Abstract
Influenza A virus segment 2 mRNA expresses three polypeptides: PB1, PB1-F2 and PB1-N40, from AUGs 1, 4 and 5 respectively. Two short open reading frames (sORFs) initiated by AUGs 2 and 3 are also present. To understand translational regulation in this system, we systematically mutated AUGs 1–4 and monitored polypeptide synthesis from plasmids and recombinant viruses. This identified sORF2 as a key regulatory element with opposing effects on PB1-F2 and PB1-N40 expression. We propose a model in which AUGs 1–4 are accessed by leaky ribosomal scanning, with sORF2 repressing synthesis of downstream PB1-F2. However, sORF2 also up-regulates PB1-N40 expression, most likely by a reinitiation mechanism that permits skipping of AUG4. Surprisingly, we also found that in contrast to plasmid-driven expression, viruses with improved AUG1 initiation contexts produced less PB1 in infected cells and replicated poorly, producing virions with elevated particle:PFU ratios. Analysis of the genome content of virus particles showed reduced packaging of the mutant segment 2 vRNAs. Overall, we conclude that segment 2 mRNA translation is regulated by a combination of leaky ribosomal scanning and reinitiation, and that the sequences surrounding the PB1 AUG codon are multifunctional, containing overlapping signals for translation initiation and for segment-specific packaging.
INTRODUCTION
Influenza A virus (IAV) is a major pathogen, capable of infecting a number of species including humans, birds, swine and horses. Its genome is contained on eight segments of negative sense viral RNA (vRNA), individually complexed with the trimeric viral polymerase (PB2, PB1 and PA) and nucleoprotein (NP) to form ribonucleoprotein (RNP) particles (1). On infection, the RNPs migrate to the nucleus where the polymerase initially transcribes the vRNA templates to produce mRNA, and later replicates the genome using positive sense cRNA intermediates (2). Subsequently, new vRNAs are exported from the nucleus (as RNPs) and packaged into progeny virus particles at the plasma membrane. As each segment encodes at least one essential gene product, a viable virus particle must contain one copy of each segment, which is facilitated via specific cis-acting packaging signals present in the terminal non-coding and coding regions of each vRNA (3). IAV strains also show considerable variation in pathogenicity, and the molecular mechanisms underlying this have not been fully elucidated. Segment 2 encodes PB1, the core component of the viral polymerase, which has been linked to inter-strain differences in pathogenicity and host range (4,5). However, the single mRNA species transcribed from the segment also encodes two further proteins that are non-essential for virus replication: PB1-F2 and PB1-N40 (6,7).
PB1-F2 is encoded by the +1 open reading frame (ORF) relative to PB1 and is initiated from AUG4 (Figure 1A). Depending on virus strain, the PB1-F2 ORF is up to 90 codons long, but in many viruses (including the recent pandemic H1N1 virus, where the gene is effectively absent), is truncated to variable extents by one or more stop codons (8,9). PB1-F2 polypeptides of 79 amino acids or longer can localize to mitochondria and the protein has been associated with pro-apoptotic and pro-inflammatory effects (6,9–12). A proportion of the protein also localizes to the nucleus where it interacts with PB1 and may influence polymerase activity (13,14). In some strains of virus, manipulating the expression or sequence of PB1-F2 altered replication and/or pathogenicity, leading to its identification as a virulence factor (6,10,14–17). However, in many cases, the presence or absence of an intact PB1-F2 ORF had little or no impact on virus replication in vitro or in vivo (7,8,17,18). Overall the contribution the protein makes to IAV pathogenesis is imperfectly understood.
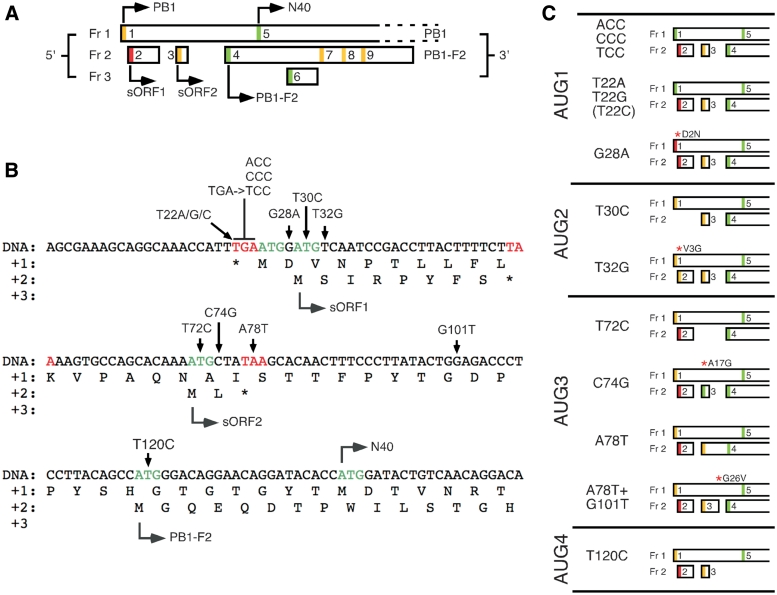
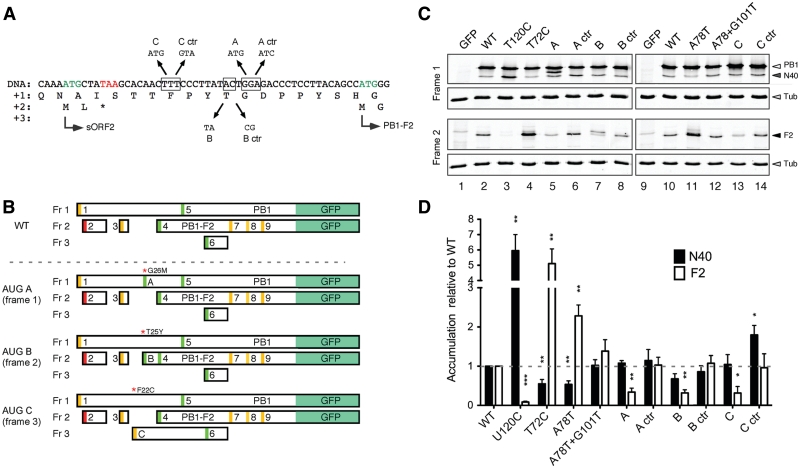
Figure 1.
Arrangement and sequence of ORFs in the 5′-end of segment 2 mRNA and mutants used in this study. (A) Schematic diagram of ORFs at the 5′-end of segment 2 mRNA with AUG codons numbered according to their position and shaded according to the strength of their Kozak consensus sequence (green, strong consensus, with A/G at −3 and G at +4; yellow, medium consensus with either A/G at −3 or G at +4; red is a weak consensus U at –3 and +4). Adapted from (7). (B) Nucleotide sequence and site of mutations used in this study. The 5′-end of segment 2 mRNA is shown in positive sense and as cDNA, since all mutations were introduced into a plasmid clone of the segment. (C) Summary of the predicted effect of the mutations used in this study on AUG strength and ORF structure (non synonymous changes in PB1 are indicated after red asterisks).
Recently, we showed that AUG5 of segment 2 is also used to initiate translation of a protein product called PB1-N40, made at ∼5% of the abundance of PB1 (7). AUG5 is in frame with AUG1, and so N40 is a truncated form of PB1, lacking the first 39 amino acids of the longer polypeptide (Figure 1A). The ‘missing’ region is important for the interaction of PB1 with PA (19), and therefore N40 should not be able to form the stable complex with PA necessary for efficient nuclear import and polymerase function (20,21). Indeed, N40 predominantly localized to the cytoplasm, and was not transcriptionally active (7). A function for PB1-N40 has not yet been identified, although PB1-N40 null viruses retaining an intact PB1-F2 ORF displayed delayed single cycle growth kinetics (7).
It has been suggested that leaky ribosomal scanning is responsible for PB1-F2 and PB1 N40 expression (6,7,17). In the scanning model of translation initiation, ribosomes bind to the 5′ end of mRNA and move along until they recognize a start codon (22). The sequence context of the AUG affects the probability that it will be recognized as a bona fide initiation codon; the Kozak consensus GCC(A/G)CCAUGG, is thought to be optimal, with a purine at −3 and G at +4 exerting the strongest effects (23,24). In support of the ribosomal scanning hypothesis, AUG1 is set in a medium strength Kozak consensus, lacking a purine at −3 (Figure 1A and B), while mutation of AUG4 has been shown to lead to up-regulation of N40 translation from AUG5 (7). However, the presence of two short ORFs (sORFs) initiated by AUGs 2 and 3 upstream of the PB1-F2 and N40 AUGs (Figure 1A and B) is suggestive of additional regulatory complexities. Furthermore, a previous study that investigated the effect of improving the Kozak consensus of AUG1 found little effect on PB1 levels in virus infected cells, and the authors suggested that start codon selection was not the primary control element for segment 2 translation (25). Thus unresolved questions remain over the control of segment 2 gene expression.
Here, we report a systematic investigation of the role of the first four AUG codons in segment 2 in directing viral protein synthesis. Our findings indicate a modified leaky scanning model in which translation initiation at internal start codons is influenced by upstream AUGs, but where sORF2 is a critical regulatory element that depresses PB1-F2 synthesis but promotes N40 translation through a reinitiation mechanism. Unexpectedly, we also found that the translational regulatory sequences surrounding AUGs 1 and 2 overlapped with sequences required for packaging of the segment into virus particles, providing an interesting insight into the evolutionary constraints acting on this section of the viral genome.
MATERIALS AND METHODS
Cells, plasmids and antisera
Human embryonic kidney 293T cells and Madin-Darby canine kidney (MDCK) cells were cultured by standard methods. For transfections, 293T cells were transfected in Optimem (Invitrogen) according to manufacturer's instructions using Lipofectamine 2000 (Invitrogen).
Plasmids pcDNA-PB2, -PA and -NP, containing cDNA copies of the influenza A/PR/8/34 (PR8) genes as well as plasmid pPolI-Flu-ffLuc containing an influenza virus-based luciferase minireplicon vRNA under the control of the human RNA polymerase I (Pol I) promoter have been previously described (7,26). Dual promoter reverse genetics plasmids for PR8 segments 1 and 3–8 and a pPol-I segment 7 clone were donated by Professor Ron Fouchier (27). A similar construct for segment 2 cloned from the NIBSC strain of PR8 is described in (7). To assess PB1-F2 expression in vitro, a CAT fragment was ligated into SmaI/XbaI digested pcDNA-PB1 in frames 1, 2 or 3. To analyse viral gene expression from transfected plasmids, nucleotides 1–380 of EF467819 were subcloned into pEGFPN1 (Clontech) as a AgeI/KpnI fragment. Site directed mutagenesis was then employed to position the green fluorescent protein (GFP) ORF into frame with either the PB1 or PB1-F2 reading frames while concurrently removing the GFP AUG codon. Additional segment 2 mutations as detailed in the results section were made using site directed mutagenesis with the wild-type segment plasmids as templates. For brevity, the sequences of the mutational oligonucleotides are not given but are available on request. All plasmids were sequence verified.
Rabbit polyclonal anti-PB1 serum V19 raised against amino acids 50–370 of PR8 PB1 has been previously described (28), as has rabbit polyclonal antiserum A2915 against PR8 NP (29). Rabbit antisera to the C-terminus of the PB1-F2 protein and to the full length PB1-F2 protein were kindly provided by Jonathan Yewdell. Rat monoclonal anti-tubulin YL1/2 was purchased from Serotec, anti-GFP mouse monoclonal JL8 from Clontech and IR800 or IR680 dye conjugated anti-rabbit IgG and anti-mouse IgG sera were purchased from LiCor.
Reverse genetics and virus titrations
Recombinant PR8 viruses were produced by transfection of plasmids into 293T cells in suspension as previously described (7,30). Rescued viruses were passaged once in MDCK cells at an input MOI of ≤ 0.001, and where indicated, once in 11-day-old embryonated eggs using an inoculum of 1000 PFU. Virus titres were determined by plaque assay on MDCK cells (30), and the presence of the desired mutations in segment 2 were confirmed by sequencing. Multiple independent rescues were performed (minimum twice, mostly 3–6 times) to ensure that a given phenotype did not result from adventitious mutations elsewhere in the virus genome. Virus infections of MDCK cells were performed at an MOI of 3–5 in serum free media for 30 min at 37°C, after which cells were overlaid with serum-containing media. Haemagglutination (HA) assays were performed as previously described (30).
Protein analyses
Coupled in vitro transcription–translation reactions were carried out in rabbit reticulocyte lysate using the Promega TNT system according to the manufacturer's instructions. SDS–PAGE followed by coomassie blue staining (to ensure equal loading of samples) and autoradiography was performed according to standard procedures. Blots were imaged using infrared fluorescence of appropriately tagged secondary antibodies and quantified using a LiCOR Odyssey scanner and software. Transcriptional activity of reconstituted RNPs was assessed using pPolI-Flu-ffLuc or pPol-I segment 7 as reporter plasmids. An amount of 50 ng of 3PNP and 20 ng of the reporter were transfected into adherent 293T cells and 48 h later, either luciferase levels from passively lysed cells were measured using a Promega GloMax luminometer or total RNA was extracted and segment 7 mRNA levels determined by reverse-transcriptase primer extension.
RNA analyses
The vRNA content of virus particles was determined by silver staining as previously described (30,31). Quantitative RT–PCR (qRT–PCR) for segments 2, 3, 5 and 7 was also performed on RNA extracted from equal PFU using the QIASymphony system (Qiagen) as previously described (30,31). Reverse transcriptase primer extension analysis of RNA from infected or transfected cells was performed as described (32), with the exception that SuperscriptIII (Invitrogen) was used and reverse transcription was performed at 50°C. Reaction conditions, primer and probe sequences are available upon request. Quantification was performed by densitometry of scanned X-ray films using Image J (Research Services Branch, NIH). Values were corrected with respect to a loading control (cellular 5S ribosomal RNA) and normalized to those of WT virus.
RESULTS
Segment 2 mRNA is known to encode three polypeptides: PB1 and PB1-N40 in frame 1, and PB1-F2 in frame 2 (6,7), translated from AUGs 1, 5 and 4 respectively (Figure 1A). The sub-optimal Kozak consensus flanking AUG1 prompted the hypothesis that the PB1-F2 ORF is accessed by leaky ribosomal scanning (6) and consistent with this, we previously showed that there was increased translation from AUG5 in the absence of AUG4 (7). However, there are two intervening start codons in frame 2 between AUG1 and the PB1-F2 ORF that initiate short sORFs with minimal protein coding capacity; eight and two codons respectively (Figure 1A and B). Nevertheless, both these AUGs are highly conserved, being present in >99% of the available segment 2 sequences, similar to the conservation shown by AUGs 4 and 5 for PB1-F2 and N40 respectively (Table 1). Notably, the termination codons for these sORFs are also highly conserved in that although three of the four overlapping PB1 codons in frame 1 are not themselves highly conserved at the RNA level, a stop codon is almost always (>99.9% for both sORF1 and 2) maintained in frame 2 (Table 2). This degree of conservation is suggestive of functional importance, potentially for the regulation of translation of the downstream PB1-F2 and N40 cistrons. Accordingly, we set out to further delineate the mechanisms controlling translation from segment 2 mRNA by systematically introducing mutations into AUGs 1–4 and their flanking regions that would be predicted to alter their usage.
Table 1.
AUG codons in the 5′-end of PR8 segment 2
| AUGa | Kozak contextb,d | Strengthc | MPDd | Frame/ ORFe | % AUGf |
|---|---|---|---|---|---|
| 1 | AUU UGA AUG G | Med | 0.03 | 1/PB1 | 100 |
| 2 | GAA UGGAUGU | Weak | 0.03/0.01 | 2/sORF-1 | 99.2 |
| 3 | CAC AAAAUG C | Med | 0.03 | 2/sORF-2 | 99.3 |
| 4 | ACA GCCAUG G | High | 0.01 | 2/PB1-F2 | 99.7 |
| 5 | UAC ACC AUG G | High | 0.05 | 1/PB1 | 99.6 |
aAUG number from 5′-end (Figure 1A).
bKozak consensus sequence (ideally GCCRCCAUGG) surrounding the AUG.
cThe initiation context of the AUG is scored as ‘weak’, ‘medium’ or ‘high’, depending on the identity of the critical nucleotides at positions −3 and +4.
dPB1 codons showing evidence of RNA-level conservation (41) are underlined and the normalized mean pairwise distance (MPD) score given. Low values indicate high conservation. Values calculated from an expanded database of 8686 unique full length segment 2 sequences available on Genbank in January 2011.
eThe reading frame and ORF which the AUG initiates or is in frame with.
fThe % of virus sequences that contain the particular AUG.
Table 2.
Conservation of segment 2 5′-sORF stop codons
| Noa. | Codonb 10 | Codon 11 | No |
|---|---|---|---|
| sORF 1 Stop codon: | |||
| 3660 | TTG (L) | AAA (K) | 6590 |
| 3309 | CTA (L) | AAG (K) | 2073 |
| 1491 | TTA (L) | AGA (R) | 10 |
| 218 | CTG (L) | AGG (R) | 3 |
| 2 | ATA (I) | AAT (N) | 1 |
| 2 | GTA (V) | ||
| 1 | CTT (L) | GTG (V) | 1 |
| 1 | TTT (F) | GAA (E) | 4 |
| 1 | TCG (S) | GAG (E) | 4 |
| 1 | CCG (P) | ||
| MPDc: | 0.87 | 0.73 |
| No. | Codon 18 | Codon 19 | No. |
|---|---|---|---|
| sORF 2 Stop codon: | |||
| 8677 | ATA (I) | AGC (S) | 6688 |
| 3 | GTA (V) | AGT (S) | 1995 |
| 1 | CTA (L) | AAC (N) | 1 |
| 2 | ATT (I) | GGT (G) | 1 |
| 2 | ATC (I) | TGC (C) | 1 |
| 1 | ACA (T) | ||
| MPD: | 0 | 0.77 |
aNumber of viruses with the listed codon. Codons are grouped into those that do and do not maintain the sORF stop codon (above and below the line respectively).
bPB1 codon number and (encoded amino acid).
cNormalized mean pairwise distance of the PB1 codon. Low values reflect high conservation in the data set (41).
The sequences surrounding AUG1 are highly conserved, and conform to a moderately strong initiation consensus (Table 1). Upstream residues were modified to each of the other possibilities at the crucial −3 position in the T22A, T22G and T22C mutants (Figure 1B), with the A/G changes but not the U→C alteration expected to result in increased ribosome recognition of the AUG (23,24) (Figure 1C). A further set of segment 2 AUG1 variants were produced on the background of the previous mutants in which the upstream residues at positions −1 and −2 were changed to match the canonical Kozak consensus (ACC, CCC, TCC mutants). The initiation context of AUG1 was also weakened by mutating the G at the +4 position to an A (G28A); this change also resulted in a non synonymous D2N change in the predicted PB1 translation product (Figure 1B and C). Similar approaches were taken to probe the function of AUG2 and 3 in regulating translation. The context of these initiation codons was improved by mutating the +4 nt to G (T32G and C74G respectively, which caused V3G and A17G changes in the PB1 ORF, respectively). Additionally, AUGs 2 and 3 were individually destroyed (T30C and T72C respectively) without altering the PB1 amino acid sequence. Two further mutations were made to examine the importance of AUG3/sORF2. A78T removes the stop codon from sORF2, and thus the predicted frame 2 protein product from this construct is an N-terminal fusion to PB1-F2. To investigate if the length and position of sORF2 was important (as in the case of a reinitiation event), a stop codon was re-introduced prior to AUG4, in the A78T+G101T mutant. This mutant left only 15 nt between the stop codon and AUG4, and also produced a G26V alteration in the PB1 sequence. Finally, AUG4 was removed by a T120C alteration, as used by many previous studies to ablate PB1-F2 expression (6,10,13,14,16,17,33,34). The positions of all mutations are shown on the PR8 segment 2 sequence in Figure 1B and their predicted effects on AUG context and ORF structure are summarized in diagrammatic form in Figure 1C.
Segment 2 polypeptide synthesis in vitro
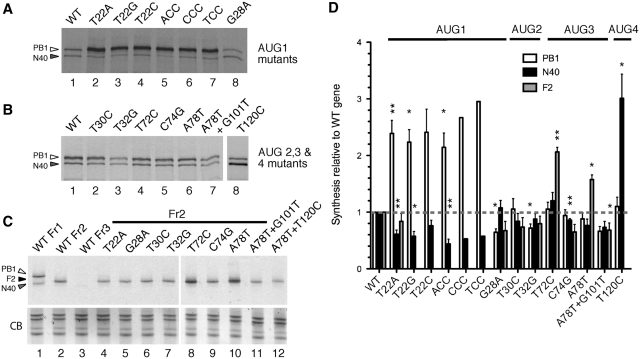
Initially, PB1 and N40 protein synthesis by the mutants was investigated using coupled in vitro transcription and translation (IVT) in rabbit reticulocyte lysate. As expected (7) wild-type (WT) PR8 segment 2 expressed both PB1 and N40, with preferential usage of AUG1 (Figure 2A, lane 1). All changes tested in the −3 to −1 positions relative to AUG1 lead to notable increases in the expression of PB1 (lanes 2–7). Quantification of replicate experiments showed these increases to be at least 2-fold relative to the WT gene (Figure 2D). Although this was the predicted outcome when the U at the −3 position was swapped to a purine, a similar effect when it was replaced with another pyrimidine was unexpected (23). Consistent with a role for leaky scanning in accessing downstream AUGs, a concomitant reduction in N40 levels was observed in all cases (Figure 2D). Conversely, weakening the Kozak consensus of AUG1 through replacement of the +4 G nucleotide downregulated PB1 expression, and although the absolute amount of N40 remained similar (Figure 2A, lane 8, quantification in Figure 2D), its ratio relative to PB1 was increased 1.7-fold compared to wild-type segment 2. In contrast, mutations affecting AUGs 2 or 3, whether by altering their context (T32G and C74G) or by destroying them (T30C and T72C) had little effect on the ratio of PB1 to N40 (Figure 2B; quantification in Figure 2D). Similarly, increasing the length of sORF2 (A78T+G101T) or fusing it with the PB1-F2 cistron (A78T) did not substantially alter relative use of AUGs 1 and 5. However, loss of the PB1-F2 AUG4 through the T120C mutation increased N40 synthesis from AUG5 by nearly 3-fold (Figure 2B, lane 8, Figure 2D). This recapitulated our previous observation for a construct in which AUG4 was mutated and its surrounding Kozak consensus disrupted [ΔAUG; (7)].
Figure 2.
Expression of segment 2 polypeptides in vitro. SDS–PAGE and autoradiographic analysis of aliquots of rabbit reticulocyte lysate supplemented with 35S-Methionine programmed with (A, B) full length clones of WT or mutant segment 2 clones as indicated or (C) segment 2 clones fused to the CAT gene in frames 1, 2 or 3 as shown. The migration of polypeptides of interest is indicated. The lower panel in (C) shows a portion of the gel stained with Coomassie Blue dye (CB) as a loading control. (D) Levels of PB1, PB1-N40 and PB1-F2 were quantified by densitometry and normalized to those of the WT construct. The mean ± SEM [n ≥ 3, with the exception of T22C (n = 2), TCC and CCC (n = 1)] are plotted. Asterisks indicate levels of significance based on P-values from a one sample t-test with the test value set to 1; *P < 0.05; **P < 0.01; ***P < 0.001.
The small size (∼10 kDa) of the PB1-F2 polypeptide made it difficult to visualize directly in IVT reactions. Accordingly, we utilized a set of constructs in which the CAT gene was fused downstream of AUG5 to increase the size of the polypeptide products. Examination of IVT reactions programmed with plasmids containing the CAT gene inserted into WT segment 2 in each of the three reading frames showed the expected set of polypeptides: frame one produced PB1 and PB1-N40 fusion proteins, frame 2 produced a PB1-F2 fusion while frame 3 lacked an obvious polypeptide product (Figure 2C, lanes 1–3 respectively). The quantity of the frame 2 product synthesized showed partial correlation with the presence and number of upstream AUG codons. For example, improving the Kozak consensus of AUG1 had only a small effect on PB1-F2-CAT synthesis (T22A: Figure 2C, lane 4; quantification in Figure 2D) and nor was F2 synthesis increased by the G28A mutation that weakened the context of AUG1 (Figure 2C, lane 5). Similarly, alteration of AUG2 by the T30C and T32G mutations had little effect (lanes 6 and 7). In contrast, the level of PB1-F2 was up-regulated 2-fold when AUG3 was removed via the T72C change (lane 8). The C74G mutation, predicted to improve the Kozak consensus of AUG3 caused a slight (<2-fold) reduction in F2 expression that was not statistically significant (lane 9). These results suggested that while AUG2 was of little translational significance, AUG3 was recognized well by ribosomes and directly regulated the level of PB1-F2 expression. Supporting this, when sORF2 was fused to the F2 ORF through removal of the intervening stop codon in the +2 frame by the A78T mutation, increased levels of a slightly longer (presumed) fusion protein were seen (lane 10). Thus overall, the data suggested that expression of PB1-F2 and PB1-N40 in vitro could be partially but not wholly explained by leaky ribosomal scanning. AUGs 1, 3 and 4 were clearly functional and exerted a significant influence on use of downstream start codons. The poor context AUG2 however, was apparently not used.
Generation of viruses with altered segment 2 translational regulatory sequences
Next, we wished to examine the behaviour of the mutant segment 2 genes in the context of virus infection. However, four of the mutants had non-synonymous changes in PB1 (G28A; D2N, T32G; V3G, C74G; A17G and A78T+G101T; G26V).
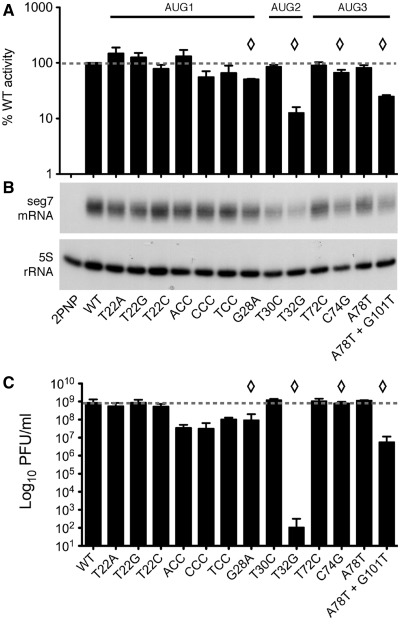
We therefore first tested the ability of the mutant PB1 polypeptides to support viral gene expression in ‘minireplicon’ assays (26,35). To reconstitute active viral RNPs, plasmids encoding the three influenza A virus polymerase proteins and nucleoprotein were co-transfected with a further plasmid that expressed a synthetic vRNA molecule encoding luciferase in antisense from an RNA polymerase I promoter. The luciferase levels in transfected cells therefore represent a measure of the transcriptional activity of the polymerase complex. When luciferase values were normalized to those obtained with WT PB1, all of the segment 2 mutants with unaltered PB1 sequences gave values that fluctuated around the 100% mark (between 62 and 129% of normal), while a sample from cells lacking PB1 gave <1% output (Figure 3A). However, three of the four non-synonymous changes in PB1 were deleterious to transcriptional activity. The T32G (V3G) and A78T+G101T (G26V) polymerases were the most impaired, with luciferase readings of 13 and 20% of WT respectively. The G28A mutant (D2N) was marginally impaired, producing luciferase activity at 54% of WT. Only the C74G mutant (A17G, a relatively conservative change) supported transcriptional activity in the range of that observed with the mutants with synonymous changes in PB1 (72%). The reduced ability of the T32G and A78T+G101T mutants to support virus gene expression was also seen when authentic segment 7 was used as a RNP substrate and M1 and M2 accumulation analysed by western blot (data not shown) or when unspliced segment 7 mRNA accumulation was measured by reverse transcriptase-primer extension assay (Figure 3B).
Figure 3.
Transcriptional activity and growth properties of the mutant segment 2 PB1 proteins and virus. (A, B) 293T cells were transfected with plasmids encoding PB2, PA, NP, and WT or mutant PB1 proteins (or as a negative control, lacking PB1; 2PNP), along with (A) a synthetic vRNA analogue containing a luciferase gene and the luciferase activity determined 48 h later or (B) a plasmid expressing authentic segment 7 vRNA followed by reverse transcriptase-primer extension analysis of segment 7 mRNA accumulation or (as a loading control), 5S rRNA. The data in (A) are the mean ± SEM of 4–6 independent experiments performed in duplicate, normalized to the values obtained using WT PB1. Mutants with non-synonymous changes in PB1 are indicated with diamonds. (C) The indicated viruses were rescued in 293T cells and amplified in MDCK cells before being titred by plaque assay on MDCK cells. Data shown are the mean ± SEM of at least two independent rescues.
WT and the 13 mutant viruses were therefore generated by transfection of cells with plasmids encoding the eight segments in cDNA form (27,30). Notwithstanding the reduced transcriptional activity associated with some of the non-synonymous changes in PB1, it was possible to rescue all of the mutants. Virus stocks were amplified in MDCK cells and titred as a first assessment of virus fitness (7,30,31). For the mutants with non-synonymous changes in PB1, the endpoint titres showed a strong correlation with polymerase activity in the minireplicon system. The most deficient virus in this system (T32G) rescued only once out of four attempts in 293T and MDCK cells, where it showed a six log10 growth defect relative to wild-type virus (Figure 3C). The A78T+G101T mutant was successfully rescued five times out of seven tested and reached endpoint titres of ∼1% of WT. All other mutants rescued on every attempt (between two and six times each). The G28A mutant, which had a 2-fold reduction in transcriptional activity, had a growth defect of ∼10-fold. Only the C74G virus grew similar levels as WT virus. However, not all viruses with unaltered PB1 coding sequences grew normally. Three of the mutant viruses with alterations around AUG1 (ACC, CCC and TCC), showed growth defects of between 8- and 20-fold relative to WT virus. In contrast, the T22A, T22G and T22C viruses grew normally, despite also having mutations to the upstream Kozak consensus of AUG1 that showed similar perturbations to segment 2 translation in the IVT system to the ACC, CCC and TCC changes. Similar relative results were also obtained when virus stocks were passaged in embryonated eggs, although here the T32G virus grew better, to ∼106 PFU/ml or 0.5% of the WT control (data not shown).
Segment 2 polypeptide synthesis in infected cells
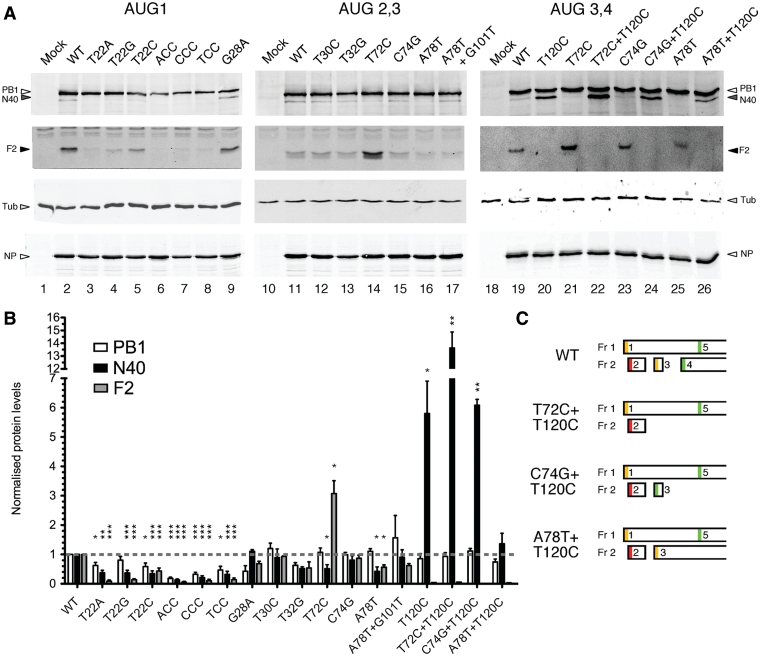
Low level expression of the replicative machinery is a general feature of many viruses and in some cases, mutations that result in overexpression of the viral polymerase have been shown to be deleterious to virus fitness (36,37). It was therefore possible that the poor replication of the ACC, CCC and TCC mutants resulted from overexpression of PB1, although as noted above, the T22A, T22C and T22G viruses did not show growth defects. More generally, we wished to compare segment 2 protein expression from recombinant and authentic viral settings. Accordingly, PB1, N40 and PB1-F2 accumulation in MDCK cells infected with the panel of viruses was examined at 8 h post-infection (p.i.) by western blotting. To provide numerical data, the levels of each protein from replicate experiments were quantified and normalized to levels in cells infected with WT PR8. The results obtained could be divided into those that were in concordance the previous in vitro analysis, and those that differed. In agreement, weakening the Kozak consensus of AUG1 (G28A) reduced PB1 accumulation relative to N40 and F2 expression (Figure 4A, compare lanes 2 and 9; quantification in Figure 4B). Similarly, mutation of AUG2 had little effect on either the quantity (T30C) or the relative ratios (T32G) of the segment 2 polypeptides (Figure 4A, lanes 12 and 13), with the lower levels of all three polypeptides seen with the latter virus being plausibly ascribed to the reduced polymerase activity of the mutant PB1 protein, as NP accumulation was also reduced. Also in agreement, loss of AUG4 increased N40 synthesis by ∼6-fold (T120C; lane 20), as we previously observed for a similar mutant virus (7).
Figure 4.
Expression of segment 2 polypeptides in virus-infected cells. (A) Lysates harvested at 8 h post-infection from MDCK cells infected (or mock infected) with the indicated viruses were analysed by SDS–PAGE and western blotting for the indicated proteins. Tubulin was used as a loading control. (B) Levels of PB1, PB1-N40 and PB1-F2 were quantified using LiCOR software and normalized to levels from WT infected cells. The mean ± SEM from at least two independent experiments are shown. (C) Predicted ORF structure of viruses with alterations to AUG3/sORF2 and AUG4 are shown. Asterisks indicate levels of significance based on P-values from a one sample t-test with the test value set to 1; *P < 0.05; P < 0.01; ***P < 0.001.
The first major discrepancy between the infection and in vitro data concerned the role of AUG3 in controlling expression of the downstream ORFs. The virus lacking AUG3 (T72C) showed a statistically significant 3-fold increase in PB1-F2 accumulation relative to WT (compare lanes 11 and 14), and this was consistent both with the in vitro data and the proposed role for leaky ribosomal scanning in accessing AUG4. Unlike in vitro however, there was a concomitant (and statistically significant) ∼2-fold reduction in N40 levels, despite normal PB1 accumulation. This result was not possible to reconcile with a model where leaky ribosomal scanning was the sole contributor to N40 expression, because removal of an upstream ORF should, at worst, leave N40 levels unchanged. Instead, we hypothesized that in vivo, ribosomes terminating at the end of sORF2 are able to reinitiate at AUG5, bypassing AUG4 because of the time taken to reacquire the necessary initiation factors (22). In the absence of AUG3, more ribosomes initiate at AUG4 due to leaky ribosomal scanning, and so F2 levels increase. However, no ribosomes are available to reinitiate at AUG5, and so N40 levels decline. Supporting this hypothesis, when sORF2 was fused to PB1-F2 (A78T), N40 levels were reduced >2-fold (Figure 4A, lane 16). This virus would also be unable to express N40 by reinitiation from sORF2, due to the removal of the stop codon. However, reintroducing a stop codon using the G101T mutation reinstated N40 levels to 90% of wild-type (lane 17).
To investigate the reinitiation hypothesis further, the AUG3/sORF2 mutations were made on a ΔAUG4 (T120C) virus background (Figure 4C). These viruses were rescued and grew to comparable titre to wild-type PR8 (data not shown). Western blotting was performed on cells infected with these viruses, as well as (for comparison) from cells infected with the ‘parental’ T72C, C74G, A78T and T120C. As before, preventing potential reinitiation from AUG3/sORF2, using either the T72C or A78T mutations in the presence of an intact AUG4 reduced levels of N40 (Figure 4A, compare lanes 19, 21 and 25), while improving the predicted strength of AUG3 had little effect (lane 23). Also as before, removal of AUG4 (T120C) led to a large increase in N40 levels (lane 20). A very similar outcome was obtained when the Kozak consensus of AUG3 was improved by the C74G mutation (C74G+T120C; lane 24). When AUG3 and 4 were removed concurrently (T72+T120C), synthesis of N40 was increased even further, to ∼14-fold greater levels than with the WT virus (Figure 4B lane 22, quantification in Figure 4B). However, when sORF2 was fused to the F2 ORF in the absence of AUG4, only a small enhancement (on average, 1.4-fold over WT) of N40 synthesis resulted (lane 26). If N40 expression was purely dependent on leaky scanning to bypass AUGs 3 and 4, this combination of mutations should have behaved identically to T120C alone. Instead, the absence of AUG4 only makes a significant difference to N40 expression when either sORF2 terminates before AUG5 or AUG3 is also absent, consistent with a reinitiation mechanism for synthesis of N40.
There were also discrepancies between virus infection and in vitro data for the AUG3/sORF2 mutant A78T virus regarding PB1-F2 expression, as the mutation decreased accumulation of the protein to ∼50% of normal instead of increasing it. Equally however, there was no evidence of expression of a larger form of PB1-F2 (Figure 4A, lane 16). Here, we surmise that the larger product was unstable in infected cells, and that the remaining F2 expression came from ribosomes initiating normally at AUG4 via leaky scanning. Consistent with this hypothesis, combining the A78T and T120C mutations (the latter removing AUG4) led to the loss of all detectable PB1-F2 accumulation (Figure 4A, lane 26).
If PB1-F2 is accessed by leaky scanning but N40 is accessed by reinitiation after translation of sORF2 then the insertion of further AUG codons in the region between the end of sORF2 and the beginning of the PB1-F2 ORF at AUG4 would be predicted to decrease F2 expression (through ‘soaking up’ initiation competent scanning ribosomes) but to have little effect on N40 expression because they would be effectively invisible to scanning small subunits that had terminated after reading sORF2 but not yet had time to acquire new initiation factors. To test this hypothesis, we introduced mutations that created new strong context AUG codons in each of the three reading frames in this region (Figure 5A). To permit the analysis of mutations that were lethal to virus growth and to try and minimize the effects of differing protein stability on polypeptide accumulation, we created sets of chimaeric plasmids containing the 5′-end of segment 2 encompassing the PB1-F2 coding sequence followed by a GFP ORF such that either PB1 (frame 1) or PB1-F2 (frame 2) were fused in frame (Figure 5B).
Figure 5.
Effect of novel AUG codons on expression of segment 2 polypeptides after plasmid transfection. (A) Sequence of mutations introduced between sORF2 and AUG4. (B) Diagrammatic summary of the predicted ORF structure of plasmids encoding fusions between the 5′-end of segment 2 and GFP in either frame 1 or frame 2. (C) Lysates from 293T cells harvested 48 h after transfection with the indicated plasmids infected were analysed by SDS–PAGE and western blotting for GFP. Tubulin was used as a loading control. (D) Levels of PB1-, PB1-N40- and PB1-F2-GFP fusion proteins were quantified using LiCOR software and normalized to levels from WT infected cells. The mean ± SEM from at least three independent experiments are shown. Asterisks indicate levels of significance based on P-values from a one sample t-test with the test value set to 1; *P < 0.05; **P < 0.01; ***P < 0.001.
To validate this system, we first retested the effect of the key mutations affecting sORF2 and AUG4. Cells transfected with a plasmid encoding the WT segment 2 fused in frame 1 to GFP produced the expected ratio of PB1 and PB1-N40-derived fusion proteins, while the frame 2 fusion construct produced the predicted PB1-F2-GFP fusion protein (Figure 5C, lane 2). As in the recombinant virus setting, mutation of AUG4 with the T120C change up-regulated N40 expression and abolished F2 expression (lane 3), while mutation of AUG3 by T72C had the opposite effect of increasing accumulation of the F2-GFP fusion but significantly decreasing N40 synthesis (lane 4, quantification in Figure 5D). Similarly, fusion of sORF2 and the F2 ORF by the A78T mutation significantly reduced N40 production (Figure 5C, compare lanes 10 and 11). In contrast to virus infection, this mutation also increased accumulation of the F2-fusion polypeptides, presumably because of the greater stability conferred by the GFP moiety. Both the decrease in N40 expression and the increase in F2 synthesis were reversed by reinstating the sORF2 stop codon by the further mutation G101T (lane 12). Thus this system successfully recapitulated the regulatory effects seen in the context of authentic virus infection.
Next, we tested the effect of introducing novel AUG codons between the termination codon of sORF2 and AUG4. Insertion of new AUG ‘A’ into the PB1 ORF by mutating glycine codon at position 26 to ATG resulted in the production of a prominent novel frame 1 product (‘PB1-N26’) as well as a significant reduction in the synthesis of the PB1-F2 fusion protein (Figure 5C, compare lanes 2 and 5, quantification data in 5D). N40 synthesis was however unaffected. These effects were specific to the creation of a new AUG codon, since mutation of codon G26 to ATC left expression of PB1-F2 unaltered (lane 6). Similarly, when new AUG codon ‘B’ was introduced into frame 2 by mutation of PB1 codon T25 to TAT (with the 5′-A→T change avoiding the simultaneous introduction of a stop codon in frame 3; Figure 5A), N40 expression was not significantly altered while F2 accumulation was substantially reduced, partially at the expense of a slightly longer N-terminally extended form (lane 7). Again, the effect was specific to the AUG codon rather than mutation of codon 25 per se, because its mutation to CGT (B ctr) left F2 expression unchanged. N40 expression was also insensitive to the introduction of an AUG codon into frame 3, whereas F2 accumulation was reduced >2-fold (codon ‘C’; lane 13). Once again, the paired control mutation had no affect on PB1-F2 synthesis, although unexpectedly, this change increased N40 accumulation (lane 14). Overall therefore, PB1-F2 levels were sensitive to the presence of start codons in all three frames following sORF2, whereas PB1-N40 levels were not significantly affected. These data show a fundamental difference in how AUG codons 4 and 5 are accessed: ribosomes can be diverted away from AUG4 by the insertion of new upstream AUG codons in the ‘UTR’ following sORF2, but AUG5 is insensitive to this approach. The simplest explanation consistent with the data is that AUG4 is primarily accessed by leaky ribosomal scanning that bypasses AUGs 1–3, while AUG5 is reached by reinitiation of ribosomes that have recently terminated synthesis after translation of sORF2.
Packaging of mutant segment 2 vRNAs into virus particles
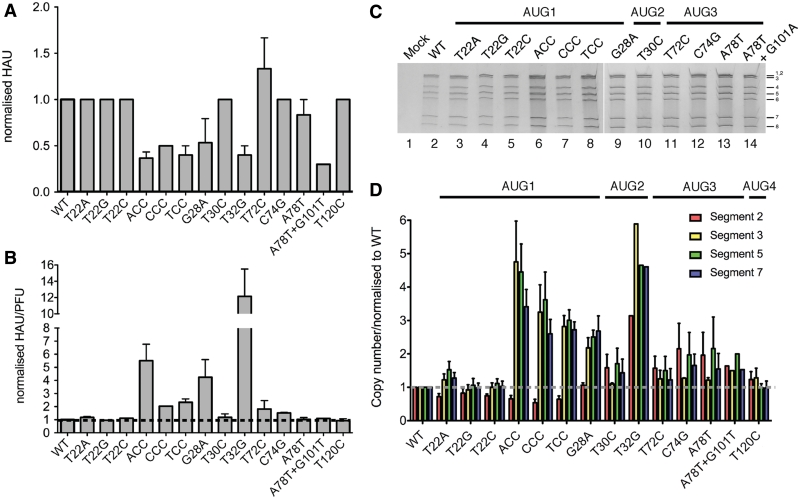
The other major source of divergence between the in vitro data and that observed from the virus infections was seen with the mutants where the Kozak consensus of AUG1 was up-regulated. While the levels of N40 and PB1-F2 were predictably reduced, in all cases, the cells infected with these mutants also underexpressed PB1 relative to the WT virus (Figure 4A, lanes 2–8), showing on average 20–80% reductions in PB1 accumulation (Figure 4B). This was most pronounced for the triple AUG1 mutants, ACC, CCC and TCC, which produced 18, 32 and 47% of the PB1 levels of WT virus respectively. This was in marked contrast to the in vitro translation data, where PB1 levels were increased ∼2-fold over WT (Figure 2). However, it should be noted that when the ratios of the three segment 2 polypeptides were considered, their relative amounts changed as predicted for leaky ribosomal scanning: N40 to PB1 levels were reduced between 2-fold (T22G) and 1.5-fold (ACC) in the AUG1 up mutants while PB1-F2: PB1 ratios decreased on average by ∼3-fold. Furthermore, transfection experiments confirmed that the AUG1 up mutations produced elevated amounts of PB1 in a cellular environment when introduced via plasmid (data not shown). We therefore considered the alternative hypothesis that in the background of authentic viruses, the AUG1 mutations also perturbed segment specific packaging. It is well established that the terminal unique coding and non-coding regions of all segments (including the regions of segment 2 under investigation here) contain specific packaging signals (3,38–42). In this hypothesis, the growth defect of the AUG1 mutants and their failure to express normal, let alone elevated quantities of PB1 could be explained by reduced delivery of segment 2 to the infected cells because of underincorporation of the segment into virions.
First, we measured virus particle formation by the panel of mutant viruses by HA assay. This showed only small fluctuations in particle assembly and release, with even the most replication deficient virus, T32G, showing on average, only a 4-fold drop in HA titre (Figure 6A). These data were then used to derive the proportion of infectious virus particles by calculating the ratio of HAU to PFU. By this measure, most mutants possessed values similar to that of the WT virus; the ACC, G28A and T32G viruses however had notably higher particle to infectivity ratios, indicating a large number of defective virions (Figure 6B).
Figure 6.
Particle:PFU ratio and vRNA content of WT and mutant virus particles. (A) Virus release was measured by HA assay. Values are plotted after normalization with respect to WT virus and are the mean ± SEM of at least two independent isolations, with the exception of T22G, T22C, CCC and A78T+G101T, which were only analysed once. (B) The ratio (normalized with respect to the WT virus) of HAU:PFU values are plotted, to provide a measure of the proportion of infectious virus particles. (C and D) RNA was extracted from equal PFU of the indicated viruses and (C) analysed by urea–PAGE and silver staining. The migration of individual segments is indicated. (D) Extracted RNA was analysed by one step qRT–PCR for segments 2, 3, 5 and 7. The copy numbers obtained for the mutant viruses were normalized to that of the WT virus to derive a relative segment copy number:PFU ratio. Data plotted are the mean ± SEM from at least two independent extractions, and for each extraction, the qRT–PCR reaction was performed in triplicate, with the exception of T32G and A78T+G101T, where RNA was extracted from a single rescue (the mean of triplicate determinations is plotted).
To examine genome packaging in the segment 2 mutant viruses directly, vRNA was extracted from equal plaque titres of virus. The segments were resolved and detected by Urea-PAGE and silver staining, and in all cases the expected pattern of seven vRNA segments were seen (Figure 6C; under these conditions, segments 1 and 2 comigrate). Obviously greater quantities of RNA were recovered from the ACC and TCC viruses (compare lanes 2, 6 and 8), a finding suggestive of an increased genome copy: PFU ratio and thus consistent with a raised virus particle: PFU ratio (30,31). However, the inability of this gel system to reliably separate the three largest genome segments hampered direct analysis of segment 2. In addition, the poor growth of the T32G virus made it difficult to extract sufficient vRNA to detect by this procedure (data not shown). We therefore used quantitative RT–PCR (qRT–PCR) to examine the copy number of segments in the mutant viruses. RNA was again extracted from equal PFU of virus and one step RT–qPCR was performed for segments 2, 3, 5 and 7. The amounts of each segment from the mutant viruses were normalized to that of the WT virus to derive a segment copy number:PFU ratio. The T30C AUG2 mutant and all AUG 3 and 4 mutants had similar levels of each of the segments tested to the WT virus, and also had equivalent amounts of each segment within each virus (Figure 6D). In contrast, most of the AUG1 up-mutants underincorporated segment 2. In addition, the ACC, CCC, TCC up-mutants as well as the AUG1 G28A and AUG2 T32G down-mutants showed several fold increases in the relative amounts of the other three segments. Since vRNA was extracted from equal numbers of infectious virus particles, these results are consistent with a specific packaging defect for segment 2 resulting in a higher number of defective virions and thus a higher segment copy number:PFU ratio of the other segments (30,31,42). This is consistent with the hypothesis that the failure of the AUG1 mutants with an improved Kozak consensus to express elevated quantities of PB1 in infected cells results from lower delivery of the segment by infecting virions.
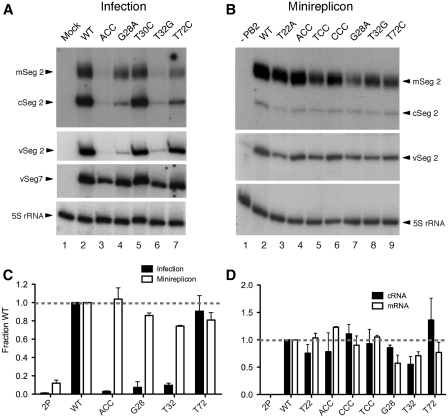
To further test this hypothesis, we analysed segment 2 RNA accumulation in cells infected with the ACC (as a representative of an AUG1 up-mutant), G28A and T32G viruses in comparison with WT and two mutant viruses (T30C and T72C) with no obvious packaging defects. All three RNA species (m-, c- and vRNA) were readily detectable in samples from cells infected with the WT, T30C and T72C viruses (Figure 7A, lanes 2, 5 and 7). However, the three viruses with apparent defects in segment 2 vRNA packaging produced much reduced quantities of vRNA and (with the exception of G28A), m- and cRNA also (lanes 3, 4 and 6). This defect was particularly apparent for segment 2, as more consistent levels of segment 7 vRNA were seen for all the viruses (Figure 7A). When replicate experiments were quantified, the three viruses with potential packaging defects produced <10% of the normal amount of segment 2 vRNA (Figure 7B).
Figure 7.
Accumulation of viral RNA in infected and transfected cells. Total cellular RNA extracted from 293T cells was analysed by reverse-transcriptase primer extension with radiolabelled oligonucleotides specific for the indicated RNAs followed by urea–PAGE and autoradiography. (A) RNA was harvested at 9 h post-infection from cells infected (or mock infected) with the indicated viruses. (B) RNA was extracted from cells 48 h after transfection with plasmids to recreate viral RNPs around the indicated segment 2 vRNAs. Lane 1 shows the background levels of vRNA produced from the segment 2 plasmid by cellular RNA PolI in the absence of a full viral polymerase complex. (C and D) Accumulation of the indicated RNAs was quantified by densitometry and normalized to the value obtained with the WT after correction with respect to the 5S ribosomal RNA loading control. The mean ± range of two independent experiments are plotted.
Although the above data were consistent with reduced delivery of the vRNA by the infecting viruses, we also considered the possibility that the AUG1/2 Kozak mutations perturbed the function of the viral RNA promoter (either the 3′-end of vRNA or the 5′-end of cRNA). This could lead to a reduction in segment 2 vRNA levels with a potential secondary effect of reducing the quantity available to be packaged into virions. Although the mutations lie well outside of the conserved promoter region, there are precedents for sequence alterations in the non-unique regions of a segment affecting RNA synthesis (30,43,44). To examine this possibility in isolation, the amount of segment 2 produced from RNPs reconstituted by transfection was measured. Wild-type PB2, PB1, PA and NP were transfected into cells with the reverse genetics plasmids encoding the mutant segment 2 vRNAs. The segment 2 plasmids would be transcribed by RNA Polymerase I to produce a negative sense segment 2 transcript that would be encapsidated, transcribed and replicated by the WT RNP proteins. In addition, mutant PB1 protein would be also be expressed from the vRNAs with non synonymous changes to the PB1 gene (G28A, T32G), but the addition of wild-type PB1 would be expected to at least partially compensate for this. Thus this system allowed us to examine viral RNA production from the mutant segments in isolation from potentially confounding issues of segment delivery and PB1 protein function. Forty-eight hours post-transfection, RNA was harvested and primer extension analysis for segment 2 v, m and cRNA was performed. Omitting PB2 from the transfections determined the baseline levels of segment 2 vRNA that were expressed from the pPolI promoter (Figure 7C, lane 1). In the presence of the full 3PNP complex, the mutant segment 2 constructs were transcribed and replicated to broadly similar extents (Figure 7C). When replicate experiments were quantified, the AUG1 mutants and the T32G AUG2 mutant accumulated vRNA to >75% of the WT level, in clear contrast to the >10-fold reductions they exhibited in the context of virus infection (Figure 7B). Similarly, all mutants expressed m- and cRNA to reasonable levels, with, on average, no change of >2-fold compared to the WT (Figure 7D). These data argue against a defect in the promoter sequence of the viral RNA being solely responsible for the reduced levels of viral RNA seen in the context of infection and support instead the hypothesis that mutations around AUG1 not only affect translation initiation of PB1 and downstream cistrons, but also affect genome packaging.
DISCUSSION
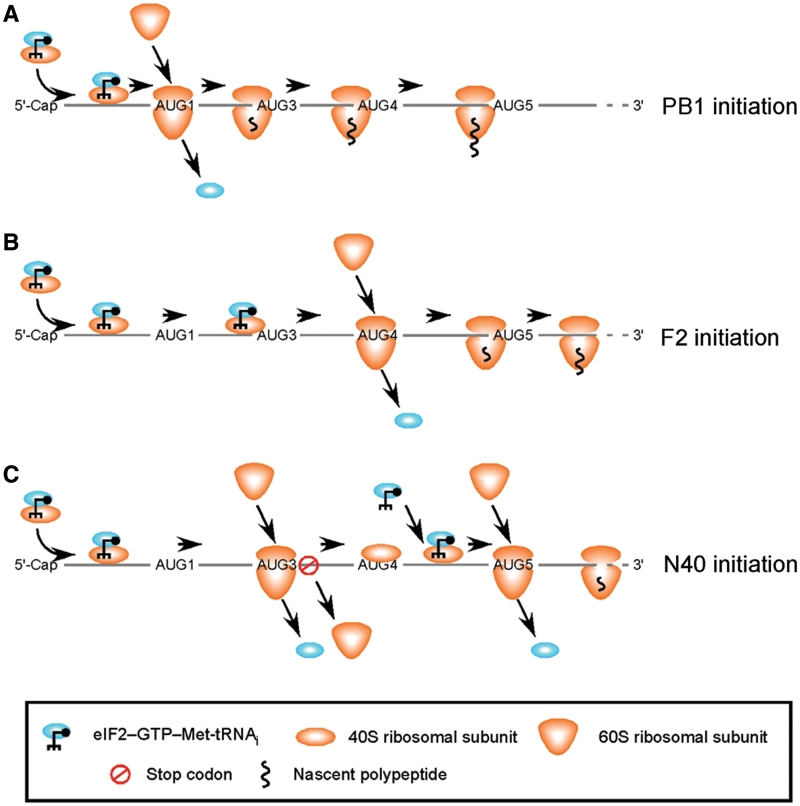
The single known species of segment 2 mRNA produces three proteins: PB1, PB1-F2 and PB1-N40. PB1 is an essential protein, encoding the potential antiviral drug target of an RNA polymerase, while PB1-F2 modulates pathogenicity in some host–virus combinations and the function of N40 is unknown. Despite representing the only known functionally tri-cistronic influenza virus mRNA, the mechanisms that control protein expression from the segment have not been fully elucidated. Here, we confirm the hypothesis that leaky ribosomal scanning has a role in mediating expression of PB1-F2 and PB1-N40. However, this mechanism does not fully explain segment 2 translation and we also identify ribosomal reinitiation after sORF2 as important for PB1-N40 expression.
Our data further refine the model for segment 2 protein expression. PB1 translation occurs via the canonical pathway of eukaryotic translation initiation (22) in which a preinitiation complex consisting of an eIF2α-ternary complex (eIF2-TC) attached to a 40S ribosomal subunit scans 3′-wards from the 5′-cap structure, recognizes AUG1 and commences translation after loss of the initiation factors and recruitment of the 60S subunit (Figure 8A). The simplest explanation for PB1-F2 expression is that it occurs via leaky ribosomal scanning, in which the preinitiation complex misses the moderate context AUGs 1 and 3 and the poor context AUG2 before initiating translation at the strong context AUG4 (Figure 8B). AUG3/sORF2 evidently plays an important role in down-regulating use of AUG4, as its loss through the T72 mutation substantially increased PB1-F2 accumulation, in vitro and in the context of virus infection. In contrast, the presence of AUG3/sORF2 up-regulated N40 expression in infected cells, a finding inconsistent with leaky scanning. Instead, we think this is best explained via leaky ribosomal scanning to bypass AUGs 1 and 2 followed by initiation at AUG3, almost immediate termination at the end of the two codon sORF2 and continued scanning of the 40S ribosomal subunit. The 40S subunit then scans past the strong context AUG4 but has time to reacquire an eIF2-TC before reaching the strong context AUG5 whereupon translation initiation occurs (Figure 8C). The distances between the sORF2 stop codon and AUGs 4 and 5 (40 and 63 nt respectively) are consistent with previously characterized instances of reinitiation (22,45–47). In some circumstances, changes in levels of eIF2-TC during conditions of cell stress (as for example when virus infection activates PKR) are known to regulate expression of downstream ORFs accessed via reinitiation strategies (22). It is therefore interesting to speculate that segment 2 translation might be further regulated during the course of infection.
Figure 8.
Model for translation of segment 2 polypeptides. PB1 translation occurs by canonical initiation at the first AUG. The majority of PB1-F2 translation occurs via leaky scanning to bypass AUGs 1–3. In contrast, reinitiation after termination at the end of sORF2 is a major contributor to PB1-N40 translation. See text for further details.
The distance between the extended sORF2 in the A78T mutant and the N40 AUG (39 nt) is similar to that between the normal sORF2 and the PB1-F2 AUG, so we would not rule out the possibility that reinitiation after sORF2 translation also contributes to F2 expression. However, shortening the intercistronic distance between sORF2 and the F2 ORF to 18 nt (a distance predicted to be too short to allow efficient reacquisition of an eIF2-TC) in the A78T+G101T mutant did not significantly reduce F2 accumulation, so we do not think it plays a major role.
Another major conclusion from this study is that the 5′-end of segment 2 mRNA itself has a number of overlapping functions. These include coding sequences critical for PB1 function, regulation of expression of downstream ORFs and also regions important for vRNA packaging. This has practical implications by reinforcing that this region represents an attractive target for therapeutic intervention, either by anti-viral drugs [e.g. those targeting the PB1–PA protein interface; (48)] or through T-cell epitope immunization (49), because the chance of finding escape mutations that maintain all functions of the protein/RNA sequence is likely to be lower than in a less functionally intricate area of the virus genome. Understanding the overlapping functional requirements also provides an interesting perspective on the evolutionary selection pressures that could be operating in this region of the influenza genome.
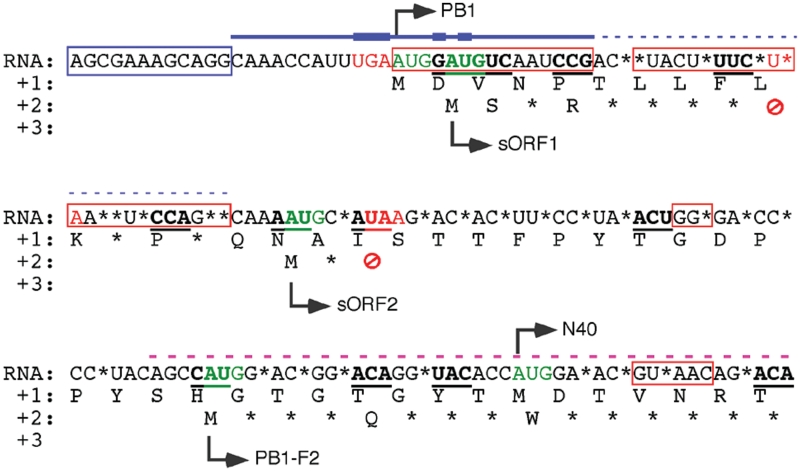
Packaging signals have been previously mapped to the general area of the 3′-end of segment 2 vRNA (38–41) (summarized in Figure 9) but this is the first study to show that the same nucleotides also contribute to translational regulatory sequences. This finding echoes our previous finding that sequences important for directing packaging of segment 7 overlap other cis-acting signals for mRNA splicing (30) and further demonstrates the functional complexities contained within sections of the influenza A virus genome. Examining the sequence of the 5′-end of segment 2 (in mRNA sense) using the criterion of reporting sequences that are conserved in >95% of the available isolates makes it evident that the primary selection pressure acting on the region is PB1 function (50). By this admittedly simple measure, only two amino acid residues (positions 12 and 14) are not conserved, in obvious contrast to PB1-F2 or sORF1 and sORF2. At the nucleotide level, as previously noted (50), it is clear that the majority of sequence polymorphisms are found at the third base position of the PB1 gene (Figure 9). Consistent with this, experimental evidence shows that the majority of the 14 N-terminal amino acids as well as (where tested) residues further downstream, are important for one or more functions of PA binding, polymerase activity and virus replication (21,51–53) (and data presented in this study). However, although over half of the first 41 codons of PB1 show some variability at the wobble position, it is notable that the primary translational signals within this region are much more highly conserved, with all five start codons showing >99% conservation and only one of the two stop codons (that of sORF1) showing apparent variation (Figure 9, Tables 1 and 2). Even here, as discussed, the variation is such that >99.9% of viruses maintain either a UGA or UAA stop codon (Table 2).
Figure 9.
Functional constraints on the 5′-end of segment 2. The 5′-end sequences (mRNA sense) of segment 2 are shown. Asterisks denote residues conserved in <95% of isolates. AUG codons are highlighted in green, stop codons in red and the beginning of ORFs are shown by arrows. Underlined codons in bold are those with evidence of RNA-level conservation (41). The blue box indicates the conserved promoter sequence, red boxes indicate PB1 sequences known from mutagenic and/or structural evidence to be important for polymerase function. The blue line indicates sequences important for segment-specific vRNA packaging, with the thick lines showing data from this study (Figure 6), medium weight from (38) and thin dashed line from (39). The purple dashed line indicates a region suggested to contain a human T-cell epitope (49).
That AUG1 is essential for PB1 expression is obvious; the moderate Kozak consensus surrounding it has presumably evolved to allow expression of one or more of the downstream ORFs via leaky scanning. This sequence element appears be additionally selected for via the contribution these nucleotides make to the segment 2 specific packaging signal. However, in light of the theory that RNA viruses gain additional genes through selection of unused or poorly expressed ORFs (54) and that a selective advantage for PB1-F2 or N40 is not always obvious (7–9,17,18,55), it is not clear which functional element came first. AUG2 or sORF1 seems to be of no significance as a translational element since modulation of the AUG made no difference to protein expression in vitro or in virus infected cells. Similarly, removal of the stop codon had no effect on segment 2 protein expression, genome packaging or virus replication (data not shown). The AUG may be retained because PB1 function requires an aspartate residue at position 2 [this study; (21,51,52,56)] and because the wobble position of codon 2 has become fixed through its secondary role in the segment packaging signal. Retention of the stop codon is more difficult to explain, although positions 10 and 11 require leucine and lysine respectively (21,51,52,56) and of the twelve possible permutations of this codon pair, only two do not result in a termination codon.
AUG5 may be maintained either because methionine 40 is essential for PB1 function and/or because expression of N40 supplies a selective advantage in vivo, for reasons as yet unknown. However, an isoleucine change at position 40 does not obviously inhibit PB1 transcriptase activity or inhibit virus growth in vitro or in eggs (6,7), perhaps favouring the latter hypothesis. AUG4 is presumably conserved at least in part to allow expression of PB1-F2, although it is less obvious what maintains it in the large number of viruses (9,57) that do not possess an intact F2 ORF. It does not seem to contribute to a packaging signal, so one possibility is that it is retained as a ‘ribosome sink’ to prevent overexpression of N40. AUG3 and the stop codon for sORF2 also seem likely to be conserved for a regulatory role: depressing PB1-F2 synthesis and/or permitting N40 expression. Since neither PB1-F2 or PB1-N40 is required for virus replication in cell culture, elucidating which (if either) of these roles is more important for maintaining virus fitness (as well as the wider question of their function in virus pathogenicity) will require either animal experiments and/or more sophisticated model systems for virus replication in vitro. Understanding the mechanisms that underlie F2 and N40 expression informs the design of virus mutants that could answer these questions.
FUNDING
Medical Research Council (grant numbers G0700815 to P.D. and G0701220 to E.C.A.); National Institutes of Health-Oxford/Cambridge Research Scholars Program (to B.W.J., P.D., and J.K.T.); and Intramural program of the NIH and the NIAID. Funding for open access charge: Lab funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Drs Nicole Robb, Ervin Fodor and John Kash for advice and discussion.
REFERENCES
- 1.Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 2.Neumann G, Brownlee GG, Fodor E, Kawaoka Y. Orthomyxovirus replication, transcription, and polyadenylation. Curr. Top. Microbiol. Immunol. 2004;283:121–143. doi: 10.1007/978-3-662-06099-5_4. [DOI] [PubMed] [Google Scholar]
- 3.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J. Gen. Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 4.Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7:440–451. doi: 10.1016/j.chom.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 2008;62:403–424. doi: 10.1146/annurev.micro.62.081307.162746. [DOI] [PubMed] [Google Scholar]
- 6.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 7.Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 2009;83:8021–8031. doi: 10.1128/JVI.00826-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hai R, Schmolke M, Varga ZT, Manicassamy B, Wang TT, Belser JA, Pearce MB, Garcia-Sastre A, Tumpey TM, Palese P. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 2010;84:4442–4450. doi: 10.1128/JVI.02717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zell R, Krumbholz A, Eitner A, Krieg R, Halbhuber KJ, Wutzler P. Prevalence of PB1-F2 of influenza A viruses. J. Gen. Virol. 2007;88:536–546. doi: 10.1099/vir.0.82378-0. [DOI] [PubMed] [Google Scholar]
- 10.McAuley JL, Chipuk JE, Boyd KL, Van De Velde N, Green DR, McCullers JA. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog. 2010;6:e1001014. doi: 10.1371/journal.ppat.1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamarin D, Garcia-Sastre A, Xiao X, Wang R, Palese P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005;1:e4. doi: 10.1371/journal.ppat.0010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs JS, Malide D, Hornung F, Bennink JR, Yewdell JW. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J. Virol. 2003;77:7214–7224. doi: 10.1128/JVI.77.13.7214-7224.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazur I, Anhlan D, Mitzner D, Wixler L, Schubert U, Ludwig S. The proapoptotic influenza A virus protein PB1-F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell Microbiol. 2008;10:1140–1152. doi: 10.1111/j.1462-5822.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- 14.McAuley JL, Zhang K, McCullers JA. The effects of influenza A virus PB1-F2 protein on polymerase activity are strain specific and do not impact pathogenesis. J. Virol. 2010;84:558–564. doi: 10.1128/JVI.01785-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3:1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAuley JL, Hornung F, Boyd KL, Smith AM, McKeon R, Bennink J, Yewdell JW, McCullers JA. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 2006;80:7976–7983. doi: 10.1128/JVI.00415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wanitchang A, Kramyu J, Jongkaewwattana A. Enhancement of reverse genetics-derived swine-origin H1N1 influenza virus seed vaccine growth by inclusion of indigenous polymerase PB1 protein. Virus Res. 2010;147:145–148. doi: 10.1016/j.virusres.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Perez DR, Donis RO. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J. Virol. 1995;69:6932–6939. doi: 10.1128/jvi.69.11.6932-6939.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J. Virol. 2004;78:9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez DR, Donis RO. Functional analysis of PA binding by influenza a virus PB1: effects on polymerase activity and viral infectivity. J. Virol. 2001;75:8127–8136. doi: 10.1128/JVI.75.17.8127-8136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 25.Maeda Y, Goto H, Horimoto T, Takada A, Kawaoka Y. Biological significance of the U residue at the -3 position of the mRNA sequences of influenza A viral segments PB1 and NA. Virus Res. 2004;100:153–157. doi: 10.1016/j.virusres.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Mullin AE, Dalton RM, Amorim MJ, Elton D, Digard P. Increased amounts of the influenza virus nucleoprotein do not promote higher levels of viral genome replication. J. Gen. Virol. 2004;85:3689–3698. doi: 10.1099/vir.0.80518-0. [DOI] [PubMed] [Google Scholar]
- 27.de Wit E, Spronken MI, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 2004;103:155–161. doi: 10.1016/j.virusres.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 28.Digard P, Blok VC, Inglis SC. Complex formation between influenza virus polymerase proteins expressed in Xenopus oocytes. Virology. 1989;171:162–169. doi: 10.1016/0042-6822(89)90523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J. Gen. Virol. 2007;88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 2008;82:11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hutchinson EC, Wise HM, Kudryavtseva K, Curran MD, Digard P. Characterisation of influenza A viruses with mutations in segment 5 packaging signals. Vaccine. 2009;27:6270–6275. doi: 10.1016/j.vaccine.2009.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robb NC, Smith M, Vreede FT, Fodor E. NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 2009;90:1398–1407. doi: 10.1099/vir.0.009639-0. [DOI] [PubMed] [Google Scholar]
- 33.Le Goffic R, Bouguyon E, Chevalier C, Vidic J, Da Costa B, Leymarie O, Bourdieu C, Decamps L, Dhorne-Pollet S, Delmas B. Influenza A virus protein PB1-F2 exacerbates IFN-{beta} expression of human respiratory epithelial cells. J. Immunol. 2010;185:4812–4823. doi: 10.4049/jimmunol.0903952. [DOI] [PubMed] [Google Scholar]
- 34.Chevalier C, Al Bazzal A, Vidic J, Fevrier V, Bourdieu C, Bouguyon E, Le Goffic R, Vautherot JF, Bernard J, Moudjou M, et al. PB1-F2 influenza A virus protein adopts a beta-sheet conformation and forms amyloid fibers in membrane environments. J. Biol. Chem. 2010;285:13233–13243. doi: 10.1074/jbc.M109.067710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang TS, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J. Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dinman JD, Ruiz-Echevarria MJ, Peltz SW. Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents. Trends Biotechnol. 1998;16:190–196. doi: 10.1016/S0167-7799(97)01167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plant EP, Rakauskaite R, Taylor DR, Dinman JD. Achieving a golden mean: mechanisms by which coronaviruses ensure synthesis of the correct stoichiometric ratios of viral proteins. J. Virol. 2010;84:4330–4340. doi: 10.1128/JVI.02480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, Huang T, Ly H, Parslow TG. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 2008;82:229–236. doi: 10.1128/JVI.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Y, Hong Y, Parslow TG. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 2005;79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gog JR, Afonso Edos S, Dalton RM, Leclercq I, Tiley L, Elton D, von Kirchbach JC, Naffakh N, Escriou N, Digard P. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marsh GA, Rabadan R, Levine AJ, Palese P. Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 2008;82:2295–2304. doi: 10.1128/JVI.02267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bergmann M, Muster T. Mutations in the nonconserved noncoding sequences of the influenza A virus segments affect viral vRNA formation. Virus Res. 1996;44:23–31. doi: 10.1016/0168-1702(96)01335-4. [DOI] [PubMed] [Google Scholar]
- 44.Zheng H, Palese P, Garcia-Sastre A. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology. 1996;217:242–251. doi: 10.1006/viro.1996.0111. [DOI] [PubMed] [Google Scholar]
- 45.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fouillot N, Tlouzeau S, Rossignol JM, Jean-Jean O. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 1993;67:4886–4895. doi: 10.1128/jvi.67.8.4886-4895.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang WL, Su TS. Translational regulation of hepatitis B virus polymerase gene by termination-reinitiation of an upstream minicistron in a length-dependent manner. J. Gen. Virol. 1998;79(Pt 9):2181–2189. doi: 10.1099/0022-1317-79-9-2181. [DOI] [PubMed] [Google Scholar]
- 48.Ghanem A, Mayer D, Chase G, Tegge W, Frank R, Kochs G, Garcia-Sastre A, Schwemmle M. Peptide-mediated interference with influenza a virus polymerase. J. Virol. 2007;81:7801–7804. doi: 10.1128/JVI.00724-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan PT, Heiny AT, Miotto O, Salmon J, Marques ET, Lemonnier F, August JT. Conservation and diversity of influenza A H1N1 HLA-restricted T cell epitope candidates for epitope-based vaccines. PLoS One. 2010;5:e8754. doi: 10.1371/journal.pone.0008754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holmes EC, Lipman DJ, Zamarin D, Yewdell JW. Comment on “large-scale sequence analysis of avian influenza isolates”. Science. 2006;313:1573. doi: 10.1126/science.1131729. author reply 1573. [DOI] [PubMed] [Google Scholar]
- 51.He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, et al. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature. 2008;454:1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 52.Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature. 2008;454:1127–1131. doi: 10.1038/nature07225. [DOI] [PubMed] [Google Scholar]
- 53.Biswas SK, Nayak DP. Influenza virus polymerase basic protein 1 interacts with influenza virus polymerase basic protein 2 at multiple sites. J. Virol. 1996;70:6716–6722. doi: 10.1128/jvi.70.10.6716-6722.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belshaw R, Pybus OG, Rambaut A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007;17:1496–1504. doi: 10.1101/gr.6305707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trifonov V, Rabadan R. The Contribution of the PB1-F2 protein to the fitness of Influenza A viruses and its recent evolution in the 2009 Influenza A (H1N1) pandemic virus. PLoS Curr. 2009;1:RRN1006. doi: 10.1371/currents.RRN1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wunderlich K, Mayer D, Ranadheera C, Holler AS, Manz B, Martin A, Chase G, Tegge W, Frank R, Kessler U, et al. Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication. PLoS One. 2009;4:e7517. doi: 10.1371/journal.pone.0007517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnitzler SU, Schnitzler P. An update on swine-origin influenza virus A/H1N1: a review. Virus Genes. 2009;39:279–292. doi: 10.1007/s11262-009-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]