Abstract
Nuclear organization of chromatin is an important level of genome regulation with positional changes of genes occurring during reprogramming. Inherent variability of biological specimens, wide variety of sample preparation and imaging conditions, though pose significant challenges to data analysis and comparison. Here, we describe the development of a computational image analysis toolbox overcoming biological variability hurdles by a novel single cell randomizing normalization. We performed a comparative analysis of the relationship between spatial positioning of pluripotency genes with their genomic activity and determined the degree of similarity between fibroblasts, induced pluripotent stem cells and embryonic stem cells. Our analysis revealed a preferred positioning of actively transcribed Sox2, Oct4 and Nanog away from the nuclear periphery, but not from pericentric heterochromatin. Moreover, in the silent state, we found no common nuclear localization for any of the genes. Our results suggest that the surrounding gene density hinders relocation from an internal nuclear position. Altogether, our data do not support the hypothesis that the nuclear periphery acts as a general transcriptional silencer, rather suggesting that internal nuclear localization is compatible with expression in pluripotent cells but not sufficient for expression in mouse embryonic fibroblasts. Thus, our computational approach enables comparative analysis of topological relationships in spite of stark morphological variability typical of biological data sets.
INTRODUCTION
In the last years, nuclear organization of chromatin has increasingly come into focus as an important level of genome regulation (1,2). During differentiation, distinct positional changes of specific genes upon transcriptional activation have been reported. One of the first correlations between gene repositioning and its activation level was observed in mouse B-lymphocyte maturation. Brown et al. (3) showed that gene repositioning away from centromeric regions could be seen upon activation. In this study, distances were not measured but co-localization with the centromeric region was evaluated. A more recent study, on Drosophila, reported distance measurements taking chromatin mobility into account and also concluded a close association of silenced genes with centromeric heterochromatin (4). Similar observations have been reported for relocation of genes to the nuclear lamina (5). However, it is unclear whether proximity to heterochromatin, in general, regulates gene (in)activity (6,7). The comparison of different biological data sets is complicated by variability of their framework parameters such as drastic changes of nuclear morphology during differentiation. Up to now, software for distance measurement has not taken into account such morphological changes or do not measure in 3D (8,9). The application of sophisticated normalization procedures becomes mandatory to produce meaningful and unbiased objective data evaluation. Prominent morphological and transcriptional changes occur upon gain and loss of pluripotency, making it an ideal test system for studying repositioning of genes during cellular differentiation. The generation of induced pluripotent stem (iPS) cells requires the expression of the pluripotency genes Oct4 and Sox2 (10). Additional factors such as Nanog have been shown to increase the efficiency of the process (11). Accordingly these three genes are upregulated at the transcriptional level in embryonic stem (ES) cells and iPS cells.
In this study, we describe a novel method to investigate whether changes in gene positioning within the nucleus correlate with their transcriptional status and/or genomic context. Our computational analysis includes a series of filters to segment the objects to be analysed, in our application, the nuclear periphery, the chromocenters and the gene loci. This is followed by a single cell-based normalization procedure, which permits the comparison of data sets exhibiting large morphological variability. In brief, using this approach, we could show that gene position relative to heterochromatin does not correlate with silencing, but internal gene positioning is compatible with expression in pluripotent cells and may be influenced by the surrounding gene density in mouse embryonic fibroblasts (MEFs).
MATERIALS AND METHODS
Biological system and microscopy
Cells and culture conditions
Transgenic MEF cells were kindly provided by R. Jaenisch (12) (Whitehead Institute, Cambridge, MA, USA). These cells had been infected with lentiviral vectors containing the four reprogramming factors Oct4, Klf-4, Sox2 and c-Myc expressed in a doxycycline (DOX)-inducible manner (13). Non-induced cells were used as controls. MEF cells were grown in cell culture flasks in Dulbecco's Modified Eagle's Medium (DMEM) GlutaMAX (Invitrogen) supplemented with 10% fetal calf serum (FCS) (PAA Laboratories) and seeded on gelatine-coated coverslips 36–48 h before fixation. Reprogramming towards iPS was achieved by adding DOX (2 ng/ml) (Sigma-Aldrich) to the medium for ∼4 weeks. After induction cells were cultured in DMEM GlutaMAX supplemented with 15% FCS, leukaemia inhibitor factor (LIF) (1000 U/ml; Chemicon/Millipore), non-essential amino acids (0.1 mM; PAA Laboratories) and β-mercaptoethanol (1 µM) (Merck). Distinct colonies of cells were observed after a few weeks. Reprogramming on a single cell level was assessed by positive immunofluorescence (IF) staining against the murine pluripotency marker SSEA-1 (mouse IgM anti SSEA-1, Millipore).
Mouse embryonic stem (ES) cells of the CCE line (129/Sv-derived mouse ES cell line) (kindly provided by C. Bonifer, Leeds, UK) were cultured without feeder cells on gelatinized glass slides under the same culture conditions as described for iPS cells. Medium was changed daily and cells were split before individual colonies touched each other. Pluripotency was tested on a single cell level by positive staining for the SSEA-1 marker as for iPS cells (Supplementary Figure S1).
DNA probes
Specific bacterial artificial chromosome (BAC) clones were used for the delineation of the genomic regions covering the pluripotency transcription factors Oct4 (MMU 17; clone RP23-75C13), Sox2 (MMU 3; RP23-425G5) and Nanog (MMU 6; RP23-474F18). The DNA probes were labelled with biotin-coupled dUTPs by nick-translation and labelled DNA probes were suspended at a final concentration of 50 ng/µl together with a 20-fold excess of unlabelled mouse COT-1 DNA (Invitrogen) in 50% formamide/10% dextransulfate/2xSSC (saline-sodium citrate buffer) following standard protocols. Hybridization solely of the endogenous gene loci was ensured by specific visualization of the addressed genes on the expected chromosomal position.
3D immuno-FISH analysis
Cells were fixed with 4% paraformaldehyde for 10 min. Subsequent treatment included permeabilization with 0.5% Triton X-100/PBS for 30 min at room temperature (RT), incubation in 20% glycerine/PBS for 30 min, repeated freezing/thawing steps in liquid nitrogen, protein degradation with 0.1 N HCl and storage in 50% formamide/2 × SSC at 4°C over night (14). This approach allows the preservation of morphological structures of single nuclei as well as of whole iPS colonies. Probes and cell samples were allowed to pre-anneal for 2–3 h at RT and thereafter denaturated simultaneously for 3 min at 76°C. Hybridization was performed in a 37°C water bath for 3 days. Stringent washing was done in 0.1 × SSC at 62°C for 3 × 5 min followed by incubation in 4% BSA/PBST (bovine serum albumin in PBS with 0.05% Tween) blocking solution for 10 min. Hybridized probes were detected with Cy5-conjugated streptavidin (1:200 in 2% BSA/PBST) (Rockland) together with immunodetection of SSEA-1 using an anti-SSEA-1 mouse monoclonal antibody (IgM) for 1 h at RT and subsequent incubation with Alexa594-conjugated goat anti-mouse IgM antibody (1:500) (Sigma-Aldrich). DNA counterstain was performed with 4′,6-diamidino-2-phenylindole (DAPI) (200 ng/ml) (Sigma-Aldrich) for 10 min and samples were mounted in Vectashield Antifade Medium (Vector Laboratories).
Image acquisition
Only SSEA-1-positive cells were used for image acquisition. 3D image stacks of single nuclei were acquired using a Leica SP5 confocal laser scanning microscope (Leica Microsystems) with a Plan-Achromat 63× oil objective (voxel size 50:50:200 nm [x:y:z]).
Computational analysis
User interface
The user interface enables the easy adjustment of various settings to respond to different image qualities (Supplementary Figure S2) and step-by-step audio visual tutorial Supplementary Movie 1.
Vid: viewer identification determines the active window.
DNA col.channel: define which channel contains the DNA counterstain from which the chromocenters and nuclei are detected.
BAC col.channel: define which channel contains the gene or other point-like signals from which distances should be measured.
Nuclear_threshold: depending on the fluorescence image the threshold for the nuclear periphery can be adjusted, the output can be simply controlled visually.
minNucPixelSize: in order to filter out false signals a minimal size for a nucleus can be set.
fill nuc holes: DNA counterstain does not stain nucleoli which results in very dark nucleoli which may be wrongly segmented as periphery. Using this setting, small holes within the nucleus, resulting from incorrect segmentation can be filled.
BAC minPixelsize: this setting defines the smallest size of the reference structure for visualization.
Baseline: using this setting the minimal fluorescent intensity of the wanted signal is set. By adjustments, larger weakly stained unwanted particles can be filtered out.
nSim (number of simulations): sets the number of simulated points in the nucleus. Depending on the size of the nucleus this number might need to be adjusted
nBins (number of bins): defines how many bins should be used for the output graph.
nucID (nucleus identification): If more than one nucleus is present per image, this identifies the nucleus used for further analysis. The number of the relevant nucleus can be entered in this field.
z-min overlap: values larger than 0 allow a certain degree of z overlap to occur without merging two separate nuclei.
Batch process multiple files for CC (chromocenter) segmentation: this button opens up a window to perform preprocessing on the raw data image files. The program automatically detects similar file names in order to carry out batch processing. The median and radius value for Gaussian blur can be set at this point. If 8 bit images are used, the ‘fix saturation value’ box with a saturation value of 255 should be kept to avoid problems due to overexposure in the image. The batch processing can be started to apply all filters to all selected files resulting in new binary (3D) image files containing the segmented nuclei and chromocenters (Supplementary Figure S3).
Load CC and show CC outlines: load preprocessed chromocenter segmentation and show chromocenter outline overlaid to the microscopic image. Additionally the gene signal centroid is marked.
Filter and label nuclei: the raw data are processed to segment nuclei, which are outlined and numbered on the viewer window.
Draw nuclei cut lines: if nuclei are very close to each other automated segmentation might fail. This button enables one to manually draw a separating line between two adjacent structures.
Paint cut: this function enables one to draw into the image without changing the raw data. This can be used in order to mask false signals that are disturbing correct segmentation.
If manual segmentation has been performed it can be documented and recalled by the save and load button.
Calc volumes: The volumes of segmented chromocenters and nuclei are printed.
Simulate 3D points: Performs the measurement and automated simulation of the random points. Output can be directly pasted into excel and contains the real distance to chromocenters and nuclear periphery and normalized quantile values.
Mark 3D sect: Visualizes the random points in each section.
Parameter settings for measurements
For the segmentation of the nucleus, a constant threshold of 40 was used across almost all data sets. Only for very light and dark images, a small deviation was required. The quality of segmentation was visually controlled.
For the segmentation of the chromocenters, a series of filtering steps (Supplementary Figure S3) was performed followed by basic global thresholding (BGT) algorithm. These settings were kept constant throughout all data sets.
The settings for gene signal detection (BAC minPixelSize and baseline) were adjusted individually for each image depending on background noise and signal intensities. The settings varied between 40 and 70 for the pixel size and between 40 and 100 for the baseline.
If segmentation was altered by manual correction, the altered image was stored to keep it for control.
To account for chromatic aberration, xyz correction (20 nm × 50 nm × 950 nm) based on bead measurements was performed. This is especially important for dyes with large spectral distances such as DAPI and Cy5, which were used in our experiments.
Image data processing, measurements and normalization
Our analysis software is geared towards optimized throughput when analysing multiple 3D data sets. The programme is implemented using the Priithon image analysis platform (http://priithon.googlecode.com), which allows fast interactive visualization and comparison of the non-normalized data versus the resulting segmentation shown as outlines.
First, the image processing and ‘chromocenter’ segmentation steps illustrated in Supplementary Figure S3 are applied to a group of files. An unsharp mask type filter is applied using the same user-specified parameters (a 4 × 4 pixel median filter and a 6 × 6 uniform filter) for all data sets. To get a robust measure for thresholding chromocenters, the volume is projected along Z using the maximum intensity method before applying the BGT algorithm (15). Here, the threshold is iteratively calculated from the intensity histogram so that the image is split into bright and dark pixels in such a way that their respective mean intensity is symmetrically far from the chosen threshold.
Nuclei are segmented in a similar way to chromocenters. First, a 3 × 3 pixel median filter and a 5 × 5 uniform filter are averaged and applied to each section of the volume. After this smoothing step, the data were thresholded using a user-specified constant threshold as described in the parameter settings. Then, we perform a 2D hole filling step using a specified radius to solve the problem posed by nucleoli or other unstained regions within the nucleus, followed by segmentation and enumeration of 3D-connected structures, in this case nuclei. Fully automated and unsupervised segmentation of nuclei often fails especially in the case of cell colonies, which are densely packed. For this case, our program provides the possibility to manually draw multiple cut-lines. These are section-wise spline-interpolated polygons (Bézier curves), which can be saved and reloaded to make these changes reproducible and documentable. The cut-lines assist the segmentation algorithm by forcing a larger area to be split into two or more nuclei. The user can furthermore specify a fractional parameter determining to what degree overlap is allowed.
‘Gene loci’ are calculated based on the fluorescence intensity distribution of the respective channel. A user-specified base intensity is subtracted to account for background signal and electronic amplification offsets. Pixels with higher intensities are weighted and used for the centroid position calculation with sub-pixel resolution.
The ‘distance analysis’ is done on a nucleus-by-nucleus basis. Random points are generated uniformly within the 3D nucleus. To improve performance for distance measurements, distance maps are pre-calculated for the chromocenter and for the nuclear envelope distances, respectively. This is done using the Euclidean Distance Transform based on the 3D-grid given by the raw data voxel size.
The acquired data were collected and documented using Excel. All distances given in micrometre from one gene and one cell type were binned in steps of 0.1 µm and their relative frequency calculated. Quantile normalized distances were binned in 0.25 steps. Graphs were created by plotting the relative frequency as y-axis and the bins as x-axis.
To test for divergence from a random distribution (relative frequency of 25% for each bin) the chi-squared value was calculated using Excel.
RESULTS AND DISCUSSION
Signal detection and segmentation are compromised by biological and technical parameters
For monitoring gene position in 3D during induced pluripotency, we measured the shortest distance between the gene locus and nuclear landmarks i.e. (1) the nearest pericentric heterochromatin (chromocenter) surface and, (2) the nuclear periphery (Figure 1a). Data were compared between MEF, iPS and ES cells in order to determine differences and similarities (Figure 1b). These measurements require the definition of the gene position and correct segmentation of compartments of interest (chromocenters and periphery). To achieve high throughput, signal detection and segmentation were automated. The first step consists of the segmentation of chromocenters and nuclear periphery. Grey scale image stacks are processed using a series of software filters (Supplementary Figure S3) resulting in a binary (segmented) image for heterochromatic regions (chromocenters) and the nuclear outline (nuclear periphery). The settings of the filtering step are set once and can be applied for a complete set of images stacks and are automatically saved. A challenging problem during segmentation is thresholding since the apparent size of chromocenters is drastically changed by this procedure (Figure 1d). To minimize subjective bias on threshold levels with subsequent significant impact on absolute distance measurements (lower threshold leading to shorter distances and vice versa), we have applied a basic global threshold approach calculating the threshold for each image individually used minimizing bias by using fix thresholds. The detection of the relevant fluorescence signals (FISH gene signals) was also automated in order to increase image analysis throughput. Gene signals can vary in size and intensity based on hybridization efficiency and antibody sensitivity. We, therefore, added a feature to set the size and intensity of signals in order to decrease false detection of background noise. The settings can be chosen individually for each image stack to ensure best possible definition of gene signals. After segmentation the signal is reduced to a 3D pixel with sub-pixel localization precision, which is subsequently used for measurements.
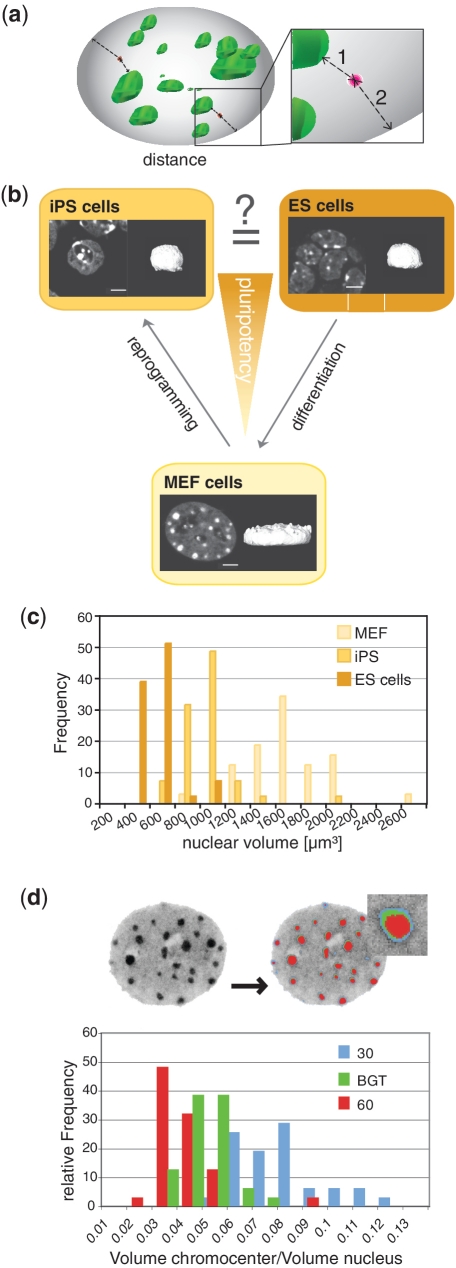
Figure 1.
Correlation of subnuclear gene topology versus gene expression is hampered by drastic morphological variability. (a) 3D representation of a mouse mammalian nucleus (grey) with chromocenters highlighted in green (DNA staining with DAPI) and gene loci visualized by FISH in red. Nearest 3D distances are measured from the signal of interest (gene locus) to the chromocenter (1) and to the nuclear periphery (2). (b) Schematic representation of the pluripotency state of MEF, iPS and ES cells (ES). Confocal optical mid section of a DAPI stained representative MEF (lower), iPS (left) and ES (right) cell nucleus and corresponding 3D reconstructions highlighting their morphological variability. Scale bar 5 µm. (c) Bar histograms of nuclear volumes measured in MEFs, iPS and ES cells (n = 30). (d) The ratios between chromocenter versus nuclear volumes were compared using either BGT depicted in green, or fluorescent intensity threshold levels of 30 (blue) and 60 (red), respectively, and their relative frequency plotted. The largely diverging volumes demonstrate the dependency of these measurements from a given threshold setting (n = 133).
Distance measurements and threshold independent single cell-based normalization
Distance measurements were performed using the gene signal channel (Cy5) and the binary segmented image from the DNA (DAPI) channel to calculate the shortest distance in 3D from the signal to the nearest chromocenter surface and towards the nuclear periphery. The non-normalized distance plots are shown in Supplementary Figure S4. The direct comparison of absolute measurements and distance analyses was complicated by the drastic changes of nuclear shape and size concomitant with the process of cellular reprogramming. Primary fibroblasts, commonly used as the source for reprogramming, have flat ellipsoid nuclei with length diameters that significantly exceed those of the much smaller spherically shaped ES and iPS nuclei, respectively, but have much smaller z-diameters (Figure 1a and b). Flat and spherically shaped nuclei thus yield shorter distance distributions to the periphery exclusively due to their shape. To solve this problem, we established a single cell-based normalization. This enabled the determination of changes of gene positioning in relation to gene expression and to elucidate similarities or differences between the three cell types examined irrespective of their diverging nuclear morphologies. We utilized a Monte Carlo approach: for each cell the software created 10 000 random points within the nucleus in 3D (Supplementary Movie 2) and calculated the distance of each point to the nearest relevant structure resulting in a cell-specific reference distribution (Figure 2a). The number of simulations was determined by balancing the requirement for speed and data reproducibility (Supplementary Figure S5). Since chromocenters are known to harbour very few genes, we further adapted the simulation to exclude chromocenters from our reference distribution. As this additional step did not alter the outcome, we neglected it in our further evaluation (Supplementary Figure S6).
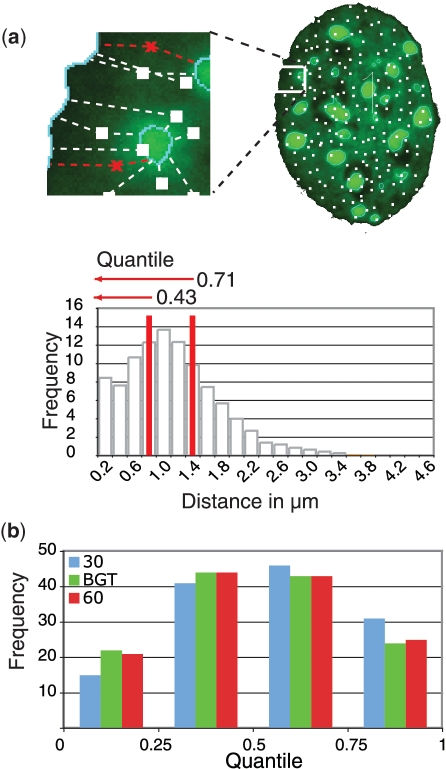
Figure 2.
Single cell-based normalization overcomes threshold dependent variability. (a) For data normalization, 10 000 random points were set throughout the 3D nuclear volume (Supplementary Movie 2) and all distances from these points (white) to the chosen nuclear landmarks measured. The target gene locus measurement (red) is set in relation to the random distribution obtained by the simulation. The fractions of random point measurements, which are smaller or equal to the gene locus distance measurements, are defined as quantile. (b) Gene loci distance measurements in MEF were normalized to a random distribution and the resulting quantiles plotted for each threshold setting (n = 133).
Combining this reference distribution with our non-normalized gene to heterochromatin distance measurement, we obtained a quantile. This is defined as the fraction of reference distances smaller or equal to the non-normalized measurement and can take any number between zero and one. A quantile of zero represents a real measurement, which is shorter than all randomly distributed points, and a quantile of one means that the real measurement is more distal than all simulated points. By this, all measured distances are set into the context of their individual cell (Figure 2a) and give an unbiased view of any deviation from a random distribution.
Using quantiles has several major advantages. First, each quantile between zero and one is equiprobable in value if measurements are sampled from the random background distribution. A deviation from an equal distribution (each quantile bin in a frequency of 25%) indicates a non-random positioning. Deviations from random positioning can be determined by Pearson's chi-square test. Second, they are independent from morphological variations and distributions. Last but not least, a major advantage lies in threshold robustness (Figure 2b), a challenge for quantitative image analysis (16). The same cells were analysed using two fixed thresholds (30 and 60) and our calculated basic global threshold for segmentation. The differences in frequency of quantiles are minimal (Figure 2b), which emphasizes the superiority of this normalization method.
When assembling quantiles obtained from numerous nuclei in a frequency distribution, one can expect three types of distribution: (i) no preference for any position within the nucleus results in an uniform distribution of quantiles (Supplementary Figure S7A); (ii) an accumulation of small quantiles would represent short distances (Supplementary Figure S7B), while (iii) accumulation of larger quantiles would indicate large distances to the respective reference nuclear landmark (Supplementary Figure S7C). The comparison of quantile distributions in data sets from different cell types allows the disclosure of differences in gene positioning (Supplementary Figure S7D–F).
Internal nuclear gene positioning is correlated with expression and gene relocation is restricted by high gene density
Using our normalization, we compared the gene positions within the three different cell types. In Figure 3a, we show the quantile distribution as relative frequencies, i.e. data points represent all quantiles in the intervals 0–0.25, 0.25–0.5, 0.5–0.75 and 0.75–1.
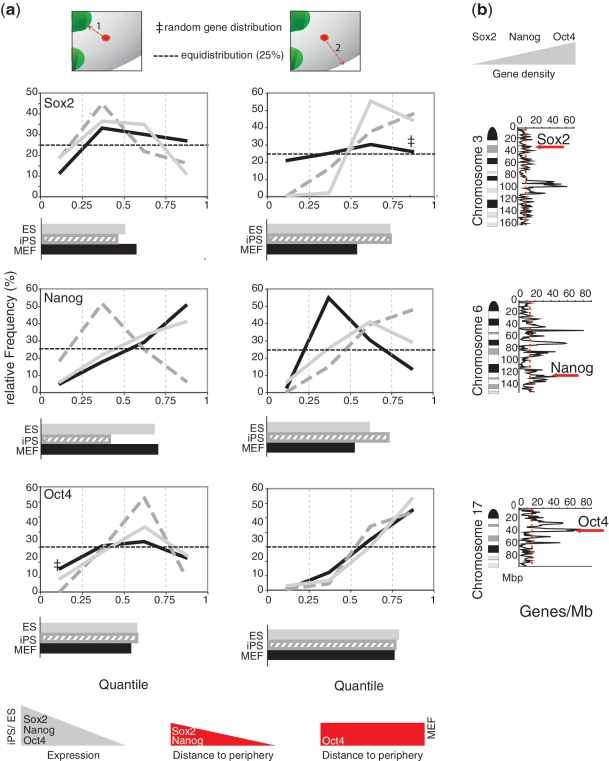
Figure 3.
Gene repositioning during gain/loss of pluripotency relative to chromosomal context and to distance from heterochromatin. Distances of pluripotency genes Sox2, Nanog and Oct4 to chromocenters and to the nuclear periphery in MEF, ES and iPS cells were normalized as shown in Figure 2A. (a) Quantile distribution relative to chromocenters (1) and to nuclear periphery (2) with histogram of the average quantiles underneath. A summary of expression and re-localization of the genes in the nucleus is added at the bottom. (b) Position of the genes within the corresponding mouse chromosome ideogram (Giemsa banding) with gene density plotted along the chromosome.
We observed only two cases of a random subnuclear distribution for the pluripotency genes using our randomizer tool. In MEF cells, Sox2 positioning did not differ from a random pattern with regard to the nuclear periphery and Oct4 relative to the distance to the nearest chromocenter (Figure 3a, double dagger). All other distributions differed highly (P < 10−4 two-tailed t-test) from randomness.
Sox2 and Oct4 did not change their distance to chromocenters between cell types but Nanog showed reduced distances in iPS cells (Figure 3a). It is not clear why Nanog exhibits different positioning in iPS cells but might be influenced by partial reprogramming of the cells. We did not observe a general change of distance to chromocenters indicating that they may not act as silencing compartments as proposed before. (3,4).
Nanog and Sox2 exhibited larger quantiles, reflecting larger distances from the nuclear periphery in iPS and ES cells where they are actively transcribed, compared to MEFs where these genes are silent (Figure 3a). In contrast, no relocation of Oct4 was detected when comparing its positioning in MEFs and iPS/ES (Figure 3a). In all three cell types, Oct4 occupied an internal nuclear position. This observation is consistent with previous data on human and mouse ES cells and a few differentiated cells (17–19), where an internal Oct4 position is maintained. Taken together these results do not support the hypothesis that the nuclear periphery acts as a general transcriptional silencing compartment (5) since in MEFs no relocation to the periphery is observed. An alternative hypothesis would be that an internal nuclear position is compatible with gene expression although not sufficient for it.
All three genes show a preferential internal nuclear localization in iPS and ES cells but differ in MEFs, which could reflect their chromosomal context. Oct4 is located within a chromosomal region of particularly high gene density (75 genes/Mb), whereas Nanog resides in a region of intermediate gene density (33 genes/Mb) and Sox2 in a region of low gene density (six genes/Mb) (Figure 3b). The very high gene density around Oct4 might restrict its relocation. The interior positioning in the active state (Figure 3a) may facilitate spatial interactions of the extended transcription network of these genes (20).
Although there are many similarities between the three genes regarding their nuclear (re)localization, there are also differences even when comparing iPS versus ES cells. One possible explanation for these differences, could be the inclusion of iPS cells in our data sets that were only partially reprogrammed although SSEA-1 positive.
Importantly, this computational analysis tool can be used for unbiased analysis of any topological change occurring in cells during cell cycle, differentiation or other physiological processes. Accompanying changes are taken into account as any observed data sets are tested against computer generated random distributions. This approach thus allows a rigorous test for functional topological relationships in cells and tissues.
Software
The software and audio video tutorial can be downloaded via: http://www.cardoso-lab.org/publications/Randomizer.zip
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (Cr59/28-1 to M.C.); (Ca198/7-1, Ca198/3-3, SFB740/TPA1 to M.C.C.); E-Rare EuroRETT network (BMBF) to M.C.C. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank R. Jaenisch for the generous gift of the transgenic MEF cells and C. Bonifer for the ES cells. We are indebted to H. Leonhardt for bringing together biology and informatics, J. Völger for her help, T. Cremer for the initial start-up of 3D-FISH analyses in iPS cells and D. Illner for experimental support. K.L.J., M.C.C. and M.C. designed the project; S.H. developed the software; J.M.S. and H.H. developed the statistical methodology; D.S. and N.S. carried out the cell culturing and FISH experiments; K.L.J. analysed the data; K.L.J and B.B. supported the software development; K.L.J., S.H., M.C. and M.C.C. wrote the manuscript.
REFERENCES
- 1.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin. Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 3.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 4.Harmon B, Sedat J. Cell-by-cell dissection of gene expression and chromosomal interactions reveals consequences of nuclear reorganization. PLoS Biol. 2005;3:e67. doi: 10.1371/journal.pbio.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 6.Deniaud E, Bickmore WA. Transcription and the nuclear periphery: edge of darkness? Curr. Opin. Genet. Dev. 2009;19:187–191. doi: 10.1016/j.gde.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Joffe B, Leonhardt H, Solovei I. Differentiation and large scale spatial organization of the genome. Curr. Opin. Genet. Dev. 2010;20:562–569. doi: 10.1016/j.gde.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Shirley JW, Ty S, Takebayashi SI, Liu X, Gilbert DM. FISH Finder: A High-Throughput Tool for Analyzing FISH Images. Bioinformatics. 2011;27:933–938. doi: 10.1093/bioinformatics/btr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gue M, Messaoudi C, Sun JS, Boudier T. Smart 3D-FISH: automation of distance analysis in nuclei of interphase cells by image processing. Cytometry A. 2005;67:18–26. doi: 10.1002/cyto.a.20170. [DOI] [PubMed] [Google Scholar]
- 10.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 11.Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- 12.Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cremer M, Grasser F, Lanctot C, Muller S, Neusser M, Zinner R, Solovei I, Cremer T. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol. Biol. 2008;463:205–239. doi: 10.1007/978-1-59745-406-3_15. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez RC, Woods RE. Digital Image Processing. 3rd edn. Upper Saddle River, N.J.: Prentice Hall; 2008. [Google Scholar]
- 16.Ronneberger O, Baddeley D, Scheipl F, Verveer PJ, Burkhardt H, Cremer C, Fahrmeir L, Cremer T, Joffe B. Spatial quantitative analysis of fluorescently labeled nuclear structures: problems, methods, pitfalls. Chromosome Res. 2008;16:523–562. doi: 10.1007/s10577-008-1236-4. [DOI] [PubMed] [Google Scholar]
- 17.Bartova E, Krejci J, Harnicarova A, Kozubek S. Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differentiation. 2008;76:24–32. doi: 10.1111/j.1432-0436.2007.00192.x. [DOI] [PubMed] [Google Scholar]
- 18.Hepperger C, Mannes A, Merz J, Peters J, Dietzel S. Three-dimensional positioning of genes in mouse cell nuclei. Chromosoma. 2008;117:535–551. doi: 10.1007/s00412-008-0168-2. [DOI] [PubMed] [Google Scholar]
- 19.Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J. Cell Sci. 2005;118:3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- 20.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.