Abstract
Among non-canonical DNA secondary structures, G-quadruplexes are currently widely studied because of their probable involvement in many pivotal biological roles, and for their potential use in nanotechnology. The overall quadruplex scaffold can exhibit several morphologies through intramolecular or intermolecular organization of G-rich oligodeoxyribonucleic acid strands. In particular, several G-rich strands can form higher order assemblies by multimerization between several G-quadruplex units. Here, we report on the identification of a novel dimerization pathway. Our Nuclear magnetic resonance, circular dichroism, UV, gel electrophoresis and mass spectrometry studies on the DNA sequence dCGGTGGT demonstrate that this sequence forms an octamer when annealed in presence of K+ or NH4+ ions, through the 5′-5′ stacking of two tetramolecular G-quadruplex subunits via unusual G(:C):G(:C):G(:C):G(:C) octads.
INTRODUCTION
G-quadruplexes are unique structures formed by Hoogsteen-type base pairing between four guanines and involving chelation of a metal ion. G-quadruplexes are typically formed by intramolecular folding of guanine-rich oligonucleotide (GRO) sequences or by intermolecular association of two or four sequences leading to the formation of dimeric or tetrameric complexes (1). The biological importance of these structures is 3-fold: (i) the occurrence of short G-rich sequences able to fold into G-quadruplex structures at the ends of telomeric DNA in eukaryotic chromosomes (2,3); (ii) the high prevalence of G-rich sequences in a large number of eukaryotic and prokaryotic genomes, and the increasing number of G-quadruplexes arising from these sequences (2,4,5); and (iii) their presence in the scaffold of several aptamers that have the ability to selectively bind to biologically relevant proteins and small molecules (6–10).
G-quadruplexes have the ability to form an array of conformations differing in structural features such as the molecularity, the relative orientation of the strands, the size of the loops connecting the strands and the glycosidic conformation of guanosine residues (syn or anti) (11). Further structural diversity arises from the quadruplex capacity to accommodate A-, T- and C-tetrads (12–14), as well as tetrads formed by modified residues (15,16). Moreover, in the last few years, several papers describing the higher order packing of intramolecular (17–19) or intermolecular (20,21) DNA quadruplexes have also been published. Particular attention has been devoted to the understanding of mechanisms responsible for the self-organization of human telomeric DNA into higher ordered G-quadruplex structures (17,19,22). Knowledge of such mechanisms is precious for shedding light on the in vivo role of quadruplex DNA as well as for the design of supramolecular DNA structures provided with novel functionalities. Understanding and controlling higher order structure formation is also of prime importance for in vitro studies of oligonucleotides derived from biologically relevant sequences, and for controlling the aggregation state of oligonucleotide aptamers aimed at therapeutic applications.
One of the identified structural moieties required for quadruplex dimerization is the presence of a stretch of guanines at the 5′- or 3′-termini of parallel G-quadruplex-forming GROs. These types of GROs have been reported to form higher ordered ‘dimer G-quadruplexes’ by either the association of interlocked slipped strands (23) or by end-to-end stacking (20,23). The presence of even a single base other than G at 5′- or 3′-termini has been reported to prevent the dimerization. In this context, we report here the results of our structural investigations on the sequence CGGTGGT under different conditions of oligodeoxynucleotide (ODN) concentration, pH and salts. We observe that the sequence folds into a ‘dimeric G-quadruplex’ (from now on 2Q), which is in fact an octamer of the sequence, coexisting in solution with minor amounts of other DNA secondary structures and with the single-stranded molecule. This higher ordered structure is obtained by head-to-head stacking between two G(:C):G(:C):G(:C):G(:C) octads, each belonging to one of the two ‘monomeric G-quadruplexes’ (1Q), as ascertained from the results described below.
MATERIALS AND METHODS
DNA sample preparation
DNA oligonucleotides TGGGGT, CTGTGTT, TGGTGGC, CGGTGGT and CGGGGT were chemically synthesized on an 8909 DNA/RNA synthesizer (Applied Biosystem), or purchased from Eurogentec (Belgium) for mass spectrometry experiments. DNA concentration expressed in strand molarity was calculated using a nearest-neighbor approximation for the absorption coefficients of the unfolded species. Ammonium acetate was obtained from a 5 M solution from Fluka (Molecular Biology grade). Stock solutions of 6 mM of each ODN were obtained by dissolving the lyophilized samples in 150 mM NH4OAc or 100 mM and 1.0 M K+ buffers (10 mM KH2PO4 supplemented with 90 mM or 990 mM KCl, respectively). Hundred percent D2O (Armar chemicals) or H2O/D2O (9:1, v/v) was used for the preparation of 6 mM solutions to be used for NMR experiments. All ODN samples were annealed by heating at 90°C for 15 min and then cooling and storing at 5°C. The pH was adjusted to 7.0 or 4.5 using HCl/KOH or AcOH/NH4OH aqueous solutions before the annealing procedure.
Polyacrylamide gel electrophoresis
The molecular size of structures were probed using non-denaturing polyacrylamide gel electrophoresis (PAGE) as previously described (24). ODN samples of 10 µM, obtained by diluting the annealed stock solutions just before the experiment, were loaded on a 15% polyacrylamide gel supplemented with 20 mM of KCl or NH4OAc. Glycerol of 10% was added just before loading. The gels were run at 26°C at constant voltage (100 V) for 2.5 h. The bands were visualized by UV shadowing and after ‘Syber green’ coloration.
Electrospray mass spectrometry
A Q-TOF Ultima Global mass spectrometer (Waters, Manchester, UK) was used to characterize the samples in ammonium acetate, and to perform a kinetics experiment. ODN samples (80 µM single-strand concentration), obtained as described in the PAGE paragraph with the addition of 10% methanol, were infused at 4 µl/min in the electrospray (ESI) source that was operated in negative ion mode (capillary voltage = −2.2 kV). Temperatures of ESI source and nitrogen desolvation gas were 80°C and 100°C, respectively. The source voltages were optimized for the best compromise to observe the single strand, tetramer and octamer: cone = 80 V, RF Lens 1 = 80 V, collision energy = 10 V and source pressure = 3.06 mbar. For the kinetics experiments, the reference strand dT6 was added to the dCGGTGGT strand, so as to follow the intensities of the reactant (dCGGTGGT single strand, noted G1), the intermediates (dimer = G2, trimer = G3, tetramer = G4 = 1Q) and the product (octamer = G8 = 2Q) relative to that of the reference. The method for obtaining relative response factors of the monomer, the tetramer 1Q and the octamer 2Q compared to the reference is described elsewhere (25), and the details of its application in the present case are given in Supplementary Data. In addition, a Bruker Apex-Qe9.4 T ESI-FTICR mass spectrometer was used to confirm the charge states from the isotopic distributions by recording high-resolution data, and to confirm the number of ammonium cations retained in each complex. The instrument voltages were chosen so as to minimize collision-induced dissociation and therefore keep the non-covalent complexes intact, including the inner ammonium cations: cap exit = −22 V, skimmer = −15 V, octapole offset = −4 V and coll cell trap = +4 V.
Circular dichroism
Circular dichroism (CD) spectra were measured at 25°C on a Jasco J-715 spectropolarimeter equipped with a Peltier Jasco JPT423S, using a 0.1-cm path length quartz cuvette with a reaction volume of 500 µl. CD spectra were recorded at different time points after the annealing on 40 µM ODN samples annealed at strand concentration of 200 µM or 6.0 mM. Spectra were averaged over three scans. A buffer baseline was subtracted from each spectrum and the spectra were normalized to have zero at 360 nm.
Nuclear magnetic resonance
Nuclear magnetic resonance (NMR) data were collected on a Varian UNITYINOVA 500 MHz spectrometer equipped with a broadband inverse probe with z-field gradient, and on a Varian UNITYINOVA 700 MHz spectrometer equipped with a triple resonance cryoprobe. The data were processed using the Varian VNMR software package. One-dimensional NMR spectra were acquired as 16 384 data points with a recycle delay of 1.0 s at 5, 25 and 65°C. Data sets were zero filled to 32 768 points prior to Fourier transformation and apodized with a shifted sine bell squared window function. Two-dimensional NMR spectra were all acquired at 25°C using a recycle delay of 1.2 s. NOESY spectra were acquired with mixing times of 100, 200 and 300 ms. TOCSY spectra were recorded with the standard MLEV-17 spin-lock sequence and a mixing time of 80 ms. For the experiments in H2O, water suppression was achieved by including a double pulsed-field gradient spin-echo (DPFGSE) module (26,27) in the pulse sequence prior to acquisition. In all 2D experiments, the time domain data consisted of 2048 complex points in t2 and 400 fids in t1 dimension. NMR samples were prepared at 6 mM strand concentration in 0.6 ml of 100 mM or 1.0 M K+ buffer obtained as above described. Sequence-specific resonance assignments for CGGTGGT were obtained by using NOESY and TOCSY spectra following the standard procedure (28,29). The self as well as sequential NOE connectivities from H8 to H1′ and H2′/H2″ were traced for all the nucleotides. The connectivities followed the standard patterns as for B-DNA, apart from noticeable exceptions between C1 and G2, and complete assignments of base and sugar protons was obtained.
Molecular modeling
The starting structure was generated using the builder module of Insight II (2005) program (Accelrys, Inc) on an Intel PC workstation running Red Hat Enterprise Linux. The model was parameterized according to the AMBER force-field, and calculations were performed using a distance-dependent macroscopic dielectric constant of 4.0 and an infinite cut-off for non-bonded interactions to partially compensate for the lack of solvent. The details of model building and energy minimization are given in Supplementary Data.
RESULTS
dCGGTGGT forms an octamer
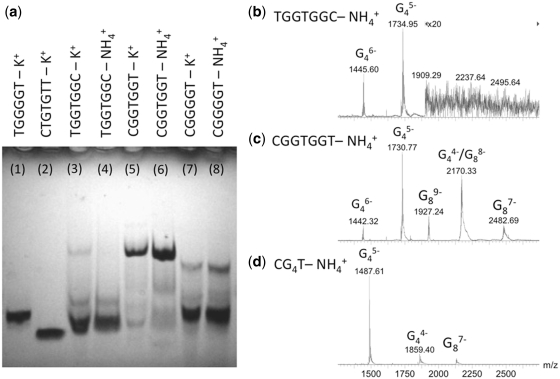
The formation of a higher order assembly than a tetramer by the DNA sequence dCGGTGGT was first noticed in native PAGE experiments. Figure 1a shows PAGE experiments on the following DNA sequences: TGGGGT annealed in 0.1 M potassium buffer (lane 1) used as a tetramolecular quadruplex ([dTGGGGT]4) size-marker, CTGTGTT (lane 2) used as a single-stranded 7-mer marker, TGGTGGC (lanes 3 and 4), CGGTGGT (lanes 5 and 6) and CGGGGT (lanes 7 and 8). The sequence TGGTGGC migrates similarly to [dTGGGGT]4; whereas, the sequence CGGTGGT shows a major band migrating much slower. We conclude that TGGTGGC forms prevalently a tetrameric assembly, in agreement with the reported tetramer G-quadruplex formation for this sequence (30), whereas CGGTGGT forms a higher order assembly. The polarity of the sequence therefore appears crucial in the higher order structure formation. The migration profile of dCGGGGT has also been investigated to determine whether the additional central thymine played any role in the higher order structure formation. Two bands are observed for dCGGGGT, the faster one migrating as the tetrameric [dTGGGGT]4, and the slower one migrating like the structure formed by CGGTGGT.
Figure 1.
Stoichiometry of self-assemblies determined by (a) PAGE for sequences TGGGGT (lane 1), CTGTGTT (lane 2), TGGTGGC (lanes 3 and 4), CGGTGGT (lanes 5 and 6) and CGGGGT (lanes 7 and 8), and by (b–d) ESI-MS for TGGTGGC (b), CGGTGGT (c) and CGGGGT (d). In the peak annotation Gnz-, n indicates the number of strands and z indicates the charge.
As the PAGE results are similar in NH4+ and in K+ cations, we therefore used ESI-MS, which can only be carried out in NH4+, to determine unambiguously the strand stoichiometry of the higher order assemblies formed by CGGTGGT and CGGGGT. The ESI-MS spectra of TGGTGGC, CGGTGGT and CGGGGT, each separately annealed at 6 mM in 150 mM NH4OAc (pH = 7.0) and diluted to 80 µM final strand concentration, are shown in Figure 1b–d, respectively. All three sequences show peaks corresponding to a tetramolecular assembly (noted G4), but the spectra of CGGTGGT and CGGGGT also display peaks corresponding to an octameric assembly (noted G8). We therefore assign the slow migrating PAGE band to an octameric self-assembly. In terms of relative intensities, although native PAGE and ESI-MS experiments were performed on samples obtained by dilution of the same annealed stock solution, the octamer is the darkest band in the gels of CGGTGGT (lanes 5 and 6), whereas the ESI-MS peaks of the octamer have lower intensities than the tetramer peaks. The analysis of the electrospray response factors in the kinetics ESI-MS experiments (see below and Supplementary Figure S2) indeed reveals the octamer has a lower ESI-MS response than the tetramer in the spectrometer used, and that the octamer is indeed the major species in solution.
The octamer contains stacked G-tetrads
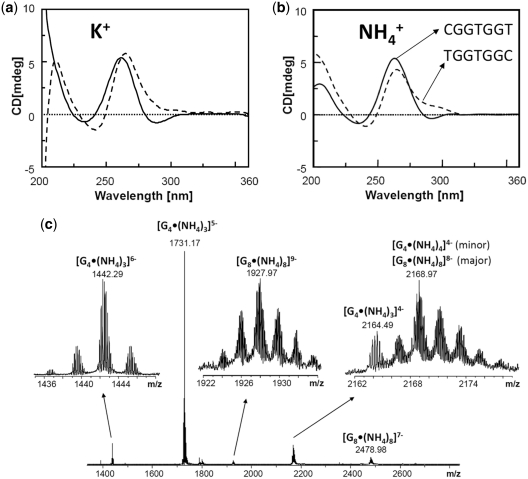
All G-rich sequences studied here are expected to form G-quadruplex structures. Note that ‘G-quadruplex’ means that the structure contains stacked G-quartets, not necessarily that the assembly is a tetramer. CD was used to probe the relative orientation of the bases in the tetramer [TGGTGGC]4 and in the octamer [CGGTGGT]8. The overall CD profiles recorded in K+ (Figure 2a) and NH4+ (Figure 2b) are in agreement with the formation of G-quadruplex structures (31,32) both in K+ and NH4+ media, showing a positive maximum at about 260 nm and a negative minimum close to 240 nm, which are characteristics of head-to-tail arrangement of guanines, as typically found in parallel oligodeoxynucleotides G-quadruplexes (33–35). Interestingly, the CD spectra of CGGTGGT also show a weak, but clearly visible negative band centered at 290 nm. This minimum suggests that additional base stacking interactions could be involved in the formation of the higher ordered quadruplex species by allowing the quadruplex multimerization via end-to-end stacking. A typical heteropolar stacking in G-quadruplexes formed by strands with no inverted polarity usually results in a positive band at 290 nm (34), but negative bands at 290 nm have been observed in octameric self-assemblies of lipophilic guanosine derivatives (36). The type of stacking present in our octamers could therefore resemble that present in those lipophilic octamers.
Figure 2.
G-quadruplex formation probed by (a and b) CD in 100 mM KCl (a) and 150 mM NH4OAc (b) of the sequences CGGTGGT (solid line) and TGGTGGC (dotted line), and by (c) counting the number of ammonium cations incorporated in the tetramer (noted G4) and the octamer (noted G8) of CGGTGGT using high resolution ESI-FTICR mass spectrometry.
Because the CD signature is peculiar and does not allow to conclude unambiguously to a G-quadruplex structure, we decided to probe cation incorporation into the octamer. Indeed, successive G-quartets incorporate monovalent cations (here K+ or NH4+) by coordination between eight guanines. High-resolution ESI-FTICRMS was used to count the number of ammonium cations per tetramer and per octamer (Figure 2c). Cation incorporation was found in all cases, confirming G-quadruplex formation both in the tetramer and the octamer. The major peaks correspond to three cations per tetramer [dCGGTGGT]4 and eight cations per octamer [dCGGTGGT]8. The number of cations is indicative of the number of stacked quartets. Interestingly, the tetramer [dCGGTGGT]4 contains three ammonium cations, whereas the tetramer [dTGGTGGC]4 contains four (data not shown), suggesting a structural difference depending on the strand polarity. However, the incorporation of eight ammonium cations in the octamer suggests that within the octamer, each tetrameric subunit is now capable of incorporating four ammoniums cations.
Influence of sample preparation procedure
In all experiments described above, the octamer was obtained by annealing the strand CGGTGGT at 6 mM single-strand ODN concentration at pH = 7.0 in 100 mM K+ or 150 mM NH4+. CD experiments carried out using a higher K+ concentration (1 M) show that higher cation concentration further favors the formation of the structure characterized by the negative band at 290 nm (Supplementary Figure S1). We also tested the influence of the solution pH, to test whether cytosine protonation might favor the formation of the new structure, but no clear influence of pH was found (see Supplementary Figure S1 and NMR results described below and in Supplementary Figure S4).
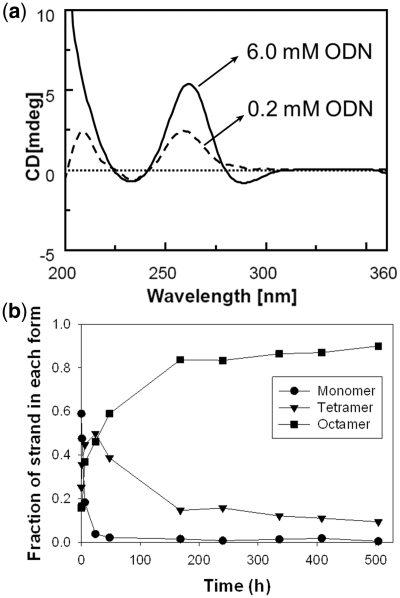
However, because of the octamer stoichiometry, we anticipated that the strand concentration upon annealing would have a great influence on the formation kinetics. Figure 3a shows the CD spectra recorded on CGGTGGT annealed at 6.0 mM or 0.2 mM ODN concentration in 1 M K+. The presence of the negative band at 290 nm was strictly dependent from ODN concentration: it is present when the sample is annealed starting from the 6 mM stock solution, whereas it is not observed when the sample is obtained by annealing the 0.2 mM ODN (Figure 3a). However, the CD spectrum of the species formed from the 0.2 mM ODN solution resembles that of a regular tetramolecular G-quadruplex. The influence of the strand concentration, therefore, indicates slow formation kinetics for the octamer.
Figure 3.
Influence of the sample preparation conditions on the octamer formation. (a) Influence of strand concentration: CD spectra of CGGTGGT in 1.0 M K+ buffer annealed, respectively, at 6 mM (solid line) and 200 µM (dotted line) single-strand concentration. (b) Kinetics of tetramer and octamer formation of 6 mM CGGTGGT in 150 mM ammonium acetate at room temperature, as determined by time-resolved ESI-MS.
To study the kinetics of octamer formation in the 6 mM stock solution, and to probe the nature of the reaction intermediates, we recorded ESI-MS spectra as a function of the time after addition of cation in the ODN solution, at room temperature. This experiment could be carried out only in ammonium acetate (150 mM). The details of the methodology, representative ESI-MS spectra at different reaction times, and the relative intensities are presented in Supplementary Figure S2. Briefly, the single-stranded dT6 strand (0.75 mM) is added to the 6 mM single-stranded dCGGTGGT in water. Quadruplex formation is initiated by the addition of ammonium acetate (final concentration: 150 mM) at room temperature. At each time point, an aliquot of the sample is diluted to 80 µM final dCGGTGGT single-strand concentration, and injected in the ESI-MS. We therefore assume that, once formed, the octamer resists the dilution step. The signal intensity of the reference is used to normalize the intensity variations of the dCGGTGGT monomer, tetramer and octamer signal, and deduce their respective response factors (see Supplementary Figure S2). Figure 3b shows the time evolution of the relative proportions of single strand, tetramer and octamer, after correction for the relative response of each species, expressed as the fraction of single strand in each form. The single strand is quickly converted into a tetramer, but the tetramer-to-octamer conversion is slower. The dimerization is therefore the rate-limiting step in the octamer formation. The results also show that, although the octamer is not the most abundant peak in the mass spectra (see Figures 1b–d and 2c), it is nevertheless the predominant species. The predominance of the octamer at long time scales also confirms that, once formed, the octamer resists the dilution step.
NMR investigation of the octamer structure
The 1H NMR spectrum of CGGTGGT in 100 mM K+ buffer at 25°C recorded 1 h after the annealing (Supplementary Figure S3, bottom left) is characterized by the presence of four well-resolved signals in the 11.0–11.6 ppm region attributable to the exchange-protected imino protons involved in the formation of Hoogsteen hydrogen bonds of four G-tetrads (37–39), in close analogy with what was previously observed for the parallel quadruplex structure formed by TGGTGGC (30). Furthermore, the count of quadruplex NMR signals is the same as what must be expected for the NMR spectrum of the corresponding single strand, thus suggesting the formation of a highly symmetric parallel G-quadruplex possessing a 4-fold symmetry. However, the aromatic region of 1H NMR spectrum is populated by more than the expected seven signals belonging to the H8/H6 aromatic protons of nucleobases. In order to discriminate whether the minor peaks were due to sample impurities or to the coexistence in solution of alternative minor conformations of CGGTGGT, we recorded the 1H NMR spectrum also at 65°C, and the high-temperature spectrum was clearly in agreement with the second hypothesis as evidenced by the presence of just seven well-defined signals in the aromatic region of the spectrum. We therefore studied the influence of time elapsed after the annealing (Supplementary Figure S3), of K+ concentration (Supplementary Figure S3), and of pH (Supplementary Figure S4) on the 1D NMR spectra. In agreement with the CD data, the NMR results confirmed that the best medium for the structural study of the new octameric structure formed by CGGTGGT requires 1.0 M K+ and long folding times (Figure 4a), and that the change of pH from 7.0 to 4.5 does not affect the folding of the structure.
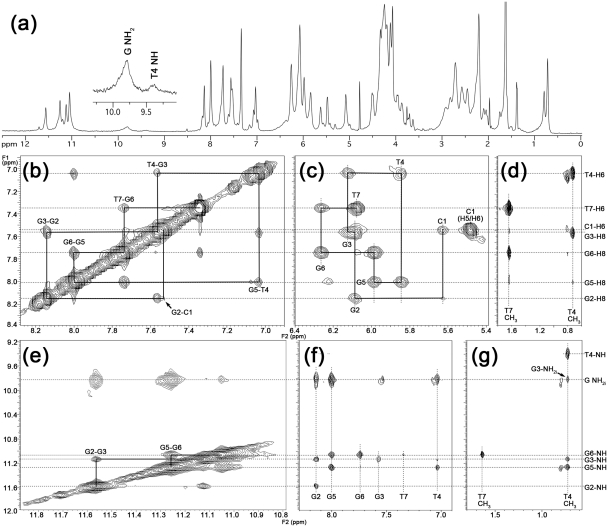
Figure 4.
Study of the octamer structure by NMR. (a) 1H-NMR spectra recorded at 25°C of CGGTGGT in H2O/D2O 9:1, annealed at 6 mM strand concentration in 1 M K+ at pH = 7.0. (b–g) Expansions of 2D NOESY spectrum of CGGTGGT (25°C, 200 ms mixing time, 1 M K+, pH = 7.0). The sequential H8-H6 and self-peaks involving H1′ protons have been labeled in (b) and (c), respectively. (d) and (g) show NOE cross-peaks involving T4/T7 methyl protons and H8/H6 or imino and amino protons, respectively. The T-tetrad diagnostic peaks between T4Me and T4NH and sequential T4Me-G3NH, T4Me-G3NH2i and T4Me-G5NH are observable in panel (g). Imino–imino NOE correlations are shown in (e). The sequential imino–imino NOE correlations have been drawn in (e). NOE correlations between aromatic H8/H6 and imino and amino protons are shown in (f).
The combined analysis of 2D NOESY and TOCSY spectra (700 MHz, 25°C, 1 M K+, 15 days after the annealing) allowed us to get the complete assignment (Table 1) of both exchangeable and non-exchangeable protons of CGGTGGT. The sequential connectivities between the aromatic protons (Figure 4b) were traced, as well as the self and sequential NOE connectivities between H8/H6 base protons and H1′, H2′, H2″and H3′ sugar protons. The intensities of intranucleotide H8/H1′ NOE cross-peaks (Figure 4c) indicate that all Gs adopt an anti glycosidic conformation. This finding together with the observation for each G-tetrad of a GiH8/GjN1H NOE cross-peak (Figure 4f), with i = j (where i and j represent the nucleotide labels), and with the observation of sequential G2N1H/G3N1H and G5N1H/G6N1H cross-peaks (Figure 4e), definitely allowed us to identify the presence of 4-fold symmetric parallel quadruplex moieties in the higher ordered structure formed by CGGTGGT when annealed in K+-containing buffer. In close analogy with what observed by Patel,P.K. and Hosur,R.V. for the quadruplex structure formed by TGGTGGC (30), we observed the formation of a central T-tetrad as ascertained by the appearance of the diagnostic T4NH/T4Me NOE cross-peak (Figure 4g) and supported by the observation of the strong shielding ring current experienced by T4 methyl protons that are significantly upfield-shifted when compared to the corresponding signals of the ‘free’ T7 (0.72 versus 1.64 ppm).
Table 1.
Proton chemical shifta (ppm) for 2Q at 25°C, pH 7.0, 1 M K+
| H6/H8 | H5/CH3 | NH | NH2ib | H1' | H2' | H2'' | H3' | H4' | H5'/H5'' | |
|---|---|---|---|---|---|---|---|---|---|---|
| C1 | 7.54 | 5.48 | 5.63 | 2.28 | 2.10 | 4.66 | 4.13 | 3.87–3.95 | ||
| G2 | 8.14 | 11.56 | 9.82 | 6.09 | 2.72 | 2.96 | 5.10 | 4.49 | 4.18 | |
| G3 | 7.56 | 11.13 | 9.80 | 6.12 | 2.55 | 2.94 | 5.07 | 4.50 | 4.30 | |
| T4 | 7.03 | 0.72 | 9.40 | 5.84 | 2.24 | 2.46 | 4.92 | 4.34 | 4.25 | |
| G5 | 8.00 | 11.26 | 9.82 | 5.98 | 2.72 | 2.84 | 5.10 | 4.49 | 4.20–4.30 | |
| G6 | 7.73 | 11.05 | 9.81 | 6.27 | 2.58 | 2.71 | 4.98 | 4.52 | 4.11–4.25 | |
| T7 | 7.34 | 1.64 | 6.08 | 2.73 | 2.21 | 4.51 | 4.08 | 4.25 |
aRelative to H2O at 4.78 ppm.
bNH2i refers to the internal (i) H-bonded amino protons of guanosines.
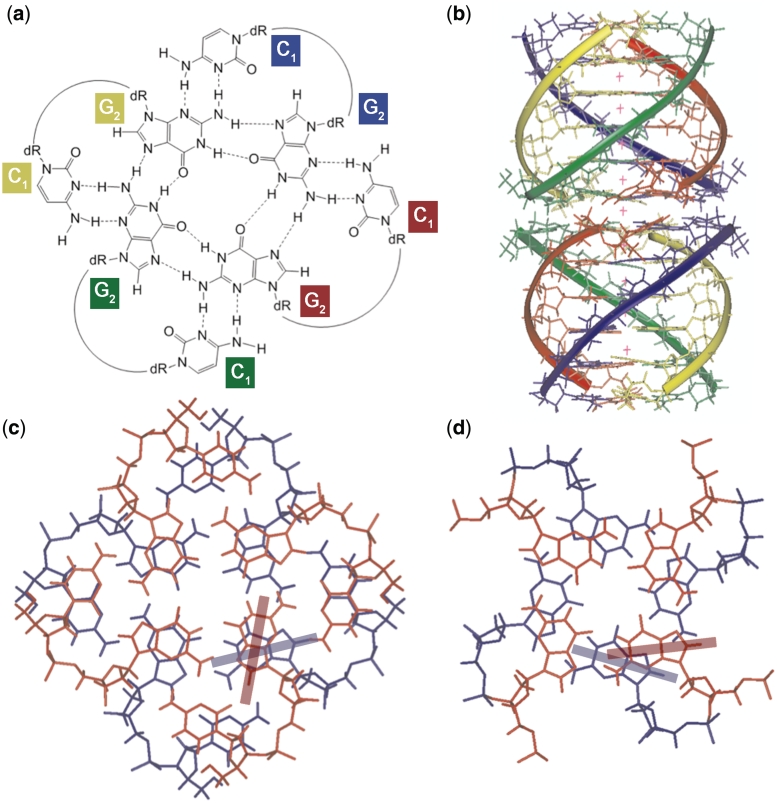
Although the overall observed NOEs account for a ‘classical’ tetramolecular parallel quadruplex, two peculiar NOE evidences drove us toward the disclosure of the higher ordered octamer structure. In Supplementary Figure S5, we show the 1D cross-section of the NOESY spectrum (200 ms mixing time) of CGGTGGT taken at 5.66 ppm (C1H1′ protons). This figure shows that the NOE between C1H1′ and C1H5 has almost the same intensity than that between C1H1′ and C1H6. These NOEs are in turn stronger than the sequential NOE C1H1′-G2H8, but weaker, as expected, than those between C1H1′ and self H2′/H2″. The relative intensities of the aforementioned NOEs involving C1H1′ require that the C1H1′-C1H5 distance is in the same range of the intranucleotide C1H1′-C1H6 distance (2.31–3.74 Å depending on the glycosyl torsion angle). This experimental distance is compatible only with the occurrence of an intermolecular dipolar coupling between C1 bases, because the corresponding intranucleotide C1H1′-C1H5 distance would be in the range 4.64–5.49 Å. This finding, together with the observation of the very weak sequential NOE between C1H6 and G2H8 (marked with the arrow in Figure 4b), disclosed that, in the octamer, each C1 base does not stack over the flanking G2 tetrad, as one may expect, but projects outside the quadruplex scaffold in such a way to allow dipolar couplings with one of the four C1 bases belonging to the other tetramer. This unusual arrangement might be favored by the formation of additional C1NH2–G2N3 and G2NH2–C1N3 hydrogen bonds, thus allowing the formation of an unusual G2(:C1):G2(:C1):G2(:C1):G2(:C1) octad (Figure 5a) in each of the two tetrameric quadruplexes (1Q).
Figure 5.
Structural model for the (dCGGTGGT)8 octamer. (a) Proposed structure of the G2(:C1):G2(:C1):G2(:C1):G2(:C1) octad. (b) Molecular model of the octamer: a different color was used for each ODN strand, and potassium ions appear as red crosses. (c) Detail of stacking of the octads. (d) Detail of stacking of G2–G3 tetrads. The transition moments of guanosines corresponding to the absorption band at approximately 250 nm are shown as colored bars in (c) and (d).
DISCUSSION
The structure adopted by the sequence dCGGTGGT must account for the following observations. It is a highly symmetrical octamer, formed from two parallel-stranded tetramers. In the octamer, each tetramer contains a central T-tetrad and the G-tetrads are stacked in head-to-tail arrangement. However, at the octamer’s interface between the two tetramers, the stacking of the G-tetrads is likely to be different, as determined from CD.
Interlocked structures like that formed by the (dGCGGTGGT)4 tetramer (40) were initially taken in consideration to build an octamer model, but were discarded due to NMR inconsistencies and symmetry-related considerations. Unresponsiveness to pH changes also led us to discard structures involving protonated cytosines. The NOEs involving C1H1′ point to the formation of a G2(:C1):G2(:C1):G2(:C1):G2(:C1) octad (Figure 5a) in each of the two tetrameric quadruplexes. The formation of the planar octad system would allow for further π–π stacking between C1 bases, thus adding more ‘hydrophobic glue’ between the two G-quadruplex tetramers that form the octamer.
Figure 5b shows the energy minimized model of the octamer formed by stacking of two tetramers, each containing a 5′- G(:C):G(:C):G(:C):G(:C) octad. The details of molecular modeling are given in Supplementary Figure S6, and additional ribbon and space-filled views of the model are shown in Supplementary Figure S6. The overall topology of each tetramer is largely similar to that of the tetramolecular quadruplex formed by TGGTGGC (see Figure 8 in reference 30), with the central T-tetrad causing a small decrease in the intertwining of the helices. Space filling views (Supplementary Figure S6) also evidence that the octamer structure is very compact, regardless the presence of two stacked octads. These are seen to determine only a slight increase in the thickness of the octamer at the dimerization interface.
Figure 5c–d also shows the head-to-head stacking between the two octads (c) and the stacking between tetrads G2-G3 (d). The stacking between tetrads G3-T4 and T4-G5 steps is shown in Supplementary Figure S7. Although C1 and G2 bases involved in the stacked octads appear slightly rotated with respect to the tetrad plane, Figure 5c shows good stacking between the six-membered rings of G2 bases and between the stacked C1 bases. The electric transition moments corresponding to the guanine absorption band at approximately 250 nm are indicated in Figure 5c and d. The stacking interactions in the model allow to explain the CD data: G2-G3 stacking resembles the standard head-to-tail stacking encountered in parallel-stranded G-quadruplexes. However, the octad stacking gives rise to a heteropolar stacking that is distinct from those typically encountered in antiparallel G-quadruplexes. We, therefore, interpret the negative CD band at 290 nm as being due to the octad stacking shown in Figure 5c (36). Interestingly, a similar weak negative CD band at 290 nm was also observed, but not discussed, by Mastugami et al. (41) in the dimer formed by d(GGAGGAGGAGGA), which contains stacked G(:A):G(:A):G(:A):G heptads at the dimerization interface.
Our results therefore suggest that, like adenine, cytosine is also capable of inducing the formation of higher order structures by the formation of octads. One must note, however, that the formation of the octamer by the sequence dCGGTGGT is slow. ESI-MS kinetics experiments in ammonium acetate provide some insight into the mechanism of octamer formation. The first step is the formation of a tetramer that incorporates only three ammonium ions. This means either that the tetramer of [dCGGTGGT]4 has either a central or a terminal tetrad disrupted, or that the tetramer contain slipped strands and hence incomplete tetrad formation (42). The rate-limiting step is the conversion of tetramer to octamer, and in contrast with the tetramer, the octamer incorporates four cations per tetrameric subunit. We therefore conclude that the rate-limiting step is the rearrangement of the tetramer into a fully aligned parallel structure that is amenable to octad formation and stacking thereof. Further experiments with various sequences are underway to probe the effect of the central T-tetrad on stability and rate of formation of octamers.
CONCLUSIONS
In summary, the present study demonstrates that the formation of a quadruplex dimer (2Q) can occur when the 5′-cytosine-ending CGGTGGT DNA sequence, but not the inverted TGGTGGC sequence, is annealed in the presence of K+ or NH4+ ions. The NMR and molecular modeling data disclosed that the formation of 2Q is allowed by the formation of an unusual octad arrangement, in each of the two 1Q units, involving C1 and G2 bases of each CGGTGGT strand. Up to now, dimerization of G-quadruplexes had only be reported via guanine–adenine heptads (41), and recently via a shared uracil tetrad (43). Our finding of G-quadruplex dimerization with 5′-cytosine containing strands adds further possibilities for the design of novel quadruplex-based DNA functional nanostructures, and further complexity to the mechanisms of folding and assembly of G-rich sequences. It is particularly important for in vitro studies of artificially designed (e.g. aptamers) or biologically relevant sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian M.U.R.S.T. (P.R.I.N. 2007); the Fonds de la Recherche Scientifique—FNRS (CC.1.5.286.09.F to V.G., research associate position to V.G.); University of Liège (starting grant FRSD-08/10 to V.G.); Walloon Region (Project FEDER FTICR). Funding for open access charge: Italian M.U.R.S.T. (P.R.I.N. 2007).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The European Commission through the COST Action MP0802 (European Network on G-quartets and Quadruplex Nucleic Acids) is gratefully acknowledged. The authors are also grateful to the Centro di Servizio Interdipartimentale di Analisi Strumentale of the University of Napoli Federico II for NMR facilities and to Dr Luisa Cuorvo and Dr Frédéric Rosu for technical assistance.
REFERENCES
- 1.Parkinson GN. In Quadruplex Nucleic Acids. London, UK: RSC Publishing; 2006. Fundamentals of quadruplex structures; pp. 1–30. [Google Scholar]
- 2.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riou J-F, Gomez D, Morjani H, Trentesaux C. In Quadruplex Nucleic Acids. London, UK: RSC Publisher; 2006. Quadruplex ligand recognition: biological aspects; pp. 154–179. [Google Scholar]
- 4.Dexheimer TS, Fry M, Hurley LH. In Quadruplex Nucleic Acids. London, UK: RSC Publisher; 2006. DNA quadruplexes and gene regulation; pp. 180–207. [Google Scholar]
- 5.Huppert J. In Quadruplex Nucleic Acids. London, UK: RSC Publisher; 2006. Quadruplexes in the genome; pp. 208–227. [Google Scholar]
- 6.Levesque D, Beaudoin J-D, Roy S, Perreault J-P. In vitro selection and characterization of RNA aptamers binding thyroxine hormone. Biochem. J. 2007;403:129–138. doi: 10.1042/BJ20061216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pileur F, Andreola ML, Dausse E, Michel J, Moreau S, Yamada H, Gaidamarov SA, Crouch RJ, Toulmé JJ, Cazenave C. Selective inhibitory DNA aptamers of the human RNase H1. Nucleic Acids Res. 2003;31:5776–5788. doi: 10.1093/nar/gkg748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang KY, McCurdy S, Shea RG, Swaminathan S, Bolton PH. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 9.Chou S-H, Chin K-H, Wang AH-J. DNA aptamers as potential anti-HIV agents. Trends Biochem. Sci. 2005;30:231–234. doi: 10.1016/j.tibs.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Girvan AC, Teng Y, Casson LK, Thomas SD, Jüliger S, Ball MW, Klein JB, Pierce WM, Barve SS, Bates PJ. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 11.Phan AT, Kuryavyi V, Luu KN, Patel DJ. In Quadruplex Nucleic Acids. London, UK: RSC Publisher; 2006. Structural diversity of G-quadruplex scaffolds; pp. 81–99. [Google Scholar]
- 12.Bhavesh N, Patel P, Karthikeyan S, Hosur R. Distinctive features in the structure and dynamics of the DNA repeat sequence GGCGGG. Biochem. Biophys. Res. Commun. 2004;317:625–633. doi: 10.1016/j.bbrc.2004.03.095. [DOI] [PubMed] [Google Scholar]
- 13.Oliviero G, Amato J, Borbone N, Galeone A, Varra M, Piccialli G, Mayol L. Synthesis and characterization of DNA quadruplexes containing T-tetrads formed by bunch-oligonucleotides. Biopolymers. 2006;81:194–201. doi: 10.1002/bip.20399. [DOI] [PubMed] [Google Scholar]
- 14.Searle MS, Williams HEL, Gallagher CT, Grant RJ, Stevens MFG. Structure and K+ ion-dependent stability of a parallel-stranded DNA quadruplex containing a core A-tetrad. Org. Biomol. Chem. 2004;2:810–812. doi: 10.1039/b314559j. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Zhang RL, Min JM, Zhang LH. Studies on the synthesis of a G-rich octaoligoisonucleotide (isoT)2(isoG)4- (isoT)2 by the phosphotriester approach and its formation of G-quartet structure. Nucleic Acids Res. 2002;30:3005–3014. doi: 10.1093/nar/gkf395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virgilio A, Esposito V, Randazzo A, Mayol L, Galeone A. Effects of 8-methyl-2′-deoxyadenosine incorporation into quadruplex forming oligodeoxyriboncleotides. Bioorg. Med. Chem. 2005;13:1037–1044. doi: 10.1016/j.bmc.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 17.Haider S, Parkinson GN, Neidle S. Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys. J. 2008;95:296–311. doi: 10.1529/biophysj.107.120501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Ishizuka T, Kurabayashi K, Komiyama M. Consecutive formation of G-quadruplexes in human telomeric-overhang DNA: a protective capping structure for telomere ends13. Angew. Chem, Int. Ed. 2009;48:7833–7836. doi: 10.1002/anie.200903858. [DOI] [PubMed] [Google Scholar]
- 20.Kato Y, Ohyama T, Mita H, Yamamoto Y. Dynamics and thermodynamics of dimerization of parallel G-quadruplexed DNA formed from d(TTAGn) (n = 3–5) J. Am. Chem. Soc. 2005;127:9980–9981. doi: 10.1021/ja050191b. [DOI] [PubMed] [Google Scholar]
- 21.Sotoya H, Matsugami A, Ikeda T, Ouhashi K, Uesugi S, Katahira M. Method for direct discrimination of intra- and intermolecular hydrogen bonds, and characterization of the G(:A):G(:A):G(:A):G heptad, with scalar couplings across hydrogen bonds. Nucleic Acids Res. 2004;32:5113–5118. doi: 10.1093/nar/gkh849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collie GW, Parkinson GN, Neidle S, Rosu F, De Pauw E, Gabelica V. Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures. J. Am. Chem. Soc. 2010;132:9328–9334. doi: 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan-Ghosh Y, Liu D, Balasubramanian S. Formation of an interlocked quadruplex dimer by d(GGGT) J. Am. Chem. Soc. 2004;126:11009–11016. doi: 10.1021/ja049259y. [DOI] [PubMed] [Google Scholar]
- 24.Guédin A, De Cian A, Gros J, Lacroix L, Mergny JL. Sequence effects in single base loops for quadruplexes. Biochimie. 2008;90:686–696. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Gabelica V, Rosu F, De Pauw E. A Simple Method to Determine Electrospray Response Factors of Noncovalent Complexes. Anal. Chem. 2009;81:6708–6715. doi: 10.1021/ac900785m. [DOI] [PubMed] [Google Scholar]
- 26.Hwang TL, Shaka AJ. Water suppression that works. Excitation sculpting using arbitrary wave forms and pulsed field gradients. J. Magn. Reson. Ser. A. 1995;A112:275–279. [Google Scholar]
- 27.Dalvit C. Efficient multiple-solvent suppression for the study of the interactions of organic solvents with biomolecules. J. Biomol. NMR. 1998;11:437–444. [Google Scholar]
- 28.Wuthrich K. NMR of Proteins and Nucleic Acids. New York, NY: John Wiley and Sons; 1986. [Google Scholar]
- 29.Hosur RV, Govil G, Miles HT. Application of two-dimensional NMR spectroscopy in the determination of solution conformation of nucleic acids. Magn. Reson. Chem. 1988;26 [Google Scholar]
- 30.Patel PK, Hosur RV. NMR observation of T-tetrads in a parallel stranded DNA quadruplex formed by Saccharomyces cerevisiae telomere repeats. Nucleic Acids Res. 1999;27:2457–2464. doi: 10.1093/nar/27.12.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 32.Mergny J-L, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardin CC, Perry AG, White K. Thermodynamic and kinetic characterization of the dissociation and assembly of quadruplex nucleic acids. Biopolymers (Nucleic Acid Sci.) 2001;56:147–194. doi: 10.1002/1097-0282(2000/2001)56:3<147::AID-BIP10011>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Mergny J-L, De Cian A, Ghelab A, Saccà B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masiero S, Trotta R, Pieraccini S, De Tito S, Perone R, Randazzo A, Spada GP. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010;8:2683–2692. doi: 10.1039/c003428b. [DOI] [PubMed] [Google Scholar]
- 36.Graziano C, Masiero S, Pieraccini S, Lucarini M, Spada GP. A cation-directed switch of intermolecular spin-spin interaction of guanosine derivatives functionalized with open-shell units. Org. Lett. 2008;10:1739–1742. doi: 10.1021/ol8003832. [DOI] [PubMed] [Google Scholar]
- 37.Jin R, Gaffney BL, Wang C, Jones RA, Breslauer KJ. Thermodynamics and structure of a DNA tetraplex: a spectroscopic and calorimetric study of the tetramolecular complexes of d(TG3T) and d(TG3T2G3T) Proc. Natl Acad. Sci. USA. 1992;89:8832–8836. doi: 10.1073/pnas.89.18.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feigon J, Koshlap KM, Smith FW. 1H NMR spectroscopy of DNA triplexes and quadruplexes. Methods Enzymol. 1995;261:225–255. doi: 10.1016/s0076-6879(95)61012-x. [DOI] [PubMed] [Google Scholar]
- 39.Feigon J. DNA triplexes, quadruplexes, and aptamers. Encyclopedia of NMR. 1996;3:1726–1731. [Google Scholar]
- 40.Mergny J-L, De Cian A, Amrane S, Webba da Silva M. Kinetics of double-chain reversals bridging contiguous quartets in tetramolecular quadruplexes. Nucleic Acids Res. 2006;34:2386–2397. doi: 10.1093/nar/gkl098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 42.Rosu F, Gabelica V, Poncelet H, De Pauw E. Tetramolecular G-quadruplex formation pathways studied by electrospray mass spectrometry. Nucleic Acids Res. 2010;38:5217–5225. doi: 10.1093/nar/gkq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sket P, Plavec J. Tetramolecular DNA Quadruplexes in Solution: Insights into Structural Diversity and Cation Movement. J. Am. Chem. Soc. 2010;132:12724–12732. doi: 10.1021/ja104889t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.