Abstract
Translation of many cellular and viral mRNAs is directed by internal ribosomal entry sites (IRESs). Several proteins that enhance IRES activity through interactions with IRES elements have been discovered. However, the molecular basis for the IRES-activating function of the IRES-binding proteins remains unknown. Here, we report that NS1-associated protein 1 (NSAP1), which augments several cellular and viral IRES activities, enhances hepatitis C viral (HCV) IRES function by facilitating the formation of translation-competent 48S ribosome–mRNA complex. NSAP1, which is associated with the solvent side of the 40S ribosomal subunit, enhances 80S complex formation through correct positioning of HCV mRNA on the 40S ribosomal subunit. NSAP1 seems to accomplish this positioning function by directly binding to both a specific site in the mRNA downstream of the initiation codon and a 40S ribosomal protein (or proteins).
INTRODUCTION
Translation of eukaryotic mRNAs commences from AUGs proximal to the 5′-end of the transcript through the recruitment of ribosomes around the cap structure (1) or to a specialized RNA element termed an internal ribosome entry site (IRES) (2,3). Many proteins that mediate cap-dependent translation have been well studied, and the roles of these proteins (designated canonical translation initiation factors) in translation have been thoroughly investigated. For instance, the cap-binding protein eIF4E (eukaryotic initiation factor 4E), in association with eIF4G and eIF3, participates in recruitment of the 40S ribosomal subunit to the 5′-ends of mRNAs (4). Some canonical translation factors that are involved in cap-dependent translation also participate in IRES-dependent translation. For example, eIF4G, eIF3 and eIF4A are required for the IRES function of picornaviruses, such as poliovirus (PV) and encephalomyocarditis virus (EMCV) (5–7). In addition to the canonical translation factors, IRES elements require proteins designated IRES trans-acting factors (ITAFs) for their activities. IRES-enhancing activities of ITAFs have been reported. For instance, La protein, poly(rC)-binding proteins (PCBPs), polypyrimidine tract-binding protein (PTB) and upstream of N-ras (UNR) protein augment PV IRES function (8–12). However, the molecular bases of the translational activation by ITAFs have remained obscure. In particular, the relationship between ITAFs and the translational machinery (ribosome) or canonical initiation factors is unclear, although some ITAFs have been suggested to activate IRES function by constructing or stabilizing IRES structures (13–16).
Hepatitis C virus (HCV), the major causative agent of virus-related liver cirrhosis and hepatocellular carcinoma in humans, is a positive-sense RNA virus. The 5′-untranslated region (5′-UTR) and the 5′ part of the core-coding region of HCV RNA contains an IRES element (17), which is the most thoroughly investigated of such elements. As such, it serves as a model system for investigating how an IRES element recruits translation machinery (40S ribosomal subunit) and understanding the functions of proteins that enhance IRES activity. The 40S ribosomal subunit binds to the HCV IRES in the vicinity of the initiation codon to yield the 48S complex, and does so without the assistance of canonical or non-canonical initiation factors (18–20). The bottom half of stem–loop III, including the pseudoknot structure and the region around the initiation codon, are necessary and sufficient for binding of the 40S ribosomal subunit (19). However, the 40S ribosomal subunit bound to the HCV IRES is not competent for translation initiation. Canonical translation factors are also required for active translation. For instance, the ternary complex eIF2/GTP/Met-tRNAi is required for precise positioning of the 40S ribosomal subunit at the initiation site. Moreover, eIF3 along with a factor (or factors) present in the 50–70% ammonium sulfate subfraction are required for joining of the 60S ribosomal subunit to the 48S complex (18). In contrast, some canonical initiation factors, such as eIF4A, 4B, 4E and 4G, are not needed for 80S complex formation on the HCV IRES (18). Additionally, several ITAFs, including La protein, heterogeneous nuclear ribonucleoprotein (hnRNP) L, NS1-associated protein 1 (NSAP1) and hnRNP D, are known to enhance the activity of HCV IRES (21–24); however, the role of these ITAFs in HCV IRES function remains obscure.
NSAP1 was originally identified as a protein capable of interacting with NS1, a protein of the mouse minute virus (25). NSAP1 is also known as hnRNP Q (26), a member of the cellular hnRNP family. Previously, we have shown that NSAP1 augments HCV IRES-dependent translation through an interaction with the A-rich region at a site downstream of the initiation codon (24). We also showed that NSAP1 functions as an ITAF on cellular IRESs in the mRNAs of BiP (immunoglobulin heavy chain-binding protein) and serotonin N-acetyltransferase [arylalkylamine N-acetyltransferase (AANAT)] (27,28). However, the molecular mechanism responsible for translational activation by NSAP1 has remained unexplained. Here, we show that NSAP1 enhances HCV IRES-dependent translation by facilitating the formation of a translation-competent 48S complex that is ready to form the 80S complex. Specifically, NSAP1 binds to the A-rich element immediate downstream of the initiating AUG of HCV mRNA, and also associates with the 40S ribosomal subunit through interactions with ribosomal proteins (r-proteins). The results of toeprinting analyses using wild-type (wt) IRES or a mutant (mt) IRES lacking the NSAP1-binding site, led us to conclude that NSAP1 facilitates HCV IRES-dependent translation by assisting the proper positioning of the 40S ribosomal subunit on the HCV mRNA. This is the first report showing that a protein that interacts with an IRES element enhances translation through a direct interaction with the 40S ribosomal subunit.
MATERIALS AND METHODS
Plasmid construction and siRNAs
A Flag-tagged expression construct of NSAP1 (pFlag-CMV2-NSAP1) was generated by treating pSK(−)–NSAP1 with NheI–Klenow–SalI and ligating the resulting fragment into pFlag-CMV2 (Sigma) treated with EcoRI–Klenow–SalI. pCDNA3.1_Fluc(m) was constructed by ligating BamHI–Klenow–NotI-treated pH374F_wt into pCDNA3.1_Fluc treated with AflII–Klenow–NotI. pACR(342–374)F_wt was generated by treating pH374F_wt with Asp718–AccI–Klenow followed by self-ligation. pGEX-4T-3-RpS constructs (RpS2, RpS3a, RpS4, RpS6, RpS8 and RpS15) were constructed by first generating each pSK(−)-RpS as follows: cDNAs corresponding to each RpS were amplified from a cDNA library by PCR using specific primer pairs. The PCR products were then treated with XbaI–BamHI and ligated into XbaI–BamHI-treated pSK(−). Finally, pGEX-4T-3-RpS plasmids were prepared by ligating each BamHI–NotI-treated pSK(−)-RpS into BamHI–NotI-treated pGEX-4T-3 (GE healthcare). The monocistronic reporters pH374F_wt and pH374F_mt were constructed by ligating the Asp718–Klenow–BamHI-treated DNA fragment (nucleotides 18–374 HCV IRES) from pRH374F_wt or pRH374F_mt, respectively, into pRH374F_wt treated with NheI–Klenow–BamHI (partial digestion). The construction of plasmids pRH374F_wt, pRH374F_mt, pH374CAT_wt and pH374CAT_mt have been described previously (24), as have the plasmids pQE31-NSAP1 (24) and pQE31-eIF4A (29).
Duplex small interfering RNAs (siRNAs) targeting exon 3 of hnRNP Q (nucleotides 768–790) and control siRNA were purchased from IDT Inc. The sequence of the siRNA targeting hnRNP Q and the control siRNA sequence were 5′-rGrGrArArCrArGrArTrTrCrTrTrGrArArGrArArTrTrTrAGC-3′ and 5′-rCrUrUrCrCrUrCrUrCrUrUrUrCrUrCrUrCrCrCrUrUrGrUGA-3′, respectively.
Purification of the 40S and 60S ribosomal subunit and recombinant proteins
40S and 60S ribosomal subunits were prepared from HeLa cells (National Cell Culture Centre) as previously described (30). The method used for purification of recombinant His-NSAP1 and His-eIF4A from Escherichia coli has been described elsewhere (24). His-NSAP1 and His-eIF4A were dialyzed against HT buffer (16.2 mM HEPES-KOH pH 7.4, 36 mM KCl, 160 mM KOAc, 1.24 mM MgOAc2, 1.6 mM DTT, 2.8 mM β-mercaptoethanol). For the purification of Flag-NSAP1, pFlag-CMV2-NSAP1 was transfected into 293T cells using a calcium phosphate method. At 56 h after transfection, cells were harvested and lysed by treating with lysis buffer (50 mM Tris–HCl pH 7.4, 300 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM β-mercaptoethanol) and briefly sonicating. Samples were centrifuged at 12 000 rpm (13 500 g) for 15 min in a microfuge, filtered with a syringe filter (0.2 µm; Millipore), and then incubated with anti-Flag M2 agarose resin (Sigma) at 4°C for 1 h. The anti-Flag agarose resin was microcentrifuged at 4000 rpm (1500 g) for 2 min and washed three times with lysis buffer. Flag-NSAP1 proteins bound to the agarose beads were eluted by adding elution buffer [150 ng/µl 3× Flag peptide (SIGMA) in lysis buffer]. Purified Flag-NSAP1 proteins were dialyzed against H100 buffer (10 mM HEPES-KOH pH 7.4, 100 mM KCl, 1 mM MgCl2, 0.1 mM EDTA, 7 mM β-mercaptoethanol).
In vitro transcription
The plasmids pRH374F_wt, pRH374F_mt, pH374F_wt, pH374F_mt and pCDNA3.1_Fluc were linearized with NotI and transcribed using T7 RNA polymerase to generate luciferase reporters for in vitro translation. The HCV IRES RNAs (nucleotides 18–374, wt and mt) used in sucrose density gradient analyses of in vitro translation reaction mixtures of rabbit reticulocyte lysates (RRLs) or 293T cell lysates were generated by incubating BamHI-treated (linearized) pH374CAT_wt and pH374CAT_mt DNAs in the presence of [α-32P]UTP and T7 RNA polymerase. RNAs used in primer extension assays were generated by in vitro transcription using reactions containing linearized (XbaI-treated) pH374CAT_wt and pH374CAT_mt. The A-rich core-coding region (ACR) RNA used in binding assays (pACRF_wt) was linearized with BamHI and then used in transcription reactions in the presence of [α-32P]UTP.
Sucrose density gradient analyses
Sucrose density gradient analyses of RRL and 293T cell lysates were performed as described previously (31), with minor modifications. In brief, translation reactions (25 µl) using RRL [17.5 µl nuclease-treated RRL, 20 µM amino acids, 4 U RNase inhibitor (Bioprince), 180 mM KCl, 100 nM RNA] or 293T cell lysates (36% 293T lysate, 40 nM RNA, 180 mM KOAc, 18.29 mM HEPES-KOH pH 7.4, 10 µM amino acids, 1.92 mM DTT, 360 µM MgOAc2, 770 µM MgCl2, 20 µg/ml tRNA, 960 µM ATP, 60 µM GTP, 9.62 mM creatine phosphate, 20 µg/ml CPK, 240 µM spermidine) were incubated at 30°C for 15 min. GDPNP (2 mM) or cycloheximide (CHX) (20 mM) was added to the reaction mixtures in certain experiments, as indicated in the text. The reaction samples were loaded onto 5–20% sucrose gradients and centrifuged at 37 000 rpm (170 000 g) in a SW41Ti rotor (Beckman) at 4°C for 2 h 40 min. Gradients were analyzed using a Brandel gradient density fractionator and an Econo UV monitor (Bio-Rad).
Cell culture and preparation of 293T cell lysates for in vitro translation
293T or HeLa cells were cultivated in Dulbecco's modified Eagle's medium (Gibco BRL) supplemented with 10% fetal bovine serum (JRH). 293T cell lysates were generated by treating 40 dishes (100 mm) of 293T cells with 600 pmol/dish of a control siRNA or a siRNA against NSAP1. After incubating for 56 h, cells were harvested and translation-competent cell lysates were prepared by washing cell pellets three times with ice-cold PBS and then incubating with 1.5 vols of cold hypotonic buffer on ice for 10 min. 293T cells were lysed by homogenization in a small dounce homogenizer (40 strokes), and then centrifuged at 12 000g (11 200 rpm) for 15 min. The supernatant was dialyzed against a buffer containing 10 mM HEPES-KOH (pH 7.4), 90 mM KOAc, 1.5 mM MgOAc2 and 2.5 mM DTT. Translation reactions using 293T lysates were performed as described previously (23) with 40 nM reporter RNA and 180 mM KOAc. Translation reactions in micrococcal nuclease (MN)-treated RRLs were performed in 180 mM KOAc as recommended by the supplier (Promega).
Primer extension inhibition (toeprinting)
Primer extension in RRL was performed as described previously (5), with minor modifications. A primer corresponding to nucleotides 69–86 (5′-GCAACTGACTGAAATGCC-3′) of the CAT-coding sequence was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas). Reaction mixtures were resolved on a 6% sequencing gel, and radioactive signals were monitored using a phosphorimager. Intensity of gel bands was quantified using Image J software.
Detection of ribosome–NSAP1 interactions
For co-migration assays (Figure 4A), 20 µl mixtures containing 1.5 pmol 40S or 60S ribosomal subunits and 6 pmol of His-NSAP1 in HT buffer were incubated at 30°C for 20 min. Samples were resolved on a 1% agarose gel, and the RNAs and proteins in the gel were electro-transferred onto a nitrocellulose (NC) membrane for 4 h using a TE42 transfer tank (Hoefer). After blocking non-specific binding of antibodies to the membrane for 20 min, western blotting was performed using a primary antibody against His and a horseradish peroxidase (HRP)-conjugated secondary antibody against mouse IgG. In Supplementary Figure S5A, 20 µl mixtures containing 10 pmol 40S ribosomal subunits and 10 pmol of either His-NSAP1 or His-eIF4A in HT buffer were used for co-migration assays.
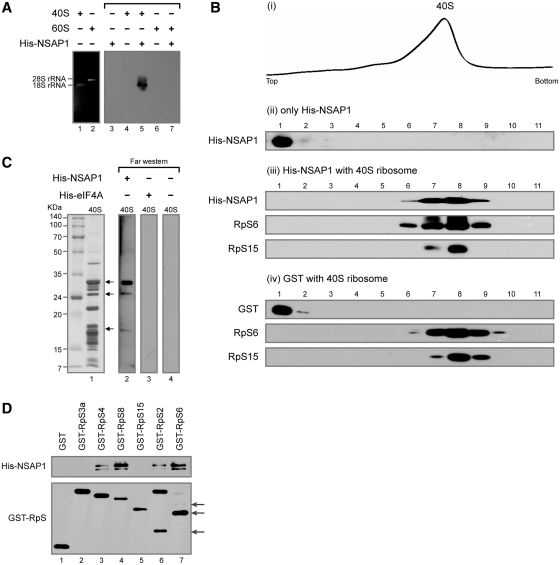
Figure 4.
NSAP1 directly interacts with the purified 40S ribosomal subunit in vitro. (A) Co-migration of 40S ribosomal subunit and NSAP1 on an agarose gel. Purified His-NSAP1 proteins were incubated with purified 40S or 60S ribosomal subunits and resolved on a 1% agarose gel. The positions of ribosomes and NSAP protein were observed by ethidium bromide staining (lanes 1 and 2) or western blotting with an antibody against the His-tag (lanes 3–7). (B) Co-migration of the 40S ribosomal subunit and His-NSAP1 in 5–20% sucrose density gradients. After centrifugation, the absorbance of the gradient at 254 nm was monitored using a Bio-Rad EM-1 UV monitor (panel i), and the gradient was fractionated into 11 samples. The presence of proteins in each fraction was monitored by western blotting (panels ii–iv). (C) Detection of NSAP1-interacting r-proteins by far-western blotting. (D) Confirmation of protein–protein interactions by GST-pull down assay using purified GST-fused r-proteins and His-NSAP1. Arrows depict proteins derived from GST-RpS2 and GST-RpS6 in protein purification process. RpS2, RpS4, RpS6 and RpS8 were shown to interact with NSAP1.
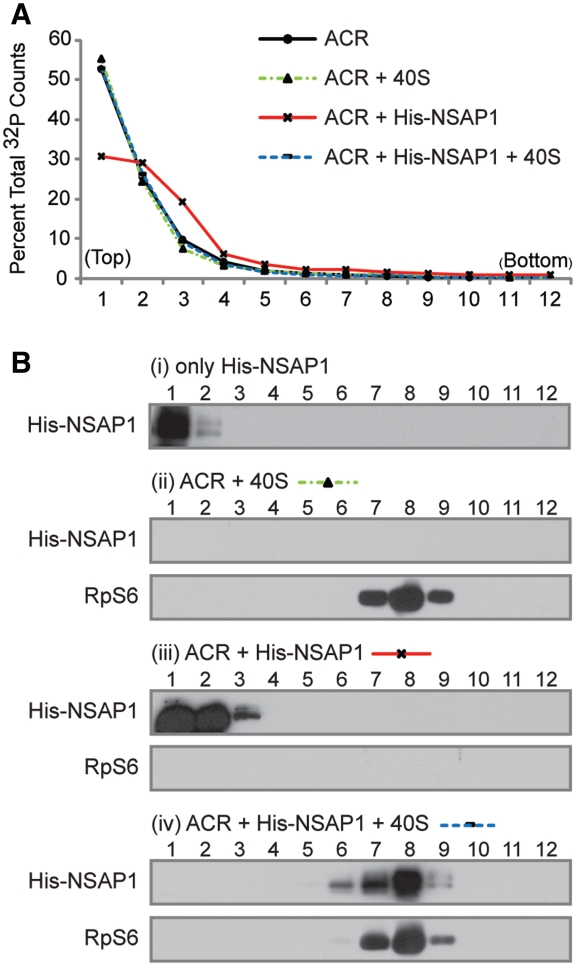
For sucrose gradient analyses (Figure 4B), 150 pmol His-NSAP1 in HT buffer was incubated with 75 pmol of 40S ribosomal subunits at 30°C for 10 min. The samples were fractionated on 5–20% sucrose gradients as described above, and then methanol precipitated to concentrate proteins. For the NSAP1–ACR interaction study (Figure 5), 140 µl mixtures containing 150 pmol His-NSAP1 and 150 pmol 32P-labeled ACR RNA in HT buffer were incubated at 30°C for 10 min, and then incubated for an additional 30°C for 10 min in the presence of 75 pmol 40S ribosomal subunit. The sucrose gradient analysis was performed as in Figure 4B.
Figure 5.
The fate of NSAP1 on the HCV IRES after interacting with the 40S ribosomal subunit. (A) Radioactivity profile of ACR (NSAP1-binding site). ACR: ACR RNA alone; ACR + 40S: ACR RNA incubated with purified 40S ribosomal subunits for 10 min; ACR + NSAP1: ACR RNA incubated with purified NSAP1 for 10 min; ACR + NSAP1 + 40S: ACR RNA incubated with His-NSAP1 for 10 min, followed by addition of purified 40S ribosomal subunits to the mixture. The reaction mixtures were subjected to sucrose gradient analyses. (B) Western blot analyses of proteins in each fraction shown in (A).
For GST-pull down assays, GST-fused r-proteins were purified from E. coli and incubated with Glutathione Sepharose 4B resin (GE) at 4°C for 1 h. After washing three times with PBS containing 0.5% Triton X-100, His-NSAP1 was added to each binding mixture, and then incubated for 2 h in the presence of RNase. The resin-bound proteins were washed three times and resolved by SDS–Polyacrylamide gel electoroporesis (PAGE).
Far-western blotting
Far-western blotting was performed as described previously (32) with minor modifications. Purified 40S ribosomal subunits were resolved by SDS–PAGE, transferred onto a polyvinylidene difluoride (PVDF) membrane and then incubated in 5% (w/v) fat-free milk in Tris Buffered Saline-Tween20 (TBST) buffer (25 mM Tris–HCl pH 7.5, 100 mM NaCl, 25 mM NaF and 0.02% Tween-20) to block non-specific protein binding. The membrane was incubated with His-NSAP1 or His-eIF4A (0.7 µg/ml in blocking solution) overnight at 4°C. After gently rinsing to remove unbound proteins, western blotting was performed using primary anti-His and secondary HRP-conjugated anti-mouse IgG antibodies.
Antibodies
The antibodies used in this study were anti-NSAP1 (Sigma, 18E4), anti-RpS6 (Cell Signaling), anti-eIF3b (Santa Cruz), anti-hnRNP D (Upstate Biotechnology), anti-actin (ICN), anti-Flag (Sigma), anti-eIF3e (Santa Cruz), anti-eIF4E (Santa Cruz), anti-eIF4AI (Santa Cruz), anti-eIF2α (Novus Biologicals), anti-HuR (Santa Cruz), anti-His (R&D), anti-RpS15 (ILAMM), anti-PABP (Santa Cruz, 10E10), anti-PTB (23), anti-eIF4G and anti-GST (33).
RESULTS
NSAP1 augments formation of translation-competent 48S complex
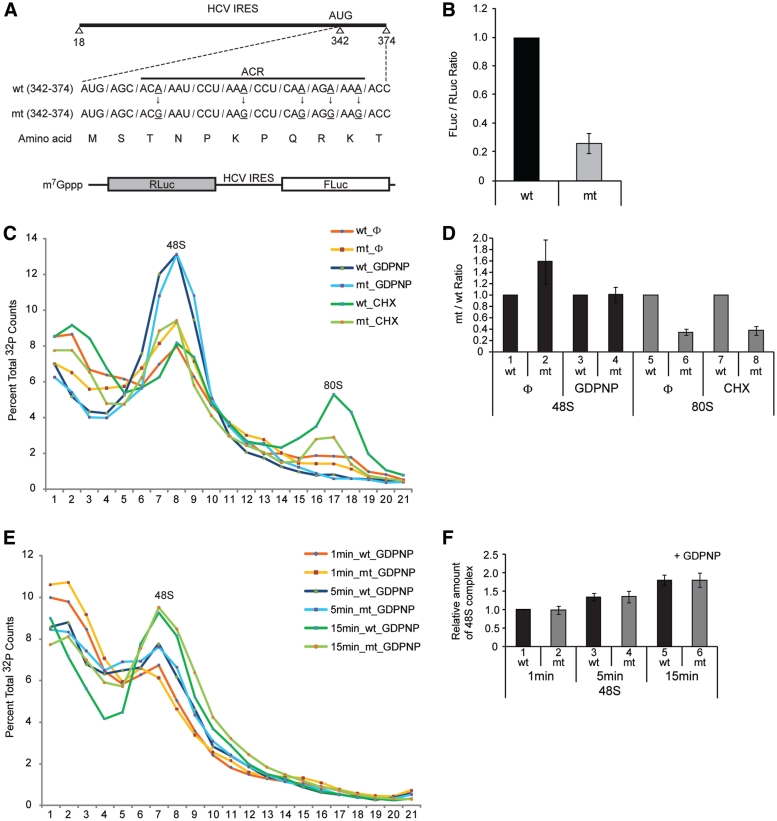
Previously, we reported that NSAP1 augments HCV mRNA translation through a specific interaction with the ACR of HCV mRNA, further showing that a mutation in the ACR (Figure 1A) that weakens the interaction with NSAP1 results in reduced translation (24). To reconfirm this, we generated dicistronic mRNAs consisting of the Renilla luciferase (RLuc) reporter gene, which is translated in a cap-dependent manner, followed by the HCV IRES (including the ACR in the HCV core-coding region) fused in-frame with another reporter gene, FLuc (Figure 1A). The efficiency of the mt IRES translation in RRL was reduced by 70% compared with that of the wt IRES (Figure 1B). The translational reduction by the mutation is not likely due to an introduction of a rare codon(s) in the mt since translational efficiency of the wt and the mt versions of mRNAs showed similar translation efficiencies when these reporter genes were translated by the cap-dependent scanning mode (Supplementary Figure S1). Moreover, the reduced translation of the mt was attributed to reduced binding of NSAP1 to the mt RNA since human NSAP1 protein (AAK59705) is highly homologous (99% identical in amino acid sequence) to rabbit NSAP1 protein (XP_002714606.1) and a large quantity of NSAP1 was detected in RRL by western blotting (data not shown).
Figure 1.
NSAP1 facilitates 80S complex formation. (A) Schematic diagram of the NSAP1-binding site on the HCV IRES. wt and mt, which contains A–G substitutions that impede NSAP1-binding of HCV IRES RNAs, are depicted. (B) Translational efficiencies of wt and mt HCV IRESs were measured in RRLs using dicistronic mRNAs (lower panel in A). The relative HCV IRES translational efficiencies are depicted. (C) Representative sucrose gradient data of three experiments. The translation reaction mixtures were incubated in RRLs at 30°C for 15 min with [32P]-labeled wt or mt HCV IRES RNAs (18–374 nt) in the presence or absence of GDPNP or CHX. Pre-initiation complex and initiation complex on the HCV IRESs were observed using 5–20% sucrose gradient analyses. Φ denotes no drug treatment. (D) Relative amount of 48S and 80S complexes on wt or mt HCV IRES quantified from panel C. The radioactivity of each complex (48S or 80S complex) was quantified by using manual planimetry after subtraction of the basal radioactivity from the radioactivity of each fraction. The mean and standard deviation values of three independent experiments are depicted by columns and bars. The complexes on wt HCV IRES were assigned a value of 1 (lanes 1, 3, 5 and 7). (E) Representative sucrose gradient data of three experiments. The translation reaction mixtures were incubated in RRLs at 30°C for 1, 5 and 15 min with [32P]-labeled wt or mt HCV IRES RNAs (18–374 nt) in the presence of GDPNP. The 48S complex on the HCV IRESs was observed using 5–20% sucrose gradient analyses. (F) Relative amount of 48S complex on wt or mt HCV IRES was measured from panel E similarly to panel D. The amount of 48S complex formed on wt HCV IRES in 1 min was assigned a value of 1.
To determine which translation steps are influenced by the mutation, we performed sucrose density gradient analyses using RRLs and 32P-labeled RNAs containing the wt or mt IRES (Figure 1C). After ultracentrifugation, sucrose gradients were fractionated from top (low-molecular weight) to bottom (high-molecular weight), and the radioactivity in each fraction was measured. The first peak (48S) represents a mixture of 48S pre-initiation complexes and binary complexes, which are composed of the 40S ribosomal subunit and the HCV IRES RNA without any translation factor (18), and the second peak (80S) represents 80S initiation complexes. In the absence of chemical treatments that inhibit the translation reaction at a specific stage of translational initiation, the proportion of wt HCV mRNA in the first peak associated with the 48S complex or in the binary complex was smaller than that of mt mRNA (Figure 1C, compare red and orange lines in the 48S peak; Figure 1D, compare lane 1 with 2). However, under the same conditions, the proportion of wt HCV mRNA in the second peak associated with the 80S complex was about 3-fold greater than that of mt mRNA (Figure 1C, compare red with orange line in the 80S peak; Figure 1D, compare lane 5 with 6). When GDPNP, a non-hydrolyzable GTP analog that specifically accumulates 48S complex, but not binary complex (31), was included in the in vitro translation reactions, the amount of complexes in the first peak on the wt HCV IRES was the same as that on the mt HCV IRES (Figure 1C, compare blue and sky-blue lines; Figure 1D, compare lane 3 with 4). We also monitored the dynamics of the first peak formation to the wt and mt HCV IRESs in the presence of GDPNP since the rate of the formation could be different between the wt and mt HCV IRES. No difference in the time required for the complex formation was observed between the wt and mt HCV IRESs (Figure 1E and F). These data show that the wt and mt IRES are equally capable of interacting with 40S ribosomal subunit and indicate that NSAP1 does not participate in the association of 40S ribosome on the HCV IRES.
Importantly, when CHX, which induces stalling of the 80S complex by interfering with translocation of the ribosome, was added to the translation mixture, there was a substantial accumulation of the 80S complex on the wt HCV IRES, but about 3-fold smaller accumulation of the 80S complex on the mt HCV IRES (Figure 1C, compare yellow–green and dark-green lines in the 80S peak; Figure 1D, compare lane 7 with 8). The reduced 80S complex formation on the mt IRES well reflects the translational reduction of the mt HCV IRES (Figure 1B and D). Under these same conditions, the amount of the first peak on the mt IRES was greater than that on the wt IRES (Figure 1C, compare yellow–green and dark-green lines in the 48S peak). These data strongly indicate that NSAP1 participates in 80S complex formation.
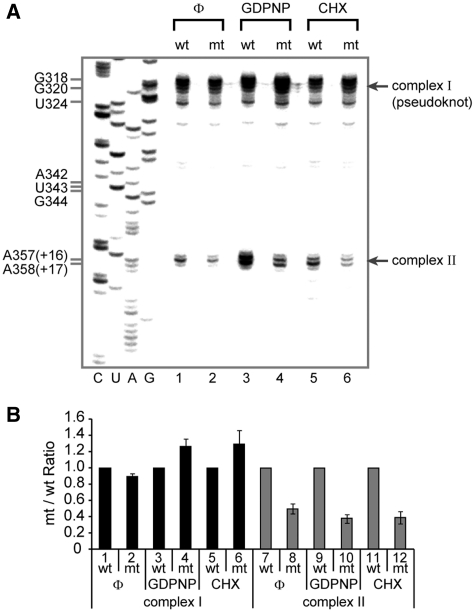
The enhancement of the 80S complex formation by NSAP1 can be acquired by two different mechanisms: NSAP1 may facilitate the formation of translation-competent 48S complex. Alternatively, NSAP1 may facilitate joining of the 60S ribosome to the 48S complex. The role of NSAP1 in HCV IRES function was further investigated using a primer extension assay with reverse transcriptase known as ‘toeprinting’, which is suitable for monitoring the status of ribosomes (integrity and quality of translation–initiation complex) on mRNA. The toeprint signal is generated by blockade of the polymerase reaction resulting from association of a ribosome with mRNA or the presence of secondary/tertiary structure in the mRNA molecule. A specific stage of the ribosome, such as the 48S and 80S complex, can be monitored by measuring the intensities of specific toeprint signals. For example, HCV RNA associated with a 40S ribosomal subunit at the initiation codon, which is translation-competent, showed toeprints at the +16 and +17 positions relative to the initiation codon (Figure 2A, complex II in lanes 3 and 4; lower panel in Supplementary Figure S2A) as previously reported (31). Blocking translation at the 80S complex stage with CHX resulted in a shift of 1 nt, from +16 to +17, in the toeprint pattern (Figure 2A, complex II in lanes 5 and 6), as previously reported (31). HCV mRNA showed another strong toeprint around the pseudoknot structure located upstream of the initiation codon (Figure 2A, complex I). The toeprints at complex I reflect HCV mRNAs associated with the 40S ribosomal subunit (as a 48S complex or a binary complex), but inadequately positioned, i.e. 40S ribosomal subunits, in which the initiation codon of the HCV RNA is not correctly positioned at the P-site even though the IRES element is associated with the 40S ribosomal subunit on the solvent side (18,31; upper panel in Supplementary Figure S2A).
Figure 2.
NSAP1 promotes the correct positioning of the 40S ribosomal subunit at the initiation codon. (A) Toeprinting analyses of wt and mt HCV IRESs in RRLs. Representative toeprint data of five independent experiments. The toeprints of wt and mt HCV RNAs in the absence of RRL are presented in Supplementary Figure S2B, which shows that the differences between toeprints are not due to mutations in the HCV RNA per se. (B) Relative amount of complexes I and II on wt or mt HCV IRES quantified from five experiments. The band intensities on the gel shown in panel A were quantified by autoradiography, and the relative amounts of the complexes on mt IRES to those on wt IRES at various experimental settings are depicted. The complexes on wt HCV IRES were assigned a value of 1 (lanes 1, 3, 5, 7, 9 and 11). The mean and standard deviation values of five independent experiments are depicted by columns and bars.
In the absence of a translation inhibitor (Figure 2A, lanes 1 and 2), the toeprint intensity of complex II on the mt HCV IRES was slightly lower than that on the wt IRES, which may indicate that more ribosomes are correctly associated with the initiation codon on the wt HCV RNA than on the mt HCV RNA. This difference was greater when stage-specific translation–initiation inhibitors were included in the translation mixtures used for toeprint analyses. The toeprint intensity of complex II on wt RNA was about 3-fold stronger than that on the mt RNA when GDPNP was included in the translation reaction mixture (Figure 2A, lanes 3 and 4; Figure 2B, lanes 9 and 10). Conversely, the toeprint intensity of complex I was stronger on mt RNA than it was on the wt RNA under the same conditions (Figure 2A, lanes 3 and 4; Figure 2B, lanes 3 and 4). These data indicate that many more ribosomes were correctly positioned on the wt HCV RNA than on the mt RNA, even though similar amounts of ribosomes were associated with HCV IRES. This was true regardless of the existence of NSAP1-binding site, as indicated by the presence of the same first peak, containing the binary complex and the 48S complex, in the sucrose gradient when GDPNP was present in the translation reaction (Figure 1C, sky-blue and dark-blue lines). Consistent with these observations, a larger amount of 80S complex formed on the wt IRES (as indicated by the +16 and +17 toeprints of complex II) than on the mt when CHX was included in the translation mixture (Figure 2A, lanes 5 and 6; Figure 2B, lanes 11 and 12). Conversely, a larger amount of complex I was observed on the mt IRES than on the wt IRES (Figure 2A, lanes 5 and 6; Figure 2B, lanes 5 and 6). Notably, the differences in the toeprints are not attributable to structural changes caused by the mutation because no changes in the toeprints of complexes I and II sites on the mt mRNAs were observed in the absence of translation mixture (Supplementary Figure S2B). Taken together, the sucrose gradient and toeprint analyses indicate that the ACR, where NSAP1 binds, plays an important role in 80S complex formation by facilitating the correct positioning of the 40S ribosomal subunit at the initiation codon.
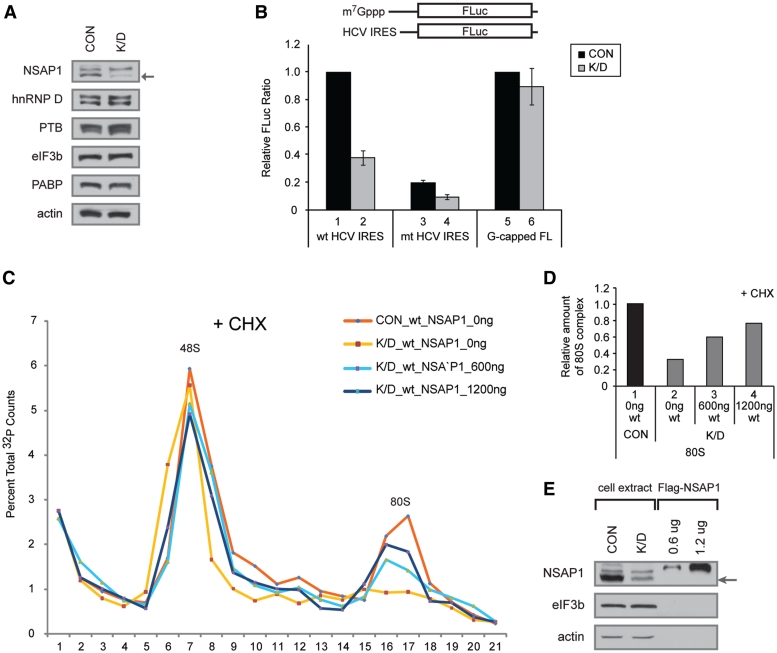
We further confirmed the importance of NSAP1 in 80S ribosomal complex formation by testing the effect of NSAP1 depletion from translation mixtures. Translation-competent cell lysates were prepared from 293T cells or 293T cells transfected with a siRNA against NSAP1 mRNA. This synthetic siRNA, which targets hnRNP Qs including NSAP1 (the smallest isoform of hnRNP Q, indicated by an arrow in Figure 3A and E), reduced the level of NSAP1 by 70% in the NSAP1-knockdown lysate (K/D) compared with the control cell lysate (CON) (Figure 3A). Using NSAP1-depleted lysates, translation of HCV mRNA in a monocistronic context was decreased by 60% (Figure 3B, compare lane 2 with 1). In contrast, translation of a cap-dependent mRNA was not affected by NSAP1 depletion (Figure 3B, compare lane 6 with 5). Translation of the mt RNA, which has mutations at the NSAP1-binding site depicted in Figure 1A, was decreased by 80% in the NSAP1-undepleted lysate (Figure 3B, compare lane 3 with 1). The level of translation reduction by the mutation in a monocistronic mRNA is similar to that in a dicistronic mRNA shown in Figure 1B. Translation of the mt HCV mRNA was further decreased by the depletion of NSAP1 (Figure 3B, compare lane 4 with 3). This additional reduction of translation by the NSAP1 depletion might be attributed to a residual NSAP1-binding at the mutated ACR or to a putative NSAP1-binding to an unidentified NSAP1-binding region in the HCV IRES. The effect of NSAP1 on formation of an 80S complex on 32P-labeled RNA corresponding to HCV IRES (nucleotides 18–374) was monitored by sucrose gradient analyses using NSAP1-depleted and -undepleted lysates, as well as NSAP1-depleted lysates supplemented with purified NSAP1. In translation mixtures treated with CHX, the 80S ribosomal complex on wt HCV IRES was clearly detectable in the presence of NSAP1 (undepleted lysates) (Figure 3C, red line; Figure 3D, lane 1), but was only marginally detectable in translation mixtures containing NSAP1-depleted lysates (Figure 3C, yellow line; Figure 3D, lane 2). Importantly, supplementing NSAP1-depleted lysates with NSAP1 proteins purified from mammalian cells (Figure 3E) induced a dose-dependent restoration of 80S complex formation on the HCV IRES (Figure 3C, sky-blue and dark-blue lines; Figure 3D, lanes 3 and 4). The addition of His-NSAP1 purified from E. coli produced a similar effect (Supplementary Figure S4A and S4B). On the contrary, the 80S complex formation on mt HCV IRES was not recovered by supplementation of recombinant NSAP1 (Supplementary Figure S3A and S3B). These results indicate that NSAP1 plays a pivotal role in the formation of 80S complexes on the HCV IRES.
Figure 3.
Depletion of NSAP1 inhibits 80S complex formation on the HCV IRES. (A) Western blot analyses of lysates from control 293T cells (CON) and 293T cells treated with an siRNA against NSAP1 (K/D). (B) Relative FLuc activity directed from monocistronic reporter RNAs bearing wt HCV IRES or m7G-capped reporter (G-capped FL) in 293T cell lysates. The luciferase activity from wt HCV IRES in control cells (lane 1 for lanes 1–4) and that from capped RNA in control cells (lane 5 for lanes 5 and 6) were assigned a value of 1. (C) The effect of NSAP1 on 80S complex formation was monitored by sucrose gradient analyses of NSAP1-depleted and NSAP1-replete lysates. The labeled RNAs were incubated in control (CON) or NSAP1-depleted lysates (K/D) in the presence of CHX, and then subjected to sucrose gradient analyses. The effect of NSAP1 supplementation on 80S complex formation was observed by adding purified Flag-NSAP1 protein (600 or 1200 ng) to NSAP1-depleted lysates. The radioactivity [32P-labeled wt IRES (18–374 nt)] in each fraction of the 5–20% sucrose gradient is depicted as a percentile in the graph. (D) Relative amount of 80S complex on wt HCV IRES quantified from panel C. (E) The amount of NSAP1 in control (NSAP1-replete) lysates, NSAP1-depleted lysates and NSAP1-depleted lysates supplemented with purified NSAP1 was monitored by western blotting.
NSAP1 associates with the 40S ribosomal subunit
To test whether NSAP1 might associate with the 40S ribosomal subunit through interactions with eIF3, a canonical translation factor needed for HCV IRES-dependent translation, we monitored protein–protein interactions between NSAP1 and eIF3b using a co-immunoprecipitation method. NSAP1 was not co-immunoprecipitated with eIF3b, indicating that these proteins do not interact each other (Supplementary Figure S5B). On the other hand, eIF3e and eIF4G were co-immunoprecipitated with eIF3b, indicating that these translation factors interact, either directly or indirectly (Supplementary Figure S5B). Similarly, a yeast two-hybrid system failed to detect protein–protein interactions between NSAP1 and other translation factors (data not shown). These data suggest that NSAP1 does not directly interact with eIF3.
We next investigated whether NSAP1 directly interacts with the 40S ribosomal subunit using purified NSAP1 protein and purified 40S ribosomal subunits. NSAP1 protein was purified by affinity chromatography from E. coli producing His-tagged NSAP1, and 40S and 60S ribosomal subunits were purified from HeLa cells using sucrose gradient centrifugation in the presence of 0.5 M KCl, as described by Merrick (34). Neither translation factors (represented by eIF3b, eIF4GI, eIF4AI, eIF2α, PABP and eIF4E) nor NSAP1 protein was detected in the purified 40S ribosomal subunit preparation (Supplementary Figure S5C), indicating that 40S ribosomal subunits were cleanly prepared. First, we monitored the interaction of NSAP1 with the ribosomal subunit in an agarose gel (Fig. 4A). The position of the 40S and 60S ribosomal subunit was visualized by staining ribosomal RNAs with ethidium bromide (Figure 4A, lanes 1 and 2). The positions of His-tagged NSAP1 were visualized by western blot analysis with an antibody against the His-tag (Figure 4A, lanes 3–7). NSAP1 proteins that were not pre-incubated with ribosomal subunits did not infiltrate the agarose gel (Figure 4A, lane 3; no NSAP1 band was detected in the agarose gel) and, as expected, the NSAP1 protein was not detected in the purified 40S and 60S ribosomal subunit preparation (Figure 4A, lanes 4 and 6). In contrast, NSAP1 proteins co-migrating with the 40S ribosomal subunit were clearly detected in samples containing both NSAP1 and the 40S ribosomal subunit (Figure 4A, lane 5). The 60S ribosomal subunit, included as a negative control, did not co-migrate with the His-tagged NSAP1, indicating that the interaction between NSAP1 and the 40S ribosomal subunit is specific (Figure 4A, lanes 6 and 7). When a negative control protein eIF4A, instead of NSAP, was incubated with 40S ribosome, it did not co-migrate with the 40S ribosomal subunit (Supplementary Figure S5A).
The interaction between NSAP1 and the 40S ribosomal subunit was further confirmed by a 5–20% sucrose density gradient analysis using purified 40S ribosomal subunits and His-tagged NSAP1 (Figure 4B). Prior to incubation with the 40S ribosomal subunits, NSAP1 proteins were enriched in the upper fraction of the sucrose gradient, corresponding to soluble proteins [Figure 4B(ii)]. After pre-incubation with purified 40S ribosomal subunits, however, all NSAP1 proteins co-migrated with the 40S ribosomal subunits [Figure 4B(i) and B(iii)]. The position of the 40S ribosomal subunit was detected by measuring optical density at 254 nm [Figure 4B(i)] and by western blot analysis with antibodies against the r-proteins RpS6 and RpS15 [Figure 4B(iii)]. A negative control protein (GST) did not co-migrate with the 40S ribosomal subunits under the same conditions [Figure 4B(iv)]. Taken together, the results of agarose gel electrophoresis and sucrose density gradient experiments lead us to conclude that NSAP1 specifically interacts with the 40S ribosomal subunit without the assistance of canonical translation factors.
As an approach to uncover the molecular basis of the interaction between NSAP1 and the 40S ribosomal subunit, we used a far-western assay to test whether the NSAP1 protein interacts with a r-protein (or proteins) (Figure 4C). For this experiment, the 40S ribosomal subunit proteins were resolved by SDS–PAGE and then transferred to a PVDF membrane. The transferred proteins were incubated with His-tagged NSAP1 or His-tagged eIF4A (negative control). The positions of NSAP1- or eIF4A-interacting proteins were visualized by western blot analysis using an anti-His antibody. Three or more bands were clearly detectible in His-tagged NSAP1 far-western blots (Figure 4C, lane 2). These proteins (Figure 4C, lane 1) were tentatively identified from a comparison of their mobility with the SDS band patterns of previously reported r-proteins (35,36), and their identities were confirmed by MALDI-TOF analyses. Multiple proteins, including RpS2, RpS3a, RpS4, RpS6, RpS8, RpS15 and RpS25, were identified by the MALDI-TOF analyses. No bands were detected in negative controls (Figure 4C, lanes 3 and 4). Interactions between NSAP1 and r-proteins were further confirmed using purified r-proteins and NSAP1 in GST pull-down assays. His-tagged NSAP1 was incubated with GST-fused r-proteins in the presence of RNase (to exclude the possibility of RNA-mediated interactions), and protein complexes were then pulled down with a GSH-Sepharose resin. All GST-fused r-proteins were precipitated by the GSH resin, as shown in the lower panel of Figure 4D. A subset of the r-proteins tested—RpS4, RpS8, RpS2 and RpS6—co-precipitated with NSAP1 protein (Figure 4D, upper panel). Taken together, these data indicate that NSAP1 directly associates with the 40S ribosomal subunit through protein–protein interactions with r-proteins.
The interaction between NSAP1 and the ACR is relieved by the association of NSAP1 with the 40S ribosomal subunit
We investigated the effect of the 40S ribosomal subunit on the NSAP1–ACR interaction using sucrose density gradient analysis. 32P-labeled RNA containing the ACR (spanning nucleotides 342–374) of HCV was enriched in sucrose gradient fractions 1 and 2 (top fractions) on a 5–20% sucrose gradient (Figure 5A). The addition of 40S ribosomal subunits to the labeled RNA did not change the profile of RNA on the sucrose gradient, indicating that the 40S ribosomal subunit does not directly interact with the ACR (Figure 5A, green dotted line). In contrast, the addition of NSAP1 shifted a portion of ACR RNAs to a heavier fraction (from fractions 1–3) in the sucrose gradient (Figure 5A, red line). This shift reflects the association of NSAP1 with the ACR RNA, which is also shown by western blot analysis of NSAP1 [compare Figure 5B(i) with B(iii)]. Surprisingly, addition of 40S ribosomal subunits to the ACR–NSAP1 complex shifted ACR RNA back to the lighter sucrose gradient fraction (Figure 5A, dotted blue line); under these conditions, NSAP1 co-migrated with the 40S ribosomal subunit in western blots, where no ACR RNA was present [Figure 5B(iv)]. The 40S ribosomal preparation was free of NSAP1 before incubation with the ACR–NSAP1 complex [Figure 5B(ii)]. These results indicate that the ribosome–NSAP1 interaction relieves the ACR–NSAP1 interaction.
DISCUSSION
Many cellular and viral RNA segments have been shown to function as IRES elements, and many cellular proteins (ITAFs), especially specific RNA-binding proteins, are known to enhance various IRES activities. However, the molecular mechanism, by which ITAFs enhance translation, is poorly understood. In particular, how ITAFs recruit translational machinery, including ribosomes and canonical initiation factors, to the IRESs or how an ITAF facilitates a particular step in translation has remained unclear. Here, we provide the first evidence for direct communication between an ITAF (NSAP1) and the translational machinery (40S ribosomal subunit). NSAP1, which binds to a specific region of mRNA (ACR), augments 80S complex formation from the 48S complex by facilitating the correct positioning of the initiation codon of HCV mRNA (Figures 1–3) associated with the solvent side of the 40S ribosomal subunit, through direct interactions with the IRES and the 40S ribosomal subunit. Direct binding of NSAP1 with r-proteins RpS2, RpS4, RpS6 and/or RpS8 seems to participate in the interaction between NSAP1 and the 40S ribosomal subunit (Figure 4C and D).
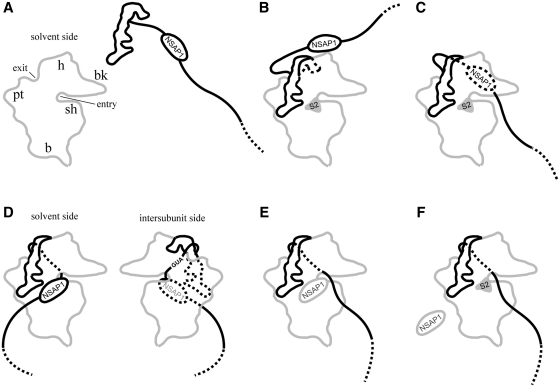
By focusing on the role of NSAP1, we can envision the sequential steps in the translation of HCV mRNA (Figure 6). HCV mRNA, which may be pre-associated with NSAP1 (Figure 6A), interacts with the solvent side of the 40S ribosomal subunit through the bottom half of stem–loop III, including the pseudoknot structure and the region surrounding the initiation codon of HCV IRES (18,37). The association of NSAP1 with the mRNA may occur before or after the interaction between the HCV IRES and the 40S ribosomal subunit. Without the assistance of NSAP1, the majority of the ribosome-associated HCV mRNAs is not properly positioned for translation initiation, as shown in lane 4 in Figure 2A (Steps B and C in Figure 6). The interaction(s) between NSAP1 and 40S ribosomal subunit r-protein(s) (RpS2, RpS4, RpS6 and/or RpS8) assists in properly positioning HCV mRNA on the 40S ribosomal subunit for the next step: joining of the 60S ribosomal subunit (Figure 6D). According to this model, correct positioning of the initiation codon-containing intervening sequence (between the ribosome-binding site and the ACR) at the 60S ribosomal subunit-binding side of the 40S ribosomal subunit is facilitated by NSAP1–r-protein(s) interactions, as shown in Figure 6D (also compare lanes 3 and 4 in Figure 2A). In this model, we used the position of the 40S ribosomal subunit protein RpS2 (equivalent to E. coli RpS5), which is located near the mRNA entrance site, because the positions of the other r-proteins (RpS4, RpS6 and RpS8) on the 40S ribosomal subunit are not yet known. Which NSAP-interacting r-protein (or proteins) actually participates in enhancing the translation of HCV mRNA, however, remains unclear (see below). After association of the 60S ribosomal subunit with the 48S complex, NSAP1 dissociates from the mRNA. This dissociation step may be necessary to allow the elongation step of translation to proceed; otherwise, NSAP1 associated with the mRNA could block threading of the mRNA through the mRNA entry site. Interestingly, the interaction of NSAP1 with the 40S ribosomal subunit triggers dissociation of NSAP1 from mRNA, as shown in Figure 5 (step E in Figure 6), allowing translational elongation of HCV mRNA. The NSAP1 on the ribosome eventually dissociates from the ribosome and is reused in the next cycle of translation (step F in Figure 6).
Figure 6.
Proposed model for the enhancement of HCV mRNA translation by NSAP1. (A) NSAP1 binds to ACR on the HCV IRES. (B) HCV IRES associates with the 40S ribosomal subunit. (C) The coding region of HCV mRNA is in motion to find the P-site in the 40S ribosomal subunit. (D) NSAP1 interacts with a r-protein (or proteins) on the solvent side of the 40S ribosomal subunit. This is predicted to result in positioning of the AUG initiation codon at the P-site of the ribosome. (E) NSAP1 is released from the ACR and translation commences. (F) NSAP1 is released from the 40S ribosomal subunit, becoming available for use in the next round of translation. (B, C and D–F) represent complexes I and II in Figure 2, respectively.
Does NSAP1 enhance translation of other mRNAs by a mechanism similar to that described here? The answer is probably yes. Several viral mRNAs of the virus family Flaviviridae, such as classical swine fever virus (CSFV), bovine viral diarrhea virus (BVDV), border disease virus (BDV) and GB virus-B (GBV-B), which are related to HCV, contain A-rich elements in the core-coding region and 5′-UTR structures, similar to those of the HCV 5′-UTR (38–43). It is likely that NSAP1 functions similarly in these mRNAs through the A-rich elements. NSAP1 might also function in some cellular mRNAs. We found that about 2% of human mRNAs contain an A-rich element (>40% adenosine) ∼30 nt downstream of the initiation codon (data not shown). It remains to be determined whether translation of these mRNAs is augmented by NSAP1 in a manner similar to that of HCV mRNA.
NSAP1 has been reported to augment translation of some mRNAs containing IRES elements, notably BiP and AANAT mRNAs (27,28). The NSAP1-binding sites of these mRNAs reside within the 5′-UTRs rather than lying downstream of the initiation codon, as exemplified by the IRES-containing mRNAs of flaviviruses (see above). Considering the position of the NSAP1-binding site, NSAP1 might enhance the translation of BiP and AANAT mRNAs through a mechanism different from the HCV mRNA translational-enhancement mechanism described here. The HCV IRES contains a 40S ribosomal subunit-binding site (the stem–loop III) that is separated from the NSAP1-binding site (ACR), whereas no known 40S ribosomal subunit-binding site exists in BiP and AANAT IRESs. Therefore, the NSAP1 associated with the 5′-UTR of BiP and AANAT mRNAs might play a role in recruiting the 40S ribosomal subunit to the IRES elements. This possibility remains to be investigated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: 21C Frontier Functional Proteomics Project (FPR08B1-220), Bio R&D Program (No. 2010-0018167), World Class University Program (No. R31-10105), NCRC (No. 2010-0028447) and BRL (No. 2010-0019706) through the National Research foundation funded by the Ministry of Education, Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell. Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 3.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang SK. Internal initiation: IRES elements of picornaviruses and hepatitis C virus. Virus Res. 2006;119:2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Boussadia O, Niepmann M, Creancier L, Prats AC, Dautry F, Jacquemin-Sablon H. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003;77:3353–3359. doi: 10.1128/JVI.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belsham GJ, Sonenberg N, Svitkin YV. The role of the La autoantigen in internal initiation. Curr. Top. Microbiol. Immunol. 1995;203:85–98. doi: 10.1007/978-3-642-79663-0_4. [DOI] [PubMed] [Google Scholar]
- 12.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 14.Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo CJ, Wolin SL. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997;89:393–402. doi: 10.1016/s0092-8674(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 16.Sandberg K, Mulroney SE. RNA Binding Proteins: New Concepts in Gene Regulation. Boston: Kluwer Academic; 2002. [Google Scholar]
- 17.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 1998;72:8782–8788. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ali N, Pruijn GJ, Kenan DJ, Keene JD, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 2000;275:27531–27540. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]
- 23.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 2008;82:12082–12093. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JH, Paek KY, Ha SH, Cho S, Choi K, Kim CS, Ryu SH, Jang SK. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 2004;24:7878–7890. doi: 10.1128/MCB.24.18.7878-7890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris CE, Boden RA, Astell CR. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol. 1999;73:72–80. doi: 10.1128/jvi.73.1.72-80.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TD, Woo KC, Cho S, Ha DC, Jang SK, Kim KT. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007;21:797–810. doi: 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho S, Park SM, Kim TD, Kim JH, Kim KT, Jang SK. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell. Biol. 2007;27:368–383. doi: 10.1128/MCB.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim WJ, Kim JH, Jang SK. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 2007;26:5020–5032. doi: 10.1038/sj.emboj.7601920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang CJ, Lo MC, Jan E. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J. Mol. Biol. 2009;387:42–58. doi: 10.1016/j.jmb.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 31.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 32.Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG. PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J. Biol. Chem. 2007;282:24514–24524. doi: 10.1074/jbc.M704406200. [DOI] [PubMed] [Google Scholar]
- 33.Kim JE, Ryu I, Kim WJ, Song OK, Ryu J, Kwon MY, Kim JH, Jang SK. Proline-rich transcript in brain protein induces stress granule formation. Mol. Cell. Biol. 2008;28:803–813. doi: 10.1128/MCB.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merrick WC. Assays for eukaryotic protein synthesis. Methods Enzymol. 1979;60:108–123. doi: 10.1016/s0076-6879(79)60011-3. [DOI] [PubMed] [Google Scholar]
- 35.Collatz E, Wool IG, Lin A, Stoffler G. The isolation of eukaryotic ribosomal proteins. The purification and characterization of the 40S ribosomal subunit proteins S2, S3, S4, S5, S6, S7, S8, S9, S13, S23/S24, S27 and S28. J. Biol. Chem. 1976;251:4666–4672. [PubMed] [Google Scholar]
- 36.Malygin AA, Shaulo DD, Karpova GG. Proteins S7, S10, S16 and S19 of the human 40S ribosomal subunit are most resistant to dissociation by salt. Biochim. Biophys. Acta. 2000;1494:213–216. doi: 10.1016/s0167-4781(00)00252-9. [DOI] [PubMed] [Google Scholar]
- 37.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 38.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muerhoff AS, Leary TP, Simons JN, Pilot-Matias TJ, Dawson GJ, Erker JC, Chalmers ML, Schlauder GG, Desai SM, Mushahwar IK. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J. Virol. 1995;69:5621–5630. doi: 10.1128/jvi.69.9.5621-5630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rijnbrand R, Bredenbeek PJ, Haasnoot PC, Kieft JS, Spaan WJ, Lemon SM. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA. 2001;7:585–597. doi: 10.1017/s1355838201000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Becher P, Orlich M, Thiel HJ. Complete genomic sequence of border disease virus, a pestivirus from sheep. J. Virol. 1998;72:5165–5173. doi: 10.1128/jvi.72.6.5165-5173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Moerlooze L, Lecomte C, Brown-Shimmer S, Schmetz D, Guiot C, Vandenbergh D, Allaer D, Rossius M, Chappuis G, Dina D, et al. Nucleotide sequence of the bovine viral diarrhoea virus Osloss strain: comparison with related viruses and identification of specific DNA probes in the 5′ untranslated region. J. Gen. Virol. 1993;74(Pt 7):1433–1438. doi: 10.1099/0022-1317-74-7-1433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.