, hedls, Rck and Dcp1a. Argonaute2 and hedls interact with the mRNA while Rck and Dcp1a interact with hedls.
In summary, we have used an interaction network in combination with temporal co-localization to identify P body proteins with functional relevance to RNAi and miRNA-regulated mRNA storage. The interaction network can be used to identify other proteins which could be tested for interactions with miRNA-regulated mRNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Health (grant number 1F32GM089102-01 to J.D.S., grant numbers GM057476 and GM36373 to P.A.S. and grant number AR51472 to N.L.K.). Funding for open access charge: GM057476 and GM36373.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Nikon Imaging Center at Harvard Medical School for use of their facilities, Jens Lykke-Anderson for Dcp1a antiserum, Jodene Moore and the HMS Systems Biology FACS facility for cell sorting, Qingqing Wang for technical assistance and Dave Savage, Mara Innis, Edwin Wintermute, Patrick Boyle and Tom Armel for their helpful comments.
REFERENCES
- 1.Pasquinelli AE, Ruvkin G. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell. Dev. Biol. 2002;18:495–512. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 2.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H2 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Alder H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphoncytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eis PA, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JD. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoecklin G, Mayo T, Anderson P. ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep. 2006;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell. Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulalio A, Behm-Ansmant I, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson CP, Bordeleau M-E, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 11.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc. Natl Acad. Sci. USA. 2008;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smolke C. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 2009;27:1099–1102. doi: 10.1038/nbt1209-1099. [DOI] [PubMed] [Google Scholar]
- 17.Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj A, van den Bogaard P, Rifkin SA, van Oudenaaren A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–841. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Rivas FV, Wohlschlegel J, Yates JR, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell. Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell. 2008;19:4469–4479. doi: 10.1091/mbc.E08-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 23.Wilhelm JE, Buszczak M, Sayles S. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell. 2005;9:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V. Similar modes of interaction enable trailer hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Biol. Cell. 2008;28:6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.