Abstract
The Type I restriction-modification enzymes comprise three protein subunits; HsdS and HsdM that form a methyltransferase (MTase) and HsdR that associates with the MTase and catalyses Adenosine-5′-triphosphate (ATP)-dependent DNA translocation and cleavage. Here, we examine whether the MTase and HsdR components can ‘turnover’ in vitro, i.e. whether they can catalyse translocation and cleavage events on one DNA molecule, dissociate and then re-bind a second DNA molecule. Translocation termination by both EcoKI and EcoR124I leads to HsdR dissociation from linear DNA but not from circular DNA. Following DNA cleavage, the HsdR subunits appear unable to dissociate even though the DNA is linear, suggesting a tight interaction with the cleaved product. The MTases of EcoKI and EcoAI can dissociate from DNA following either translocation or cleavage and can initiate reactions on new DNA molecules as long as free HsdR molecules are available. In contrast, the MTase of EcoR124I does not turnover and additional cleavage of circular DNA is not observed by inclusion of RecBCD, a helicase–nuclease that degrades the linear DNA product resulting from Type I cleavage. Roles for Type I restriction endonuclease subunit dynamics in restriction alleviation in the cell are discussed.
The Type I restriction-modification (RM) systems function to protect bacterial host cells against parasitic DNA by specifically targeting and degrading the unmodified foreign polynucleotide (1,2). Type I RM enzymes are multifunctional heteroligomers consisting of three different subunits; HsdM (methylation activity), HsdS (specific DNA sequence recognition) and HsdR (ATPase, translocase and endonuclease activity). Two HsdM subunits and one HsdS subunit (M2S1) can function independently as an adenine methyltransferase (MTase) (3,4). To cleave DNA efficiently, two HsdR subunits must also associate with the MTase (R2M2S1) (5,6). Upon encountering fully unmodified recognition sequences, the HsdR subunits begin to translocate DNA in an ATP-dependent manner, while the MTase remains bound at the recognition site. Collision of two translocating complexes promotes DNA cleavage at the random, non-specific site where the two enzymes met (7–9). It has been shown previously that the HsdR subunits of the Type I enzyme EcoR124I are weakly associated with the cognate MTase and that HsdR dissociation from the MTase limits loop translocation (6,10,11). It has been suggested therefore that the dynamic interactions of HsdR and MTase play a key role in the cellular regulation of nuclease activity (5,12). Here, we investigated whether this ‘turnover’ of HsdR subunits is a universal property of Type I enzymes. In addition, we explored the dynamic properties of the DNA–MTase interaction and the influence of another bacterial DNA processing enzyme, RecBCD.
Restriction endonucleases could potentially target any DNA in the cell that carries an unmodified recognition sequence. While the host genome is normally protected due to maintenance methylation by the cognate MTase activity, nucleotlytic DNA damage is still possible under certain circumstances (13): first, if an unmodified sequence should arise due to aberrant DNA repair; or second, when a strain acquires a new RM system where none of the host genome sites are modified. So far, there is no evidence of transcriptional regulation of the genes-encoding Type I RM systems (14). Instead they appear to have evolved restriction alleviation (RA) mechanisms that prevent cleavage of the host DNA, even if it is completely unmodified (15–19). For the Types IA and IB RM systems EcoKI and EcoAI, RA is at least partly dependent on the proteolytic complex ClpXP. When RA is induced, detectable in vivo levels of HsdR decrease dramatically. Mutant HsdRs that are unable to translocate are unaffected by ClpXP whereas restriction deficient mutants are still degraded (20). It, therefore, seems likely that induction of an SOS response due to DNA cleavage is not necessary. A straightforward mechanism is that a ClpXP target motif is revealed when HsdR begins to translocate (and is targeted before DNA cleavage occurs) (1). However, RA is not observed upon bacteriophage infection and is only observed when unmodified sites arise on the host genome, although translocation will occur in both cases. The molecular mechanism by which ClpXP distinguishes the two translocating forms of HsdR is unknown. In contrast to EcoKI and EcoAI, RA by the Type IC system EcoR124I appears to be independent of ClpXP activity (17). Instead the process of HsdR dissociation is thought to prevent cleavage of ‘self’ DNA by limiting the number of translocation events (11).
In contrast to the dynamic binding of HsdR during translocation, the association of the MTase and DNA appear more long-lived (11). Early studies of the DNA cleavage activity of Type I enzymes established a long-held view that there is no turnover during DNA cleavage (21–26). In other words, a stoichiometric concentration of enzyme and DNA are necessary for DNA cleavage. This view was reiterated by later studies that used both reconstituted and holoenzyme preparations (6,27,28). A simple explanation is that the MTase does not release its recognition site following DNA cleavage and thus the complex cannot cleave further DNA molecules. Note that the inability to release the DNA product does not prevent a Type I enzyme from carrying out its in vivo role since a single double strand DNA (dsDNA) break would be sufficient to inactivate a parasitic nucleic acid. However, contrary studies by Bianco and Hurley (29) and Bianco et al. (30) suggested that Type I enzymes could turnover during cleavage, in particular in the presence of RecBCD that digests the linear DNA product and possibly accelerates release of the MTase from the recognition site post-cleavage. These studies, therefore, proposed that the Type I enzymes were ‘true catalytic enzymes’ in that they did have the ability to turnover during DNA cleavage.
To further investigate the subunit dynamics of the Type I RM enzymes, we compared the DNA translocation and cleavage properties of EcoKI (Type IA), EcoAI (Type IB) and EcoR124I (Type IC) using both holoenzyme preparations and in vitro reconstituted enzymes. We found that EcoKI behaves in a similar way to EcoR124I, with dissociation of the HsdR subunit from complexes translocating on linear DNA but not from complexes on circular DNA. In addition, we found that the EcoKI and EcoAI MTases could turnover at DNA cleavage under conditions where HsdR was available to bind the dissociated MTase. In contrast, EcoR124I MTase did not turnover during DNA cleavage under conditions tested and we found that, in our hands, RecBCD had no effect on this result. The role of protein complex stability is discussed with reference to interpretation of in vitro data and relevance to in vivo enzyme control.
MATERIALS AND METHODS
DNA
The expression plasmids for EcoKI are described in the Supplementary Data. pLKS5 (31) was used as the substrate for all cleavage and triplex experiments. To prepare DNA for the biochemical assays, Escherichia coli (E. coli) TOP10 (Invitrogen) or HB101 (Promega) cells were transformed with the required plasmid, grown in M9 minimal medium supplemented with 37 MBq/L [3H-methyl] thymidine (PerkinElmer, MA, USA) and the DNA extracted by density gradient centrifugation in CsCl-ethidium bromide (32). To prepare linear DNA, plasmids were incubated with the appropriate Type II restriction enzyme (New England Biolabs) as instructed by the manufacturer, and the linear DNA purified by phenol/chloroform extraction followed by ethanol precipitation. DNA concentrations were determined from absorbance at 260 nm, assuming an extinction coefficient of 0.02 ml/µg•cm and a DNA molecular weight of 6.6 × 105 Da/kb.
Protein expression and purification
EcoKI HsdR and MTase were purified as separate protein pools as described in the Supplementary Data. The HsdR and MTase from EcoR124I and EcoAI were expressed and purified as described previously (6,33). The HsdR(D298E) mutant of EcoKI endonuclease was supplied by David Dryden (University of Edinburgh) and purified as a holoenzyme (5). RecBCD was supplied by Mark Dillingham (University of Bristol) and was prepared as described (34).
Triplex displacement assays
The triplex displacement assay has been described previously and was carried out using a 22 bp triplex-forming oligonucleotide (5′-dTTCTTTTCTTTCTTCTTTCTTT-3′) (35). Assays were performed with 5 nM triplex DNA (either linear or supercoiled), 4 mM ATP, 100 μM S-adenosyl methionine (AdoMet) and varying amounts of MTase, HsdR and RecBCD as indicated, in TMD buffer (50 mM Tris, pH 8, 10 mM MgCl2, 0.1 mM DTT) at 20°C. The Type I endonucleases were reconstituted immediately before use without further purification. To stabilize the proteins, 0.1% (v/v) Triton X-100 was added to TMD buffer and this was used to dilute the proteins for use in the assay, giving a final concentration of 0.005% (v/v). For the RecBCD assays, the buffer was supplemented with 3 U/ml creatine phosphokinase and 30 mM creatine phosphate. Reactions were allowed to progress for 1 h and stopped with 0.2 volumes of GSMB [15% (w/v) glycerol, 3% (v/v) Sodium dodecyl sulphate (SDS), 250 mM 3-(N-Morpholino) propanesulfonic acid (MOPS) (pH 5.5), 0.1% (w/v) bromophenol blue]. Samples were run on 1% (w/v) agarose gels (20 mM Tris–acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.1 mM Ethylenediaminetetraacetic acid (EDTA), pH 5.5) and analyzed using a Molecular Dynamics Typhoon PhosphorImager and ImageQuant software (GE Healthcare Ltd UK). Triplex displacement time-courses were carried out in an SF61-DX2 stopped flow fluorimeter as described previously (11,36). Reactants were mixed 1:1 to give the final buffer and reaction conditions above.
DNA cleavage assays
Assays were performed with 5 nM 3H-labelled DNA, 4 mM ATP, 100 μM AdoMet and varying amounts of varying amounts of MTase, HsdR and RecBCD as indicated, in TMD buffer at 37°C. The Type I endonucleases were reconstituted immediately before use without further purification. To stabilize the proteins, 0.1% (v/v) Triton X-100 was added to TMD buffer and this was used to dilute the proteins for use in the assay, giving a final concentration of 0.005% (v/v). For the RecBCD assays, the buffer was supplemented with 3 U/ml creatine phosphokinase and 30 mM creatine phosphate. Reactions were allowed to progress for 1 h and stopped with 0.5 volumes of STEB [40 % (w/v) sucrose, 0.1 M Tris–Cl, pH 8, 0.1 M EDTA, 0.1 % (w/v) bromophenol blue]. Samples were analysed by agarose gel electrophoresis and the percentage of 3H-labelled DNA in each band per lane quantified by scintillation counting (32).
RESULTS
Dynamic dissociation–reassociation of HsdR on linear DNA but not circular DNA
Upon termination of DNA loop translocation, there are three possible outcomes (11,37): (i) the entire R2M2S1 endonuclease complex could dissociate from the DNA intact; (ii) the HsdR subunits could dissociate from the MTase–DNA complex; or (iii) the HsdR subunits could release the DNA loop without dissociating from the MTase–DNA complex. For the Type IC R-M system EcoR124I, it has been shown that the HsdR subunits dissociate from the MTase following translocation termination and so turnover in the reaction (11). One conclusion from our observations was that dissociation allows the HsdR to re-bind another MTase–DNA complex and initiate another translocation event. It has also been observed that EcoR124I forms a weakly associated R2M2S1 complex (KD,app = 240 nM) which dissociates into free HsdR and a more tightly associated R1M2S1 complex (KD,app < 1 nM) (6,10). Therefore this mode of translocation termination could be related to the weak association of the EcoR124I HsdR subunits with the MTase core. We wanted to test other Type I R-M systems to determine whether HsdR dissociation could be a general mechanism for translocation termination. Unlike EcoR124I, EcoKI has been shown to form a relatively stable R2M2S1 complex (KD,app < 1 nM) (5). Therefore, one might predict that EcoKI would not demonstrate HsdR turnover in the same way as EcoR124I.
To test the subunit dynamics of Type I complexes in vitro, we compared the translocation and cleavage properties of EcoR124I and EcoKI. Assays were performed with a constant, saturating concentration of MTase relative to DNA binding sites and increasing amounts of HsdR. The endonucleases were reconstituted immediately before use from separate HsdR and MTase preparations without further purification (‘Materials and Methods’ section and Supplementary Data). The reactions were allowed to proceed for 1 h, an incubation time that is significantly beyond the end point of the reactions under saturating conditions (e.g. Figure 1B). If upon translocation termination the HsdR subunits remain bound to the MTase–DNA complex and the MTase–DNA complex does not dissociate, a 2-fold or greater molar excess of HsdR relative to the MTase–DNA concentration would be required for full displacement of the triplex or full cleavage of the DNA substrate. If, however, the HsdR subunits or the R2M2S1 complex can dissociate from the DNA, then turnover would be observed in the triplex assay at substoichiometric concentrations of HsdR. Since the cleavage of a one-site supercoiled DNA requires an R2M2S1 complex (6,27), the extent of DNA cleavage will give a measure of the amount of this complex formed and also whether it can recycle following DNA cleavage.
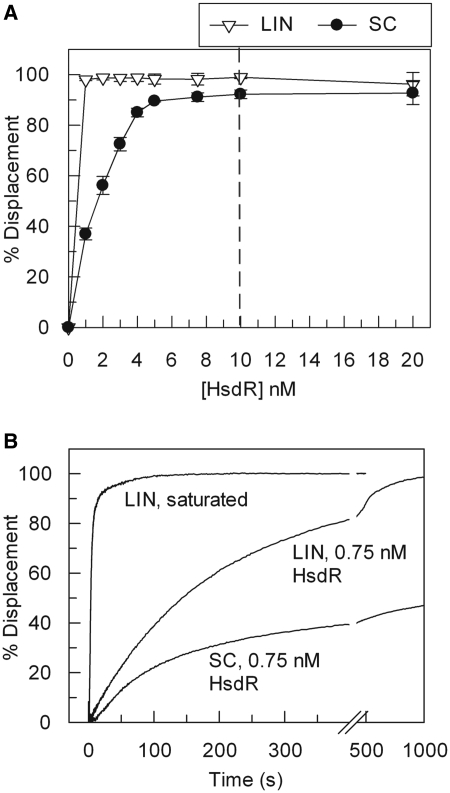
Figure 1.
‘Turnover’ of the HsdR subunit of EcoR124I during translocation. (A) Triplex displacement by EcoR124I on 5 nM one-site linear (LIN) or supercoiled (SC) DNA substrates with 40 nM MTase and varying concentrations of HsdR, as indicated. The dashed line represents where an R2M2S1 complex could form on the DNA (i.e. where the HsdR concentration is twice that of the DNA, and assuming the single recognition site is saturated with MTase). Error bars represent the standard deviations from at least two repeat experiments. (B) Time course of triplex displacement using 1 nM DNA labelled with TAMRA-triplex forming oligonucleotide (linear or supercoiled as indicated), 40 nM MTase and either 80 nM HsdR (saturated) or 0.75 nM HsdR. Enzyme and DNA were pre-incubated and rapidly mixed with ATP. Under these conditions, second order protein–protein assembly rates may affect the kinetics profiles. Hence we did not observe a ‘burst’ of triplex displacement.
We first tested EcoR124I translocation on both linear and supercoiled DNA substrates. As observed previously (11), full displacement of the triplex on linear DNA occurred at sub-stoichiometric concentrations of HsdR relative to the MTase–DNA concentration (Figure 1A). This indicates that the HsdRs are ‘turning over’ in the translocation reaction; i.e. upon translocation termination, HsdR dissociates from the DNA and can then bind another DNA and initiate another displacement event. In contrast, turnover on the supercoiled DNA was less efficient; a linear relationship between displacement and protein concentration was observed, with complete displacement requiring 5 nM HsdR. Because the R2M2S1 complex is weakly associated compared to the R1M2S1 complex (see above), 5 nM HsdR under these conditions could correspond to near complete formation of the R1M2S1 complex with negligible R2-complex (10). The triplex on a circular substrate can be completely displaced by this complex because translocation events can travel either clockwise or anticlockwise around the ring and can both, eventually, reach the triplex binding site. The reduced efficiency of triplex displacement from the circular DNA below 5 nM HsdR suggests that either the HsdR cannot dissociate from the DNA once bound or that the dissociation/turnover is extremely inefficient. Figure 1B shows a time-course for linear DNA at saturating and sub-saturating HsdR and for circular DNA at sub-saturating HsdR. The displacement profile for the linear DNA approaches full displacement at 16 min while the circular DNA saturates at ∼45%, consistent with different dissociation characteristics between the two DNA.
We then tested if EcoKI behaved in the same way as EcoR124I. Because the HsdR binding sites on the EcoKI MTase have high affinities for the cognate HsdR (see above) (5), the endonuclease would be expected to form according to a binomial distribution. So, at 5 nM HsdR and 5 nM MTase–DNA, we would expect ∼25% R2M2S1, ∼50% R1M2S1 and ∼25% M2S1. If HsdR turnover does not occur, we would only expect to observe 50% displacement on a linear substrate because, assuming random association of the HsdR and MTase, translocation by the R1M2S1 complexes would be away from the triplex 50% of the time. On a circular substrate, displacement would be higher at 75%, because the R2M2S1 complex and both forms of the R1M2S1 complex can reach the triplex. The results using linear DNA show that >70% displacement was observed with 1–3 nM HsdR, and maximum displacement was observed above 3 nM HsdR (Figure 2A). On the circular substrate, displacement was less efficient and maximum displacement was only observed with an excess of HsdR. Although EcoKI is less efficient than EcoR124I on both substrates (requiring around 3-fold more HsdR to achieve the same level of displacement), the pattern is the same; on linear DNA HsdR appears to be able to dissociate and recycle to new DNA molecules while on circular DNA such turnover is not observed.
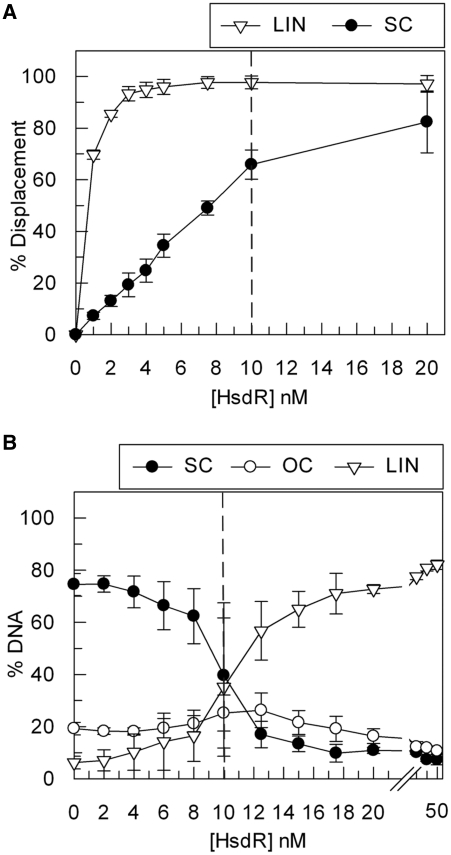
Figure 2.
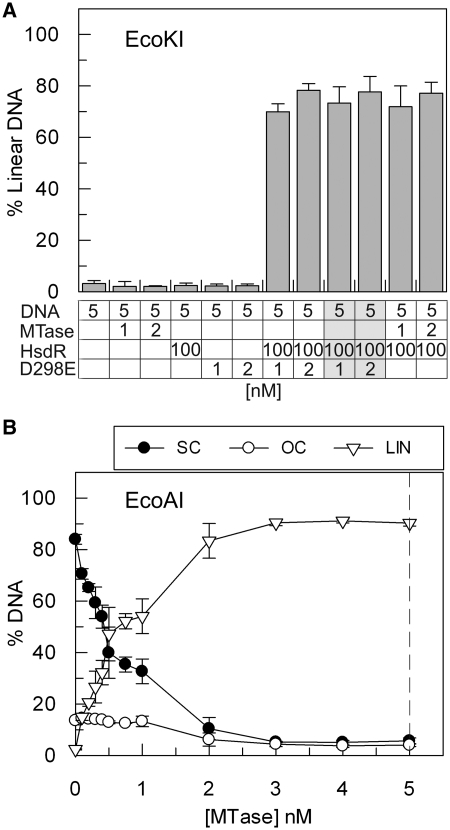
‘Turnover’ of the HsdR subunit of EcoKI during translocation and cleavage. (A) Triplex displacement by EcoKI on 5 nM one-site linear (LIN) or supercoiled (SC) DNA substrates with 5 nM MTase and varying concentrations of HsdR, as indicated. The dashed line represents where an R2M2S1 complex could form on the DNA (Figure 1). (B) Cleavage activity of EcoKI on 5 nM one-site supercoiled DNA substrate with 5 nM MTase and varying concentrations of HsdR, as indicated. OC is open circle, the intermediate of DNA cleavage cut in just one strand. The dashed lines represents where an R2M2S1 complex could form on the DNA. Error bars represent the standard deviations from at least two repeat experiments.
The DNA cleavage properties of EcoKI were also measured. Full cleavage of the circular substrate was only achieved in the presence of an ∼3-fold molar excess of HsdR over DNA sites (Figure 2B). Although turnover from circular DNA does not occur during translocation (Figure 2A), one might expect that the HsdRs could escape via the ends of the linear DNA that result from dsDNA cleavage. The apparent lack of turnover may, therefore, appear counterintuitive. However, even if turnover of the HsdR subunits could occur from the linear DNA products, the formation of sufficient R2M2S1 complexes necessary for further DNA cleavage may be limiting (R1-complexes may be forming instead). Alternatively, the HsdR subunits may become inactivated following DNA cleavage and thus unable to turnover under any circumstance. This latter suggestion is consistent with results presented below and is considered further in the ‘Discussion’ section.
Triplex displacement experiments were also performed with EcoAI, but unfortunately full displacement of the triplex and full-DNA cleavage was only achieved using a 20-fold molar excess of HsdR over DNA (data not shown). The requirement for excess HsdR could be simply due to the specific activity of the preparation of enzyme. Alternatively, the HsdRs could be dissociating so frequently that they are never translocating on the DNA long enough to actually displace the triplex or cleave the DNA. It was therefore not possible to determine whether EcoAI HsdR can turnover in the same way as EcoKI and EcoR124I.
Dissociation–reassociation of EcoKI MTase on linear and circular DNA
It is generally accepted that Type I restriction enzymes do not act ‘catalytically’ in the DNA cleavage reaction—i.e. they do not turnover in the classical sense of an enzyme which can convert multiple substrates before inactivation (21,22,25,26). For example, observations have been made that following DNA cleavage the Type I enzymes remain bound at their recognition sites where they continue to hydrolyze ATP (21–23,38). Consequently, stoichiometric amounts of enzyme relative to DNA are required to achieve full cleavage of the substrate (6,27). More recently, however, it has been suggested that Type I enzymes can turnover and one enzyme can cleave multiple DNAs (29,30). We sought to investigate this process further by examining the possibility of dissociation–reassociation by the core MTase.
We repeated our translocation and cleavage assays using a constant molar excess of HsdR relative to DNA sites and varying concentrations of MTase. We could not undertake this analysis with EcoR124I as a >6-fold molar excess of MTase is required to fully activate cleavage (see below and Figure 5). This may reflect a low specific activity of the complex following purification. In contrast, using EcoKI in the triplex assay, maximal displacement was still observed on both linear and circular DNA at a concentration of EcoKI MTase 5-fold lower than DNA sites, and ∼50% displacement was still observed at a 20-fold lower concentration (Figure 3A). In other words, a single MTase can bind the recognition site, support translocation (and triplex displacement), dissociate and then re-bind another DNA and repeat the cycle. No significant difference between the linear and circular DNA was observed, most likely as the concentration of HsdR was not limiting (see below).
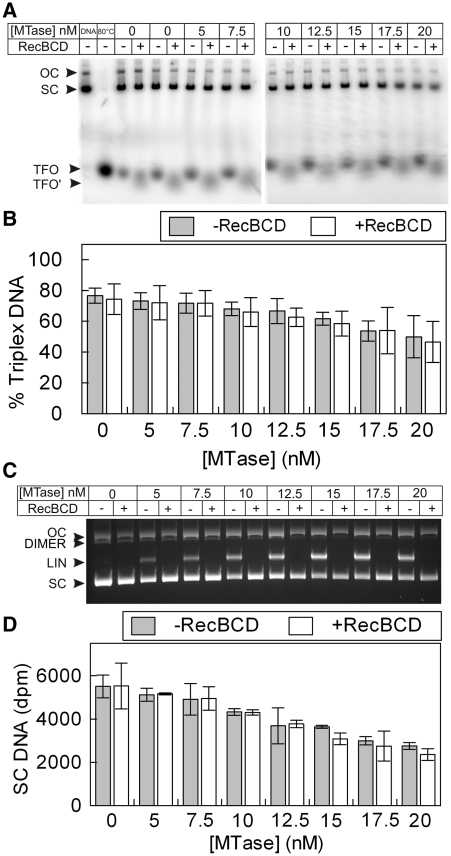
Figure 5.
‘Turnover’ of EcoR124I MTase during DNA cleavage and translocation does not occur and is not influenced by the presence of the exonuclease activity of RecBCD. (A) Triplex Displacement by EcoR124I on 5 nM one-site supercoiled DNA using 80 nM HsdR and varying concentration of MTase, as indicated, in the absence and presence of 20 nM RecBCD. An ATPase recycling system was also included (‘Materials and Methods’ section). TFO, free/displaced triplex forming oligo; TFO′, free TFO band that migrates slightly further into the gel in the presence of RecBCD; SC, supercoiled DNA bound with triplex; OC, open circle bound with triplex. Lane 1 had stop buffer added immediately while lanes 3–20 were incubated for 1 h. The triplex displacement seen in lanes 3–6 in the absence of MTase represents background triplex displacement due to the long incubation time and is independent of HsdR (35). (B) Quantification of triplex displacement presented as the percentage of triplex DNA remaining (the sum of that on SC and OC DNA). (C) Agarose gel showing cleavage activity of EcoR124I and RecBCD on 5 nM one-site supercoiled DNA (SC) under the same reaction conditions as above. DIMER—plasmid dimer which was present as a contaminant of <5% in our DNA preparations. (D) Quantification of the DNA cleavage data presented as the raw dpm counts from the supercoiled DNA in each lane determined by scintillation counting. We could not present the data as percentage cleaved in this case as the linear DNA is digested in the RecBCD lanes and cannot be accurately counted. In B or D, turnover of EcoR124I following RecBCD treatment would be seen as a decrease in the triplex or supercoiled substrates. Error bars represent the standard deviations from at least two repeat experiments.
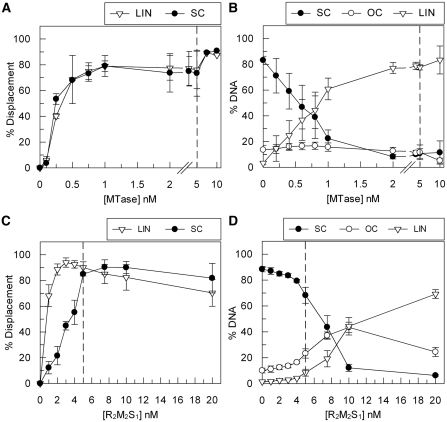
Figure 3.
‘Turnover’ of EcoKI MTase during translocation and cleavage. (A) Triplex displacement by EcoKI on 5 nM one-site linear (LIN) and supercoiled (SC) DNA substrates with an excess of HsdR (100 nM) and varying concentration of MTase, as indicated. (B) Cleavage activity of EcoKI on a 5 nM one-site supercoiled DNA substrate at 100 nM HsdR and varying concentrations of MTase, as indicated. OC is open circle. (C) Triplex Displacement by EcoKI on 5 nM one-site linear and supercoiled DNA substrates with varying concentration of the reconstituted R2M2S1 complex, as indicated. (D) Cleavage activity of EcoKI on 5 nM one-site supercoiled DNA substrate at varying concentrations of the reconstituted R2M2S1 complex, as indicated. In each graph, dashed lines indicate the concentration of MTase/R2M2S1 where one complete endonuclease is available to bind to each DNA site (Figure 1). Error bars represent the standard deviations from at least two repeat experiments.
Using the circular substrate, we also tested the DNA cleavage properties of EcoKI under the same conditions. Surprisingly, the amount of linear DNA product observed could exceed the input concentration of MTase, indicating that MTase turnover following cleavage was also occurring (Figure 3B).
A trivial explanation for these results is that the concentration of our MTase preparation is incorrect. However, our preparations compared well to enzymes from another source (Supplementary Table S1 and Supplementary Figure S1). It is, of course, possible that the concentrations of both preparations are wrong or that the calculated extinction coefficients used in determining the protein concentrations are incorrect. However, they would have to be substantially out by >5-fold. Errors in determining protein and DNA concentrations will mostly arise due to errors in determining theoretical extinction coefficients, and the typical error ranges for both are calculated as ±5% (SD), with maximum errors in the range ±10% (39–41). Even assuming the opposite limits of this range, our results are still consistent with MTase turnover.
In the above experiments, the HsdR is always in molar excess over the MTase and DNA. Since EcoKI can form a stable R2M2S1 complex at low nanomolar concentrations (5), we repeated the triplex displacement and DNA cleavage assays at the same MTase concentrations but using a reconstituted R2M2S1 complex; i.e. under conditions where the HsdR concentration was always twice that of the MTase concentration and where there would be very little free HsdR in solution. In the triplex displacement reactions, turnover of the R2M2S1 complex was observed on linear DNA, while a stoichiometric amount of the complex was required for full displacement on supercoiled DNA (Figure 3C). This result mirrors that of previous experiments varying HsdR (Figures 1 and 2). In the corresponding cleavage reaction on circular DNA, an excess of R2M2S1 complex was required for full cleavage of the supercoiled substrate, consistent with the triplex result (Figure 3D). Therefore, on circular DNA, the EcoKI MTase can only dissociate and reassociate when a molar excess of HsdR was present. This suggests that turnover of the R2M2S1 complex on circular DNA may be limited by the slow HsdR dissociation kinetics (Figures 2 and 3), even when DNA cleavage has produced a linear DNA. In turn, this suggests that the HsdR subunits are somehow inactivated following DNA cleavage. We also note that if the results in Figure 3A and 3B were due to an underestimation of the MTase concentration, this is not reflected in Figure 3D which if anything would suggest an overestimation of the concentration.
Since we have prepared our EcoKI enzymes using different expression clones and protein purification protocols than used previously by other groups (Supplementary Data), the observed MTase turnover may be unique to our enzyme preparations. To address this, we tested an EcoKI preparation that had been purified as a holoenzyme from cells expressing the complete EcoKI operon. This preparation comprises a nuclease mutant HsdR (D298E) subunit with wild-type MTase. This mutant enzyme cannot cleave DNA but has wild-type ATPase activity and is capable of DNA translocation (36,42,43). In the absence of wild-type HsdR subunits, a substoichiometric concentration of this endonuclease preparation has no DNA cleavage activity (as measured by the production of linear DNA in Figure 4A). However, when supplemented with a molar excess of wild-type HsdR, cleavage activity was observed with the same efficiency as when MTase alone was supplied. Allowing the mutant endonuclease complex to begin translocation by pre-incubating for 5 min with DNA and ATP prior to the addition of wild type (wt) HsdR did not alter this result (Figure 4A). In each case, the extent of DNA cleavage exceeded the input concentration of endonuclease/Mtase. This data indicate that the MTase within the holoenzyme can dissociate from the mutant HsdR subunits, dissociate from the DNA, acquire wild-type HsdR subunits and bind new DNA multiple times.
Figure 4.
MTase ‘turnover’ of an EcoKI ‘holoenzyme’ and of wild-type EcoAI (A) Cleavage of 5 nM one-site supercoiled DNA was measured following 60 min incubation and is presented as the percentage of linear DNA product produced. EcoKI MTase, HsdR and a nuclease mutant R2M2S1 holoenzyme (D298E) were added, at the concentrations indicated. Reactions 9 and 10 (indicated as light grey columns in the table) differ from reactions 7 and 8 as they were allowed to proceed for 5 min before the addition of excess wt HsdR (see main text). (B) Cleavage activity of EcoAI on 5 nM one-site supercoiled DNA substrate (SC) with an excess of HsdR (300 nM) and varying concentrations of MTase, as indicated. OC is open circle, LIN is linear DNA product. The dashed line represents where an R2M2S1 complex could form on the DNA (see Figure 1). Error bars represent the standard deviations from at least two repeat experiments.
To address if the phenomenon of MTase turnover is a more general property of Type I enzymes, we also tested the Type IB enzyme EcoAI using the cleavage assay. The EcoAI complex was produced as two separate HsdR and MTase preparations and then reconstituted without further purification [(44), ‘Materials and Methods’ section]. As observed with EcoKI, the one-site circular substrate DNA was fully cleaved at substoichiometric concentrations of MTase relative to the site concentration in the presence of a molar excess of HsdR, indicating turnover of the MTase (Figure 4B). Under conditions where HsdR is in excess, the MTases of both EcoKI and EcoAI are capable of dissociating from one DNA substrate, binding another and participating in additional cleavage events.
RecBCD does not induce ‘turnover’ of EcoR124I during DNA cleavage
It has been suggested that DNA translocation and processing by the helicase–nuclease activities of the recombinase RecBCD can act to promote turnover of Type I complexes (29). In that study, holoenzyme preparations were used at an apparent molar concentration—i.e. corrected due to a low specific activity—3- to 10-fold lower than the DNA. An enhancement of cleavage, therefore, assumed turnover of both the MTase and the HsdR subunits. We examined if the presence of RecBCD could alter the dissociation properties of our EcoR124I MTase preparation (Figure 5). RecBCD was obtained from Mark Dillingham (University of Bristol). Translocation and cleavage were measured at a reaction endpoint (60 min) using an excess of HsdR and RecBCD relative to DNA, and varying concentrations of EcoR124I MTase. In the absence of RecBCD, an excess of EcoR124I MTase was required for activity and turnover did not appear to occur in the presence of excess HsdR in either translocation assays (Figure 5A and B) or cleavage assays (Figure 5C and D). This is possibly because following cleavage the MTase remains bound to the DNA or it becomes inactivated. RecBCD requires a DNA end for initial binding so will not cut the plasmid substrates in our assays (Figure 5C and D). Only the linear DNA cut following Type I activity was a substrate (Figure 5C). Although RecBCD removed the linear DNA generated by the Type I enzyme, it did not increase triplex displacement or DNA cleavage levels. These results contradict those observed by Bianco and Hurley (29). Similar experiments with EcoKI have also found that RecBCD has no effect on turnover of the holoenzyme (45).
DISCUSSION
Our investigations into the in vitro subunit dynamics of the Type I RM systems have shown that both the HsdR and MTase components have the ability to turnover with respect to DNA during translocation and cleavage. The HsdR subunits of both EcoR124I and EcoKI are likely to dissociate from the MTase following translocation termination but can only dissociate from the DNA track via a DNA end. In contrast, the MTases of EcoKI and EcoAI can dissociate from the DNA independent of DNA ends but subsequent re-binding requires a source of free HsdR. Each of these events is discussed in turn, including how the dynamics may be of importance to regulation of Type I enzymes in vivo.
Recycling of HsdR subunits
On linear DNA, EcoKI and EcoR124I HsdR behave in much the same way with dissociation of the HsdR from one DNA and re-binding to another. This suggests that DNA loops formed during translocation by EcoKI disassemble in the same way as those for EcoR124I. We observed a slightly lower turnover efficiency of EcoKI compared to EcoR124I, but this may reflect differences in in vitro protein stability at low concentrations or differences in specific activity of our enzyme preparations. Nonetheless, these results show that the dissociation/re-association of motors during translocation is a common feature of Type I enzymes, and, is not a special feature of the >240-fold weaker binding affinity of the R2-complex of EcoR124I relative to the R1-complex. As noted previously, the R1-complex appears stable in the absence of ATP but readily undergoes disassembly in its presence (Figure 1A) (10,11). Similarly, the EcoKI complex is apparently stable (28) yet can also readily undergo disassembly in the presence of ATP (Figure 2). Loss of protein subunits by EcoKI (and the related EcoBI) during translocation were also inferred from early EM studies (26,46). Conformational changes in the endonuclease complex that are required for translocation must also change the affinities of the subunits within the complex.
On supercoiled DNA, in contrast, neither EcoR124I nor EcoKI demonstrated measurable levels of HsdR turnover following translocation. This difference in activity could be attributed to: (i) the topology of the DNA. On supercoiled DNA the translocating enzyme will cause a build-up of positively supercoiled DNA in the downstream DNA whereas on linear DNA no topological barrier would be present (27); or, (ii) the presence of DNA ends, which could be the route for dissociation of HsdRs during translocation on linear DNA. This would predict that the rate of dissociation from internal sites is slower than from DNA ends. A model for loop disassembly during translocation and HsdR turnover that is consistent with previous experiments and those here is presented in Figure 6.
Figure 6.
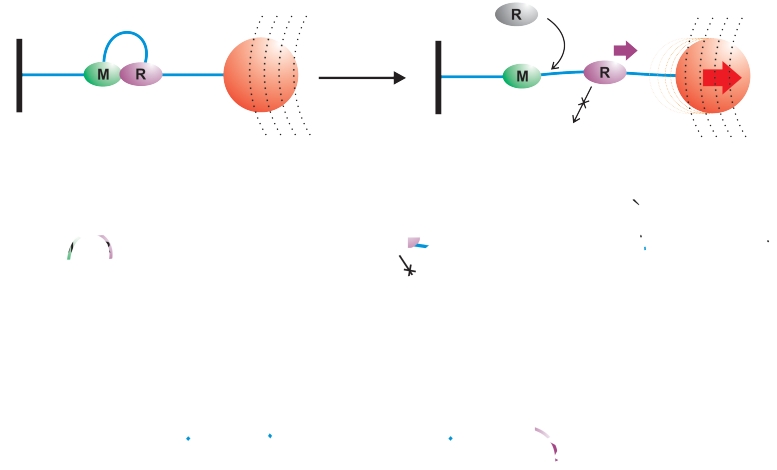
Fates of HsdR subunits on different DNA substrates in vitro following dissociation from the MTase core. DNA is represented as a blue line, the MTase (i.e. both the HsdM and HsdS subunits) as a green oval (M) and HsdR as a purple oval (R). (A) Tethered DNA in a magnetic tweezers apparatus. The coverslip surface is shown as a black line, the magnetic bead as a red circle and the magnetic field as a series of dotted lines. (B) Linear DNA in solution. (C) Circular DNA in solution.
In the model, loop translocation terminates by dissociation of the HsdR from the MTase. Subsequent turnover of HsdR is then dependent upon the in vitro experimental conditions: in the magnetic tweezers apparatus (Figure 6A), the DNA is tethered to the glass coverslip at one end and to a magnetic bead at the other. There are therefore no free DNA ends. Upon termination and release of the MTase the motor does not release the DNA but may continue translocation as far as the bead (data not shown) or surface (data not shown). Free HsdR in solution can associate with the vacant binding sites on the MTase and re-initiate translocation. However, turnover of the DNA-bound HsdR cannot occur. The ‘off-rate’ measured from this assay represents the loop lifetime, rather than dissociation from the DNA per se; in ensemble solution experiments on linear DNA (Figure 6B), loop translocation may terminate at an internal site or upon reaching the DNA end. For the former case, continued translocation of the motor would also allow dissociation from the end. The HsdR motor could therefore exit the DNA and re-bind to another MTase. The time for the HsdR to leave the linear DNA will not necessarily reflect the lifetime of the loop translocation state; in the ensemble solution experiments on circular DNA, disassembly of the translocating loop leaves the HsdR on the DNA and unable to dissociate because of the lack of free DNA ends (Figure 6C). Therefore, HsdR turnover cannot occur, although new translocation events could be initiated by HsdR from solution.
HsdR subunits do not appear to turnover following cleavage of circular DNA, despite the presence of DNA ends and the turnover of the MTase (Figures 3 and 4). This may be because: (i) the HsdR subunits remain irreversibly-associated with the DNA following cleavage, possibly at the cleavage loci; or, (ii) because a DNA cleavage event catalysed by the nuclease domain irreversibly inactivates the associated helicase domain regardless of subsequent dissociation of the HsdR from the DNA. The first model could account for the continued ATPase activity that is associated with the post-cleavage phase (21). If the HsdR subunits are associated with the cleaved DNA ends, they must still allow access by the end-dependent exonuclease activity of RecBCD (Figure 5). It could be argued that RecBCD activity re-models the HsdR interaction following cleavage, dissociating it from the DNA and facilitating turnover (29). Our experiments with RecBCD used excess HsdR and only addressed MTase turnover (see below). However, complementary experiments by Dryden and co-workers (45) demonstrate that RecBCD-catalysed digestion of EcoKI-cleaved DNA does not appear to release active HsdR to allow MTase turnover. It may be that the HsdR subunits remain tightly bound to a fragment of DNA to which RecBCD cannot gain access. The exact mechanism of dsDNA cleavage by Type I enzymes and the fate of the HsdR subunits involved remains to be clearly elucidated.
Protein complex disassembly has been suggested to determine cleavage frequency in vivo and play a role in RA (11,12). Our results here indicate that on DNA without free ends, the HsdR subunits would become trapped following translocation termination. This new observation could play an important role in RA. Under normal circumstances, translocation on the host DNA does not occur (all the cognate sites are modified by maintenance methylation). However, if unmodified sites arise due to DNA repair, translocation could occur but would lead to HsdR subunits becoming trapped on the circular genome. If following translocation termination these HsdR cannot participate in further translocation events, then the pool of available HsdR would drop. For EcoR124I, this would lead to the observed decrease in restriction while the total HsdR pool appears constant (17). For EcoKI (and EcoAI), the protease ClpXP has been implicated in RA through the targeting of the complexes translocating on the host DNA and not on bacteriophage DNA. The targeting of translocating EcoKI molecules by ClpXP may be due to kinetic limits. On linear bacteriophage DNA, if cleavage does not immediately occur, the HsdR subunits would be able to dissociate via the free DNA ends. Therefore, the window of opportunity for ClpXP to interact with the HsdR would be relatively short. On the circular host genome, HsdR subunits would have a longer lifetime in a DNA-bound state (either translocating or terminated), and in turn this would give ClpXP more time to recognize and degrade the HsdR subunits. Preliminary in vitro data suggest that ClpXP can indeed target HsdR on circular DNA but not on linear DNA (Michelle Simons, Fiona Diffin and Mark Szczelkun unpublished data).
Recycling of MTase complexes
Recent studies have reported turnover of Type I enzymes that was attributed either to specific reaction conditions, where RecBCD was used to further degrade the cleaved DNA (29), or, to particular enzyme preparations, where Type I complexes were purified as holoenzymes rather than as separate MTase and HsdR pools (30). We have also shown that MTase turnover during DNA translocation and cleavage is possible, yet it is clear that there is nothing unique about our enzyme preparation as turnover can also be observed using enzyme purified as holoenzyme by a different research group. Indeed, a number of studies have carefully shown that there is no difference between Type I enzymes that are purified as holoenzymes or reconstituted from separate protein pools (5,6,11). In our hands we could not measure any turnover of EcoR124I MTase during translocation and cleavage and, in contrast to the previous studies, we found that the inclusion of RecBCD in EcoR124I reactions did not promote additional Type I-dependent DNA cleavage.
On linear DNA, turnover of the EcoKI MTase as measured by translocation or endonuclease activity was observed under conditions where HsdR was either limiting or in excess. In contrast, turnover on the circular DNA was only observed when HsdR was in excess. This difference can be explained simply by the corresponding difference in HsdR turnover: on linear DNA, the HsdR subunits can dissociate in an active form following translocation and can therefore re-associate with DNA bound or free MTase; on the circular DNA, the HsdR subunits either remain bound after translocation termination or become inactivated following DNA cleavage (see above). Therefore, if the MTase subsequently dissociates and there are no free, active HsdR molecules in solution, additional translocation or cleavage events cannot initiate. The early analyzes of EcoKI that established the dogma that Type I enzymes do not turnover all used holoenzyme preparations where HsdR was limiting. The requirement for saturating or supersaturating amounts of enzyme during DNA cleavage seen in those studies is consistent with what we saw here. Analysis of DNA cleavage by the recently characterized Type I SP (single polypeptide) restriction enzyme LlaGI also indicated a requirement for a supersaturating concentration of enzyme (5–10 monomers per DNA) (47). The model for DNA cleavage of both linear and circular DNA by LlaGI requires the interaction of (at least) two enzymes molecules bound to two separate sites on the same DNA. Therefore, cleavage at subsaturating concentrations is severely limited by the lower probability of multiple enzymes being on the same DNA at the same time. Where binding of enzymes to distant DNA sites is not cooperative, efficient long-range interaction will always require enzyme concentrations that allow simultaneous occupancy of both sites with reasonable frequency. It is possible to address turnover in such instances by the addition of a second DNA substrate following cleavage of the first (29,30). However, because of the requirement for a high protein concentration relative to DNA (47), the ability of LlaGI monomers to carry out more than one cleavage event has yet to be assessed.
Does the turnover of the EcoKI and EcoAI MTases have any relevance to endonuclease activity in vivo? The answer to this question relies in part on the relative levels of MTase and freely available HsdR. It has been estimated that the cellular concentration of EcoKI HsdR is ∼4-fold lower than that of the MTase (48). Therefore, the turnover of individual MTase complexes during translocation and cleavage would be limited by the lack of available HsdR. It is still possible that the released MTase could go on to methylate DNA, although this was not tested here. Further limits to the production of endonuclease complexes would then occur during RA, where ClpXP-mediated proteolysis would further reduce the available free HsdR pool (15,16). An important role for turnover during cleavage of a bacteriophage genome is hard to envisage. It is unlikely to be relevant to a single infection event because: first, cleavage will most likely occur on the linear form of the bacteriophage genome early in infection and thus require the collision between two enzymes translocating from two distant sites (7). Supersaturating enzyme concentrations are therefore necessary to elicit cleavage; and second, a single dsDNA break would be sufficient to prevent infection and so multiple cleavage events are not necessary. It could be argued that an ability to turnover releases the enzyme to cut DNA during future infections, but this does not take into account the protein half-life in vivo. Other proteins, such as the alkyl DNA transferases (49), also rely on so-called suicidal enzyme mechanisms. The apparent waste of protein resources is balanced by the important role such systems play in protecting the host against cellular stress. It has also been suggested that the apparently high levels of ATPase activity of the Type I enzymes that remain on the DNA following cleavage could play a role in depleting infected cells of ATP and thus removing them, at least temporarily, from the population (50).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Wellcome Trust (084086); Biotechnology and Biological Sciences Research Council (BBSRC) doctoral training grant. Funding for open access charge: Wellcome Trust Value-in-People Award.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Dyden and Mark Dillingham for materials, and David Dryden, Mark Paget, Ralf Seidel and members of the DNA–Protein Interactions Unit for discussions.
REFERENCES
- 1.Murray NE. Type I restriction systems: sophisticated molecular machines. Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray NE. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. doi: 10.1099/00221287-148-1-3. [DOI] [PubMed] [Google Scholar]
- 3.Dryden DTF, Cooper LP, Murray NE. Purification and characterization of the methyltransferase from the Type I restriction and modification system of Escherichia coli K12. J. Biol. Chem. 1993;268:13228–13236. [PubMed] [Google Scholar]
- 4.Taylor I, Patel J, Firman K, Kneale G. Purification and biochemical characterisation of the EcoR124 type I modification methylase. Nucleic Acids Res. 1992;20:179–186. doi: 10.1093/nar/20.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dryden DTF, Cooper LP, Thorpe PH, Byron O. The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry. 1997;36:1065–1076. doi: 10.1021/bi9619435. [DOI] [PubMed] [Google Scholar]
- 6.Janscak P, Abadjieva A, Firman K. The type I restriction endonuclease R.EcoR124I: over-production and biochemical properties. J. Mol. Biol. 1996;257:977–991. doi: 10.1006/jmbi.1996.0217. [DOI] [PubMed] [Google Scholar]
- 7.Studier FW, Bandyopadhyay PK. Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szczelkun MD, Janscak P, Firman K, Halford SE. Selection of non-specific DNA cleavage sites by the type IC restriction endonuclease EcoR124I. J. Mol. Biol. 1997;271:112–123. doi: 10.1006/jmbi.1997.1172. [DOI] [PubMed] [Google Scholar]
- 9.Shulman MJ. Model for wandering restriction enzymes. Nature. 1974;252:76–78. doi: 10.1038/252076a0. [DOI] [PubMed] [Google Scholar]
- 10.Janscak P, Dryden DTF, Firman K. Analysis of the subunit assembly of the type IC restriction-modification enzyme EcoR124I. Nucleic Acids Res. 1998;26:4439–4445. doi: 10.1093/nar/26.19.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seidel R, Bloom JG, van Noort J, Dutta CF, Dekker NH, Firman K, Szczelkun MD, Dekker C. Dynamics of initiation, termination and reinitiation of DNA translocation by the motor protein EcoR124I. EMBO J. 2005;24:4188–4197. doi: 10.1038/sj.emboj.7600881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Firman K, Dutta C, Weiserova M, Janscak P. The role of subunit assembly in the functional control of Type I restriction-modification enzymes. Mol. Biol. Today. 2000;1:35–41. [Google Scholar]
- 13.Blakely GW, Murray NE. Control of the endonuclease activity of type I restriction-modification systems is required to maintain chromosome integrity following homologous recombination. Mol. Microbiol. 2006;60:883–893. doi: 10.1111/j.1365-2958.2006.05144.x. [DOI] [PubMed] [Google Scholar]
- 14.Loenen WAM, Daniel AS, Braymer HD, Murray NE. Organization and sequence of the hsd genes of Escherichia coli K12. J. Mol. Biol. 1987;198:159–170. doi: 10.1016/0022-2836(87)90303-2. [DOI] [PubMed] [Google Scholar]
- 15.Makovets S, Titheradge AJB, Murray NE. ClpX and ClpP are essential for the efficient acquisition of genes specifying type IA and IB restriction systems. Mol. Microbiol. 1998;28:25–35. doi: 10.1046/j.1365-2958.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 16.Makovets S, Doronina VA, Murray NE. Regulation of endonuclease activity by proteolysis prevents breakage of unmodified bacterial chromosomes by type I restriction enzymes. Proc. Natl Acad. Sci. USA. 1999;96:9757–9762. doi: 10.1073/pnas.96.17.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makovets S, Powell LM, Titheradge AJB, Blakely GW, Murray NE. Is modification sufficient to protect a bacterial chromosome from a resident restriction endonuclease? Mol. Microbiol. 2004;51:135–147. doi: 10.1046/j.1365-2958.2003.03801.x. [DOI] [PubMed] [Google Scholar]
- 18.Doronina VA, Murray NE. Proteolytic control of restriction by the type I restriction enzyme EcoKI. Biochem. Soc. Trans. 2000;2000:A177. [Google Scholar]
- 19.Doronina VA, Murray NE. The proteolytic control of restriction activity in Escherichia coli K-12. Mol. Microbiol. 2001;39:416–428. doi: 10.1046/j.1365-2958.2001.02232.x. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill M, Powell LM, Murray NE. Target recognition by EcoKI: the recognition domain is robust and restriction-deficiency commonly results from the proteolytic control of enzyme activity. J. Mol. Biol. 2001;307:951–963. doi: 10.1006/jmbi.2001.4543. [DOI] [PubMed] [Google Scholar]
- 21.Eskin B, Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. III. Studies of the restriction adenosine triphosphatase. J. Biol. Chem. 1972;247:6192–6196. [PubMed] [Google Scholar]
- 22.Eskin B, Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. II. Purification, subunit structure and catalytic properties of the restriction endonuclease. J. Biol. Chem. 1972;247:6183–6191. [PubMed] [Google Scholar]
- 23.Endlich B, Linn S. The DNA restriction endonuclease of Escherichia coli B. I. Studies of the DNA translocation and the ATPase activities. J. Biol. Chem. 1985;260:5720–5728. [PubMed] [Google Scholar]
- 24.Endlich B, Linn S. The DNA restriction endonuclease of Escherichia coli B. II. Further studies of the structure of DNA intermediates and products. J. Biol. Chem. 1985;260:5729–5738. [PubMed] [Google Scholar]
- 25.Yuan R, Bickle TA, Ebbers W, Brack C. Multiple steps in DNA recognition by restriction endonuclease from E. coli K. Nature. 1975;256:556–560. doi: 10.1038/256556a0. [DOI] [PubMed] [Google Scholar]
- 26.Bickle TA, Brack C, Yuan R. ATP-induced conformational changes in the restriction endonuclease from Escherichia coli K-12. Proc. Natl Acad. Sci. USA. 1978;75:3099–3103. doi: 10.1073/pnas.75.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szczelkun MD, Dillingham MS, Janscak P, Firman K, Halford SE. Repercussions of DNA tracking by the type IC restriction endonuclease EcoR124I on linear, circular and catenated substrates. EMBO J. 1996;15:6335–6347. [PMC free article] [PubMed] [Google Scholar]
- 28.Dryden DTF, Cooper LP, Murray NE. Assembly of the multifunctional EcoKI DNA restriction enzyme in vitro. Tech. Prot. Chem. 1997;8:593–601. [Google Scholar]
- 29.Bianco PR, Hurley EM. The type I restriction endonuclease EcoR124I, couples ATP hydrolysis to bidirectional DNA translocation. J. Mol. Biol. 2005;352:837–859. doi: 10.1016/j.jmb.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 30.Bianco PR, Xu C, Chi M. Type I restriction endonucleases are true catalytic enzymes. Nucleic Acids Res. 2009;37:3377–3390. doi: 10.1093/nar/gkp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley LK, Szczelkun MD. Direct and random routing of a molecular motor protein at a DNA junction. Nucleic Acids Res. 2006;34:4387–4394. doi: 10.1093/nar/gkl569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vipond IB, Baldwin GS, Oram M, Erskine SG, Wentzell LM, Szczelkun MD, Nobbs TJ, Halford SE. A general assay for restriction endonucleases and other DNA-modifying enzymes with plasmid substrates. Mol. Biotechnol. 1995;4:259–268. doi: 10.1007/BF02779019. [DOI] [PubMed] [Google Scholar]
- 33.Janscak P, Bickle TA. The DNA recognition subunit of the type IB restriction-modification enzyme EcoAI tolerates circular permutations of its polypeptide chain. J. Mol. Biol. 1998;284:937–948. doi: 10.1006/jmbi.1998.2250. [DOI] [PubMed] [Google Scholar]
- 34.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 35.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClelland SE, Dryden DTF, Szczelkun MD. Continuous assays for DNA translocation using fluorescent triplex dissociation: application to type I restriction endonucleases. J. Mol. Biol. 2005;348:895–915. doi: 10.1016/j.jmb.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 37.Seidel R, van Noort J, van der Scheer C, Bloom JGP, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction-modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 38.Yuan R, Heywood J, Meselson M. ATP hydrolysis by restriction endonuclease from E. coli. K. Nat. New Biol. 1972;240:42–43. doi: 10.1038/newbio240042a0. [DOI] [PubMed] [Google Scholar]
- 39.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 40.Cavaluzzi MJ, Borer PN. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004;32:e13. doi: 10.1093/nar/gnh015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seidel R, Bloom JG, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davies GP, Kemp P, Molineux IJ, Murray NE. The DNA translocation and ATPase activities of restriction-deficient mutants of EcoKI. J. Mol. Biol. 1999;292:787–796. doi: 10.1006/jmbi.1999.3081. [DOI] [PubMed] [Google Scholar]
- 43.Davies GP, Martin I, Sturrock SS, Cronshaw A, Murray NE, Dryden DTF. On the structure and operation of type I DNA restriction enzymes. J. Mol. Biol. 1999;290:565–579. doi: 10.1006/jmbi.1999.2908. [DOI] [PubMed] [Google Scholar]
- 44.Janscak P, Sandmeier U, Bickle TA. Single amino acid substitutions in the HsdR subunit of the type IB restriction enzyme EcoAI uncouple the DNA translocation and DNA cleavage activities of the enzyme. Nucleic Acids Res. 1999;27:2638–2643. doi: 10.1093/nar/27.13.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts GA, Cooper LP, White JH, Su T-J, Zipprich J, Geary P, Kennedy C, Dryden DTF. An investigation of the structural requirements for ATP hydrolysis and DNA cleavage by the EcoKI Type I DNA restriction and modification enzyme. Nucleic Acids Res. 2011;39:7667–7676. doi: 10.1093/nar/gkr480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosamond J, Endlich B, Linn S. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J. Mol. Biol. 1979;129:619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- 47.Smith RM, Diffin FM, Savery NJ, Josephsen J, Szczelkun MD. DNA cleavage and methylation specificity of the single polypeptide restriction-modification enzyme LlaGI. Nucleic Acids Res. 2009;37:7206–7218. doi: 10.1093/nar/gkp790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiserova M, Janscak P, Benada O, Hubacek J, Zinkevich VE, Glover SW, Firman K. Cloning, production and characterization of wild type and mutant forms of the R.EcoK endonucleases. Nucleic Acids Res. 1993;21:373–379. doi: 10.1093/nar/21.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samson L. The suicidal DNA repair methyltransferases of microbes. Mol. Microbiol. 1992;6:825–831. doi: 10.1111/j.1365-2958.1992.tb01533.x. [DOI] [PubMed] [Google Scholar]
- 50.Bickle TA. The ATP-dependent restriction endonucleases. In: Linn SM, Roberts RJ, editors. Nucleases. New York: Cold Spring Harbor Laboratory Press; 1982. pp. 85–108. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.