Abstract
Using an enhancer-associated epigenetic signature, we made genome-wide predictions of transcriptional enhancers in human B and T lymphocytes and embryonic stem cells (ES cells). We validated and characterized the predicted enhancers using four types of information, including overlap with other genomic marks for enhancers; association with cell-type-specific genes; enrichment of cell-type-specific transcription factor binding sites; and genetic polymorphisms in predicted enhancers. We find that enhancers from ES cells, but not B or T cells, are significantly enriched for DNA sequences derived from transposable elements. This may be due to the generally relaxed repressive epigenetic state and increased activity of transposable elements in ES cells. We demonstrate that the wealth of new enhancer sequences discerned here provides an invaluable resource for the functional annotation of gene-distal single nucleotide polymorphisms identified through expression quantitative trait loci and genome-wide association studies analyses. Notably, we find GWAS SNPs associated with various cancers are enriched in ES cell enhancers. In comparison, GWAS SNPs associated with diseases due to immune dysregulation are enriched in B and T cell enhancers.
INTRODUCTION
One of the most prominent features displayed by transcriptional enhancers, compared to that of promoters and insulator elements, is their cell-type-specific activities. These cell-type-specific regulatory interactions play an essential role in establishing cell type and developmental stage specific gene expression patterns in higher eukaryotes.
Several recent genome-wide expression quantitative trait loci (eQTLs) studies in humans have provided us a first glimpse of regulatory variations in the human population (1–5). Strikingly, about 70–80% of regulatory variants operate in a cell-type-specific manner and are found at larger distances from protein-coding genes, suggesting that a large proportion of these variants could be located in distal enhancers.
In terms of human diseases, a large body of previous studies has uncovered many causal and risk-conferring mutations located in transcriptional enhancers. Examples include thalassemia (6,7), preaxial polydactyly (8,9), Hirschsprung's disease (10,11), cleft clip (12) and prostate cancer (13), among others. At a genome scale, Visel et al. (14) recently performed a meta-analysis of 1200 single nucleotide polymorphisms (SNPs) identified as the most significantly trait- and/or disease-associated variants in a compendium of genome-wide association studies (GWAS) published up to March 2009 (15). Using conservative parameters that tend to overestimate the size of linkage disequilibrium blocks, they found that in 40% of cases (472 of 1170) no known exons overlap, either the linked SNP or its associated haplotype block, suggesting that in more than one-third of cases non-coding sequence variation causally contributes to the traits under investigation. The major classes of non-coding sequences include enhancers, proximal promoters, insulators and non-coding RNAs. Among these, enhancers comprise a large fraction. Therefore, it is likely that many yet-to-be-discovered causal genetic variations reside in enhancers.
Taken together, recent genome-wide mapping of regulatory variants in both healthy and diseased cells has demonstrated the abundance of enhancer sequence variation and its impact on gene expression and disease etiology. Therefore, a comprehensive set of enhancers may facilitate the identification of many causal non-coding variants. To this end, integrating genome-wide enhancer catalogs with GWAS data becomes an effective strategy for linking enhancer mutations with diseases. Likewise, integrating enhancer catalogs with eQTL data will enable us to establish regulatory relationships between enhancers and their target promoters at the systems level.
Transcription enhancers are notoriously difficult to map, which hinders studies of their biology and links to diseases. In the past, reporter gene assays, comparative genomics and transcription factor (TF) ChIP-Chip/Seq have been used to experimentally map enhancers. Computational algorithms based on DNA sequence analysis have also been developed to predict enhancers. However, significant challenges remain for the aforementioned approaches, including low through-put, lack of tissue/specific information, high cost and low accuracy. Recently, a number of studies (16–21) have demonstrated that unique chromatin modification patterns associated with enhancer elements can serve as an effective and accurate mark for cell-type-specific enhancers. Compared with previous approaches, this chromatin-signature-based approach is better suited for finding cell- and developmental-stage-specific enhancers since the activity of enhancers is often modulated by chromatin structure in a condition-specific manner.
Towards the goal of a systems-level understanding of cell-type-specific enhancers, we have used cell-type-specific histone modification maps to generate a genome-wide atlas of transcriptional enhancers in three human cell types: B and T lymphocytes and embryonic stem cells (ES cells). We corroborated the set of predicted enhancers using several complementary lines of evidence, including overlap with other genomic marks for enhancers; location bias of enhancers to cell-type-specific genes; enrichment of cell-type-specific TF binding sites (TFBSs). Our integrative analyses generated a wealth of high-confidence novel enhancers for each cell type. Most importantly, we used our set of predictions to gain insights into enhancer evolution and disease link. We first examined the connections between enhancers and mobile DNA elements (MEs). We also mapped a compendium of eQTL and GWAS SNPs onto our predicted enhancers. Our analyses led to a number of hypotheses suggesting a role of predicted enhancers in disease etiology. Further, comparative analyses of enhancers from different cells revealed unique characteristics of ES cell enhancers in terms of their evolutionary history and disease association.
MATERIALS AND METHODS
Data sources
Histone modification ChIP-Seq data
Three histone modifications, H3K4me1, H3K4me3 and H3K27ac, were used. Data sources are as following: Wang et al. (2008) for T cells (17), the ENCODE Consortium for B cells (22) and Hawkins et al. (2010) for ES cells (23).
Training set of enhancers
To create a high-confidence training set of enhancers, we selected distal p300 binding peaks (2.5 K bp away from known RefSeq TSS) mapped using ChIP-Seq in (24,25), and the ENCODE Consortium, respectively. We only used p300 peaks shorter than 2 K bp to increase the precision of identifying the center of p300 sites. From this set of distal and narrow p300 peaks, we chose those that overlapped with computationally predicted enhancers from the PreMod database (26). The resultant training sets contain 394 enhancers for T cells, 717 enhancers for B cells and 580 enhancers for ES cells.
Gene expression data
A compendium of microarray expression profiles of 20 human cell types (including T, B and ES cells) was compiled from Su et al. (27) and Mayshar et al. (28). Raw microarray data were quantile-normalized using the GC-RMA algorithm (29).
eQTL and GWAS SNP data
T cell eQTL SNPs were obtained from (1). B cell eQTL SNPs were compiled from (1–5). GWAS SNPs were obtained from (15).
Compendium of TF motifs
TF motifs were obtained from the JASPAR (30), TRANSFAC (31) and Uniprobe (32) databases. Redundant motifs were removed by manual inspection. See Supplementary Data for the description of additional data sources.
Computational framework for predicting enhancers based on their chromatin modification patterns
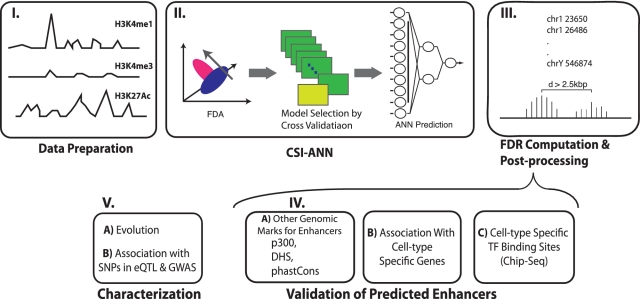
We recently developed a computational method, termed Chromatin Signature Identification by Artificial Neural Network (CSI-ANN), for identifying functional DNA elements using their chromatin signatures (18). The framework consists of a data transformation and a feature extraction step using Fisher Discriminant Analysis, followed by a classification step using artificial neural network (Figure 1). Through a series of benchmarking analyses, we showed that CSI-ANN achieves a significant performance gain over previous best method by Won et al. (33). In this article, we introduced the following improvements to the original CSI-ANN algorithm: (i) use of cross-validation during ANN model training to choose a best-trained model with a minimum mean squared error; (ii) calculation of empirical false discovery rate (FDR) for the predicted enhancers. Details of the improvement are described in the following sections.
Figure 1.
Overview of the study, which includes three major steps: prediction, validation and characterization. FDA, Fisher discriminant analysis.
ANN training and model selection
We generated a training data set for each cell type. Each training set consisted of known enhancers (data source above) and 10 times more background sequences (randomly selected genomic loci). Histone modification signals in a 2 K bp window centered on enhancer or background sequence centers were used as the input for the algorithm.
We implemented a 16-fold cross-validation procedure to select a best-trained ANN model for classification. The training data were partitioned into 16 subsets of equal size. At each iteration, 15 subsets of data were used for training the ANN by particle swarm optimization algorithm (Supplementary Data) and the 16th subset for testing. A best-trained model was selected based on the minimum mean squared error (MSE) criterion:
False discovery rate calculation
To access the statistical significance of predicted enhancers, we calculated an empirical FDR based on randomization. Briefly, for a given histone modification, ChIP-Seq tag counts within a 200 bp window were randomly re-distributed across the genome. Enhancer predictions were made using the randomized data set. FDR was calculated as the ratio of number of predictions in the randomized data relative to that of real data. For all predictions reported in this article, we used a FDR cutoff of 0.05.
Transcription factor binding site analysis
Using the program Patser (34), each predicted enhancer sequence was scanned using the compendium of 446 non-redundant TF binding motifs described above. Sites scored greater than 70% of the consensus motif score (i.e. maximal score given the motif) were regarded as binding sites of a given TF. See Supplementary Data for the description of additional methods.
RESULTS
Genome-wide prediction of transcriptional enhancers in human T, B and ES cells
A number of studies have demonstrated that high levels of H3K4me1 and H3K27Ac modifications in combination with low level of H3K4me3 modification serve as a robust epigenetic signature for enhancers (16–21). Previously, we developed a computational tool, CSI-ANN, to predict transcriptional enhancers based on their histone modification signature (18). Here, we applied our tool to genome-wide maps of the above three histone modifications obtained from human T, B and ES cells (Figure 1). At a FDR of 0.05, CSI-ANN predicted 20 214, 21 832 and 31 273 enhancers in B, T and ES cells, respectively. Overall, we predicted ∼50% more enhancers in ES cells than the differentiated cells. Supplementary Figure S1 presents the number of shared and unique enhancers among the three sets of predictions. For all three cell types, at least 50% of the predictions are cell-type-specific. On the other hand, only about 1500 predictions are shared by all three cell types. On average, 35.3%, 59.6% and 5.5% of the predicted enhancers are located in intergenic, intronic and exonic regions (Supplementary Table S1), consistent with the general observation that enhancers mostly reside in non-coding regions.
Validation of predicted enhancers
We conducted a series of computational analyses using three types of genome-scale experimental data (except for sequence conservation), including additional genomic marks for transcriptional enhancers; genes specifically expressed in the three cell types studied; and genome-wide location data of TFs known to function specifically in the three cell types studied. In the following sections, we present supporting evidence to our predictions using these external data sources.
Predicted enhancers significantly overlap with other genomic marks for enhancers
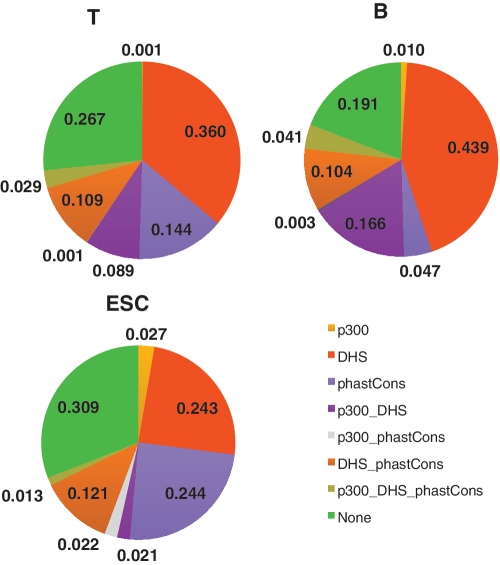
To corroborate our predictions, we first used genome-scale data of three enhancer marks: p300 binding sites, DNase I Hypersensitivity Sites (DHS) and PhastCons scores. P300 is a transcriptional co-activator that is often associated with enhancers. DHSs are nucleosome-free regions that are often associated with functional DNA elements, including enhancers. PhastCons scores quantify the conservation level of a DNA sequence using multiple sequence alignment of 17 vertebrate genomes (35). We only considered distal p300 and DHS sites [>2.5 kb from closest transcription start site (TSS)] to avoid confusion with promoters. Using distal p300 and DHS sites as true enhancer marks, the average sensitivity (fraction of known distal p300/DHS sites recovered) of enhancer prediction in B, T and ES cells are 49.3%, 45.5% and 48.5%, respectively (Supplementary Table S2). The corresponding positive predictive values (PPV, fraction of predicted enhancers supported by either distal p300 or DHS sites) are 81.3%, 79.2% and 68.8%, respectively (Supplementary Table S2). Compared to our previous analysis of HeLa cell enhancers in the ENCODE region (18), our current predictions have ∼10% lower sensitivity but ∼10% higher PPV. This is likely due to the stringent FDR we used in this analysis (5%). In addition to p300 and DHS marks, using pre-computed PhastCons scores, we found that 40.0%, 28.3% and 19.4% of our predictions are conserved in B, T and ES cells, respectively. Considering all three lines of evidence and averaging overall three cell types, 70–80% of our predictions are supported by at least one line of evidence.
Figure 2 shows the fraction of predicted enhancers supported by different combinations of enhancer marks described above. Focusing on the two most comprehensive marks, DHS site and PhastCons conservation score, enhancers from the lymphocytes were more supported by DHSs (75% and 58.7% for B and T cells, respectively) than those from ES cells (39.8%). On the other hand, ES cell enhancers were more conserved (39.8%) compared to enhancers in the lymphocytes (19.5% and 28.3% for B and T cells, respectively).
Figure 2.
Fraction of predicted enhancers supported by different combinations of genomic features. Three types of genomic markers for enhancers are used: distal p300 sites, distal DHS sites, and conserved sequences identified by the phastCons algorithm. PhastCons, conservation score based on genome comparison of 17 vertebrates.
Predicted enhancers are preferentially located near cell-type-specific genes
A hallmark of enhancers is their cell-type-specific activity. In other words, if the predicted cell-type-specific enhancers are real, then we expect that they regulate genes having cell-type-specific expression pattern. To identify genes specifically expressed in a given cell type, we used a compendium of microarray expression profiles of 20 human cell types, including B, T and ES cells. We calculated an expression specificity score for each gene using an entropy-based measure that quantifies the skewness of expression level toward a given cell type (Supplementary Methods). Using this measure, we ranked genes by their expression specificity to a given cell type and identified the top 500 genes that are the most and least specifically expressed. As a baseline comparison, we randomly selected 500 genes. We repeated the random selection ×100 and reported the average number for the analysis with random gene set. We then examined whether predicted enhancers are enriched within domains of cell-type-specific genes (‘Methods’ section). Domains are defined as the 20 K bp region centered on the TSS of a gene. All three sets of predictions are significantly enriched near genes specifically expressed in their corresponding cell type, but not near either random genes or non-specifically expressed genes (Supplementary Figure S2). We also used an alternative definition of expression domain for the enrichment analysis, i.e. a genomic region enclosed by two adjacent CTCF binding sites and contains the TSS of gene under study. We obtained the same result as with the 20 K bp expression domain definition (Supplementary Figure S3).
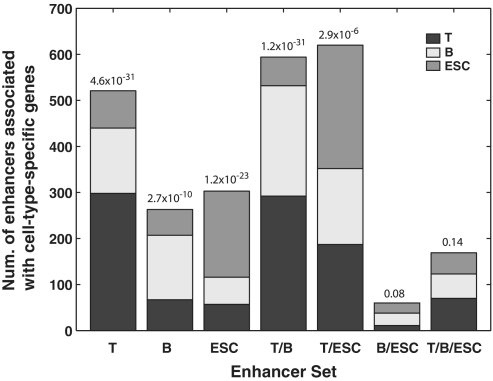
The above test examined the entire set of predicted enhancers in a given cell type. Next, we did a more stringent test to determine whether more cell-type-specific predictions are near cell-type-specific genes compared to shared predictions. It is indeed the case for all subsets of predicted enhancers (Figure 3). For instance, when examining T cell-specific enhancers, we found a significantly larger number of them were near the top 500 T cell-specific genes (298) while a smaller number of them are associated with B (142) or ES cells (81), respectively (Pearson's χ2-test P = 4.6 × 10−31). The same trend was observed for enhancers unique to B and ES cells. We also examined enhancers shared by all three cell types. As expected, we did not observe any specific association between shared enhancers and corresponding cell-type-specific genes (Pearson's χ2-test P > 0.05).
Figure 3.
Enrichment of cell-type-specific enhancers near cell-type-specific genes. Each column represents a subset of enhancers, e.g. column T represents predicted enhancers unique to T cell and column T/B/ESC represents predicted enhancers shared by all three cells. y-axis, number of enhancers in the subset that are near T-cell, B-cell and ES-cell specific genes. P-values are for Pearson's χ2-tests for equal distribution of a subset of enhancers among three sets of cell-type-specific genes.
Predicted enhancers are enriched for binding sites of cell-type-specific TFs
Cell-type-specific enhancers are bound by cell-type-specific TFs. Therefore, we predicted that binding sites of cell-type-specific TFs are enriched in our predicted enhancers, particularly in those predictions unique to each cell type. To this end, we identified a set of TFs known to play a specific role in a given cell type based on expert knowledge (36–38). We obtained ChIP-Seq data for these TFs measured in the specific cell type. We calculated the overlap between predicted cell-type-specific enhancers and binding sites of cell-type-specific TFs. Cell-type-specific enhancers are highly enriched for binding sites of cell-type-specific TFs (Figure 4). For instance, B cell-specific enhancers are enriched for binding sites of Batf, Bcl11a, Ebf, Irf4, Pax5 and Pu.1, all of which are known to be specific regulators in B cells. The same type of enrichment was also observed for T and ES cell enhancers. As a control, we examined the overlap of the predicted enhancers with two non-specific and irrelevant factors, CTCF and NRSF. Several recent genome-wide ChIP-Seq studies have shown that CTCF binding sites are largely cell type invariant (16,39). Consistent with this observation, we did not observe CTCF binding site enrichment in any subset of enhancers. NRSF is a repressor of neuronal genes in non-neuronal cell types, such as stem cells and T and B cells studied here (40). Consistent with its repressor role, we did not observe an enrichment of NRSF sites in the predicted enhancers.
Figure 4.
Enrichment of cell-type-specific TFBSs in cell-type-specific enhancers. x-axis, each column represents a set of cell-type-specific TF ChIP-Seq binding peaks. y-axis, fraction of overlap between the set of cell-type-specific TF binding peaks and T-cell, B-cell and ES-cell specific enhancers. Overlap is calculated as the ratio of the intersection of the two sets to the union of the two sets.
Taken together, the multiple complementary lines of evidence presented above suggest that the majority of our predictions could be bona fide enhancers. For genomic locations of the predicted enhancers and their supporting evidence, Supplementary Tables S3–S5.
Evolution and sequence polymorphism of T, B and ES cell enhancers
Next, to gain insights into enhancer evolution and disease link, we characterized and compared the three sets of predicted enhancers from two aspects: association with MEs and impact of enhancer sequence polymorphism on gene expression and disease etiology.
ES cell but not lymphocyte enhancers are enriched for exapted sequences derived from transposable elements
Over the years, researchers had come across numerous mobile elements that acquired a cellular role, a process termed ‘exaptation’ (41). Thanks to technology development such as ChIP-ChIP/Seq, there is a growing number of reported cases in which highly conserved MEs act as transcriptional enhancers (25–28). To gain a global perspective on this phenomenon, we conducted a genome-wide search for transcriptional enhancers derived from MEs. To this end, we used a set of 10 402 highly conserved non-coding elements of clear ME origins in the human genome identified by Lowe et al.(42). These elements have been under strong purifying selection since at least the boreoeutherian ancestor (100 Mya) (42). These sequences are at least 50 bp long and cover just over 1Mb (0.04%) of the human genome. All four characterized classes of MEs are present in this set, including long interspersed elements (LINEs), interspersed elements (SINEs), DNA transposons and long terminal repeat retro-transposons (LTRs), with LINEs and SINEs contributing the bulk of the constrained non-coding sequence.
We intersected our predicted enhancers with the set of exapted sequences. Respectively, 137, 168 and 406 predicted T, B and ES cell enhancers were found to contain at least one exapted sequence. For all three cell types, the top three ME families involved are MIR (mammalian interspersed repeat), CR1 (chicken repeat 1) and L2 (LINE2) (Supplementary Table S6). Notably, these three families are more ancient compared to younger families such as Alu and L1. This may reflect the stringent conservation criterion for identifying the exapted MEs by Lowe et al. Therefore, the actual number of ME-derived enhancers may be higher if less stringent criterion for exapted sequences is used.
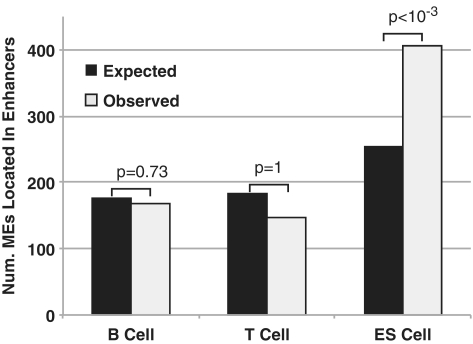
We found two arresting features about the set of enhancers containing exapted MEs. First, ES cell enhancers are significantly enriched for exapted MEs (P < 10−3) but not T or B cell enhancers (P = 1 and 0.73, respectively) (Figure 5). Further, ES cell enhancers containing exapted MEs are significantly enriched near developmental genes, but not T or B cell enhancers containing exapted MEs (Supplementary Table S7). Second, we found that for those enhancers that contain exapted MEs, the majority of them are cell-type specific (Supplementary Table S8, Pearson's χ2-test P-values are 6.4 × 10−5, 9.6 × 10−3 and 9.1 × 10−33 for B, T and ES cell enhancers, respectively). Taken together, our results suggest the hypothesis that compared to differentiated cells, a larger proportion of ES cell-specific enhancers are derived from MEs.
Figure 5.
Enhancers in ES cells, but not T and B cells, are enriched for ME derived sequences. y-axis, number of MEs found in predicted enhancers. Enrichment P-values are computed based on 1000 sets of randomly chosen sequences that match the number and length distributions of the set of real exapted sequences (Supplementary Data).
Genetic variations in predicted enhancers and their impact on gene expression
eQTLs are genomic regions that have an impact on the expression levels of nearby or distant genes, either through -cis or -trans action (43). Therefore, eQTLs that overlap with putative enhancers provide functional evidence for the enhancers. To this end, we compiled a set of eQTL SNPs (termed eSNPs here for brevity) for both T and B cells from multiple eQTL studies(1–5). This set contains 76 518 and 7235 SNPs for B and T cell, respectively. We intersected the eSNPs with our predicted enhancers. Overall, 3.64% (2786) and 1.52% (110) of eSNPs mapped to B and T cell enhancers, respectively (Table 1, Supplementary Tables S9–S10). Compared with the entire set of SNPs in the dbSNP database (∼29 million), the sets of eSNPs significantly overlap with our predicted enhancers (One-tailed proportion test P-values are 4.1 × 10−239 and 3.4 × 10−5 for B and T cell eSNPs, respectively), providing additional supporting evidence to our predictions.
Table 1.
eQTL and GWAS SNPs located in predicted enhancers
| No. of Enhancers | No. of SNPs | Median distance to closest TSS (bp) | No. of enhancers overlap TFBS | No. of SNP overlap TFBS | Median distance to closest TSS (bp) | No. of TFs involved | |
|---|---|---|---|---|---|---|---|
| eQTL SNPs (B:76 518, T:7235) | |||||||
| B | 1987 | 2786 | 16 609 | 751 | 968 | 15 928 | 193 |
| T | 110 | 110 | 5673 | 77 | 79 | 3732 | 77 |
| GWAS SNPs (3409) | |||||||
| B | 128 | 127 | 30 654 | 84 | 83 | 30 654 | 89 |
| T | 114 | 112 | 10 820 | 75 | 76 | 8640 | 95 |
| ESC | 138 | 138 | 14 907 | 84 | 84 | 20 590 | 94 |
Total number of eQTL/GWAS SNPs used in this study is shown in parenthesis.
If an eSNP is embedded in an enhancer then it is likely part of a transcription factor binding site (TFBS). The presence of overlapping TFBSs and eSNPs in a predicted enhancer not only provides strong supporting evidence for the predicted enhancer it also suggests which TFs bind the enhancer. To check the overlap between eSNPs and TFBSs, we compiled a set of 446 non-redundant TF motifs from three major motif databases, JASPAR (44), TRANSFAC (31) and Uniprobe (32). We scanned each enhancer to identify TFBS(s) that overlaps with the embedded eSNPs. Overall, 34.7% (968) and 71.8% (79) of the enhancer-associated eSNPs overlap with a TFBS in B and T cells, respectively. The corresponding TFBSs belong to 193 and 77 TFs in B and T cells, respectively (Table 1, Supplementary Tables S9 and S10).
Genetic variations in predicted enhancers and their links to disease etiology
To date, a genome-wide survey of enhancer mutations in human diseases has not been conducted. Our catalog of predicted enhancers makes it possible to systematically predict enhancers involved in human diseases. To do so, we took advantage of a comprehensive catalog of disease/trait-associated SNPs curated from over 707 GWAS studies (15). For brevity and in contrast with eQTL-associated SNPs, we term these SNPs gSNPs. In total, the current version of the catalog contains 3409 non-redundant gSNPs implicated in 424 diseases/traits. Similar to the eSNP analysis described above, we mapped this collection of gSNPs onto our full set of predicted enhancers. In total, 3.72% (127), 3.29% (112) and 4.05% (138) of the gSNPs were mapped to B, T and ES cell enhancers, respectively (Table 1, Supplementary Tables S11–S13). Compared to the entire set of SNPs in the dbSNP database, the sets of gSNPs significantly overlap with our predicted enhancers (one-tailed proportion test P-values are 2.5 × 10−14 and 3.0 × 10−8, and 2.2 × 10−7 for B, T and ES cell enhancers, respectively).
To gain a mechanistic understanding of the gSNP-associated enhancers in disease etiology, like the eSNP analysis above, we identified TFBSs overlapping the gSNPs that are embedded in predicted enhancers. Overlap with TFBSs suggests that these gSNPs could modulate the binding of important TFs responsible for the disease etiology. Among the three sets of gSNPs, 65.4% (B cell), 67.9% (T cell) and 60.9% (ES cell) overlap with a TFBS (Table 1, Supplementary Table S11–S13). Supplementary Table S14 shows the top 10 gSNPs for each cell type based on their disease association P-values from GWAS studies. As expected, B and T gSNPs are enriched for diseases due to immune dysregulation or tumor of the lymphoid system, such as rheumatoid arthritis, systemic lupus erythematosus, psoriasis and leukemia. In stark contrast, embryonic stem cell (ESC) gSNPs are enriched for various types of cancers, including breast cancer, testicular germ cell cancer, pancreatic cancer, colorectal cancer and nasopharyngeal carcinoma. The same enrichment trend was also observed in the full set of gSNPs. Overall, 32, 31 and 34 different diseases were found to have a link to B, T and ES cell enhancers, respectively. Among these, 6, 5 and 12 are cancers, respectively. The proportion of cancers that are associated with SNPs that map to ES cell enhancers (37.5%) is significant larger than those for B (17.2%) and T (16.1%) cell enhancers (one-tailed proportion test P-values are 0.05 and 0.01, respectively).
Recently, Wong et al. (45) used a compendium of gene expression profiles in ES cells and various differentiated cells to identify ES cell-specific gene modules. They then compared the ES cell gene modules to various human cancer cell expression profiles and found that many ES cell gene modules were active in various cancers. This result provided evidence for the hypothesis that ES cells and at least some cancer stem cells share a common genetic program. However, no systematic analysis was conducted to reveal the cis-regulatory elements (e.g. enhancers) for the regulation of the shared gene expression program. Our result here provides a complementary evidence for the above hypothesis by reporting a set of ES cell-specific enhancers that are enriched near cancer-related genes and harbor SNPs that are linked to cancers.
DISCUSSION
In summary, we have identified and computationally validated a large population of enhancers in human B, T and ES cells. Why there are 50% more enhancers in ES cells compared to differentiated T and B cells? The answer may lie in the unique developmental requirement and chromatin characteristics associated with ES cells. ES cells exhibit a global chromatin structure that is more ‘open’ than that found in differentiated cells (46). Therefore, one can speculate that protein–DNA interactions at ES-cell-specific enhancers may be necessary to prevent the enhancers from assembling into repressive chromatin structures during differentiation that may be resistant to activation. In support of this hypothesis, accumulating lines of evidence suggest that enhancers of cell-specific genes interact with pioneer TFs in ES cells and at other early stages of development, long before the genes are transcribed (47). For instance, the liver-specific enhancer for the Abl1 gene is occupied by the FoxD3 TF in ES cells. Other examples include enhancers for the thymocyte-specific Pctra gene and macrophage/dendritic cell-specific Ill2b gene. It has been proposed that these early protein–DNA interactions essentially prepare or poise the enhancers for later activation (20,21,47). In light of the observation that many cell-specific enhancers are occupied in ES cells, it is plausible that more enhancers are occupied in ES cells compared to differentiated cells.
Our results demonstrate that ES cells have a higher fraction of conserved enhancers than differentiated lymphocytes. A likely explanation is that the higher conservation of ES cell enhancers is simply a measure of essentiality of early embryo maintenance and development. In ES cells, there might be a higher number of critical regulators that need precise protein–DNA interactions with their cis-regulatory elements to regulate mission-critical functions, i.e. core self-renewal and pluripotency genes. Such precise regulatory control may be less critical during adult homeostasis (e.g. in B and T cells).
In mammals, retro-transposons are particularly active in germ cells and in early embryos (48). The reason for the increased ME activity is probably due to the relaxation of epigenetic control in these cells. Indeed, genome-wide loss of DNA methylation accompanies the acquisition of pluripotent states in primordial germ cells and pre-implantation embryos, which opens a window of opportunity for MEs to escape from host restraint (49,50). This suggests an adaptive strategy for transcriptional networks associated with germinal and pluripotent states. Previous studies have reported L1 mobilization occur during early embryonic development in humans, as evidenced by the retro-transposition of a transgenic L1 element observed in human ES cells (51). Our study provides additional evidence to support the hypothesis that exapted MEs play a more significant role in shaping the transcriptional network in ES cells than differentiated cells.
Our results demonstrate the power of global comparative and integrative analysis for gaining insights into cell-type-specific enhancers. Traditionally, cell-type-specific activity of enhancers is difficult to determine. One approach is to combine DNA sequence analysis with a compendium of gene expression microarray data, such as what is done in (52). The advent of global and cell-type-specific chromatin modification maps provides another effective means to identify cell-type-specific enhancers at a genome scale. Future computational analyses using additional cell types along with functional validations will significantly advance the rate and scale at which cell-type-specific enhancers are characterized in humans.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Human Genome Research Institute (Grant 1R21HG005948 to K.T.). Funding for open access charge: National Human Genome Research Institute, USA.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Pamela Geyer and Dana Levasseur for their careful reading of the manuscript. We thank the ENCODE consortium for making their data freely available in a timely manner. We acknowledge the use of ChIP-Seq data generated by the following ENCODE consortium members: Broad/Bernstein lab (histone modification data), HudsonAlpah/Myers lab (TF location data) and Duke/Crawford lab (DHS data).
REFERENCES
- 1.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez Arcelus M, Sekowska M, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, Pritchard JK. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG. Beta-globin gene inactivation by DNA translocation in gamma beta-thalassaemia. Nature. 1983;306:662–666. doi: 10.1038/306662a0. [DOI] [PubMed] [Google Scholar]
- 7.Semenza GL, Delgrosso K, Poncz M, Malladi P, Schwartz E, Surrey S. The silent carrier allele: beta thalassemia without a mutation in the beta-globin gene or its immediate flanking regions. Cell. 1984;39:123–128. doi: 10.1016/0092-8674(84)90197-1. [DOI] [PubMed] [Google Scholar]
- 8.Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 9.Furniss D, Lettice LA, Taylor IB, Critchley PS, Giele H, Hill RE, Wilkie AO. A variant in the sonic hedgehog regulatory sequence (ZRS) is associated with triphalangeal thumb and deregulates expression in the developing limb. Hum. Mol. Genet. 2008;17:2417–2423. doi: 10.1093/hmg/ddn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emison ES, McCallion AS, Kashuk CS, Bush RT, Grice E, Lin S, Portnoy ME, Cutler DJ, Green ED, Chakravarti A. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–863. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 11.Grice EA, Rochelle ES, Green ED, Chakravarti A, McCallion AS. Evaluation of the RET regulatory landscape reveals the biological relevance of a HSCR-implicated enhancer. Hum. Mol. Genet. 2005;14:3837–3845. doi: 10.1093/hmg/ddi408. [DOI] [PubMed] [Google Scholar]
- 12.Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat. Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firpi HA, Ucar D, Tan K. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics. 2010;26:1579–1586. doi: 10.1093/bioinformatics/btq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang ZB, Zang CZ, Cui KR, Schones DE, Barski A, Peng WQ, Zhao KJ. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferretti V, Poitras C, Bergeron D, Coulombe B, Robert F, Blanchette M. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–D126. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayshar Y, Ben-David U, Lavon N, Biancotti JC, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 29.Wu Z, Irizarry RA, Gentleman R, Murillo FM, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- 30.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Won KJ, Chepelev I, Ren B, Wang W. Prediction of regulatory elements in mammalian genomes using chromatin signatures. BMC Bioinformatics. 2008;9:547. doi: 10.1186/1471-2105-9-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 35.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 2005;5:497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Vega VB, Ng HH. Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008;73:203–209. doi: 10.1101/sqb.2008.73.026. [DOI] [PubMed] [Google Scholar]
- 39.Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 42.Lowe CB, Bejerano G, Haussler D. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc. Natl Acad. Sci. USA. 2007;104:8005–8010. doi: 10.1073/pnas.0611223104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 44.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 47.Smale ST. Pioneer factors in embryonic stem cells and differentiation. Curr. Opin. Genet. Dev. 2010;20:519–526. doi: 10.1016/j.gde.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamudio N, Bourc'his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity. 2010;105:92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 49.Rougier N, Bourc'his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, Moran JV. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 52.Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.