Abstract
Investigation and manipulation of mitochondrial genetics in animal and plant cells remains restricted by the lack of an efficient in vivo transformation methodology. Mitochondrial transfection in whole cells and maintenance of the transfected DNA are main issues on this track. We showed earlier that isolated mitochondria from different organisms can import DNA. Exploiting this mechanism, we assessed the possibility to maintain exogenous DNA in plant organelles. Whereas homologous recombination is scarce in the higher plant nuclear compartment, recombination between large repeats generates the multipartite structure of the plant mitochondrial genome. These processes are under strict surveillance to avoid extensive genomic rearrangements. Nevertheless, following transfection of isolated organelles with constructs composed of a partial gfp gene flanked by fragments of mitochondrial DNA, we demonstrated in organello homologous recombination of the imported DNA with the resident DNA and integration of the reporter gene. Recombination yielded insertion of a continuous exogenous DNA fragment including the gfp sequence and at least 0.5 kb of flanking sequence on each side. According to our observations, transfection constructs carrying multiple sequences homologous to the mitochondrial DNA should be suitable and targeting of most regions in the organelle genome should be feasible, making the approach of general interest.
INTRODUCTION
Mitochondrial genetic systems are essential for eukaryotic cells, as they provide a number of proteins which are involved in the structure or in the biogenesis of the oxidative phosphorylation chain. Mutations in the compact human mitochondrial DNA (mtDNA, 16.5 kb) cause numerous degenerative disorders which are currently incurable (1). Mutations in the mitochondrial genome in mammals have also been implicated in ageing (2). In plants, mitochondrial genetic processes are complex, including cis- and trans-splicing of introns (3), RNA processing and surveillance (4), as well as RNA editing (5). Mitochondrial genetics influences traits of great agronomical importance, especially cytoplasmic male sterility (CMS), which is widely used in crops for breeding and hybrid maintenance (6–8). Manipulating the mitochondrial genetic systems is thus of both fundamental and applied relevance. Unfortunately, only yeast and Chlamydomonas reinhardtii cells are amenable to mitochondrial genetic transformation using biolistic approaches (9,10). Despite the efforts of a number of laboratories, neither mammalian nor plant mitochondria could be transformed in whole cells with the current conventional methodologies. Gene therapy for diseases originating from mtDNA mutations is therefore still in its infancy (11). The lack of reverse genetic approaches makes it difficult to search for new genes and regulatory functions in the large plant mitochondrial genomes (up to 740 kb so far) in which more than half of the sequences are unassigned (12). Similarly, the expression of foreign RNAs or proteins in mitochondria could not be addressed, despite its fundamental and biotechnological interest.
Failure to transform mammalian and plant mitochondria raises fundamental questions regarding the transfer and fate of foreign DNA in the organelles. We previously demonstrated that isolated plant and mammalian mitochondria actually import double-stranded linear DNA (13,14). Sensitive to a number of effectors (13–15), this transport process appears to be mediated by a physiological mechanism which might possibly be exploitable to promote transfection of mitochondria in whole cells. In this respect, different types of mitochondriotropic vesicles have been developed to carry exogenous DNA into mammalian cells and towards mitochondria [e.g. (16,17)]. A prerequisite for such strategies to become a major breakthrough towards mitochondrial transformation is that the imported exogenous DNA functionally joins the organelle genetic system. As a first step, we previously showed that, following incorporation into isolated plant or mammalian mitochondria, a gene controlled by a mitochondrial promoter can be transcribed in organello, establishing that the imported DNA is able to recruit the components of the RNA synthesis system (13,14). The produced transcripts can be processed and matured (14,18).
To open the way for novel mitochondrial transformation approaches built on organelle natural competence, it remains to be established that exogenous DNA taken up by mitochondria can be maintained and propagated. To that end, the imported DNA has to join the organellar genome and DNA metabolism. Whether in fungi, mammals or plants, the mtDNA is packaged into membrane-associated DNA–protein particles called nucleoids (19–22). Each nucleoid might contain several copies of the mitochondrial genome and the different organisms seem to have unique sets of nucleoid-associated proteins (23). The data suggest that mitochondrial nucleoids in eukaryotic cells are centers of mtDNA maintenance and expression (20). To be maintained, exogenous DNA imported into the organelles likely has to become integrated into nucleoids. However, it has been shown that mitochondrial nucleoids do not freely exchange their DNA but rather tightly regulate their genetic content (24).
If foreign sequences introduced into mitochondria can reach the regular location of the mtDNA, two ways of maintenance and propagation can be considered, i.e. autonomous replication or integration into the mitochondrial genome following recombination. Designing exogenous replicons on the basis of the current knowledge on mitochondrial replication mechanisms and sequence requirements remains difficult. The early model of mtDNA replication in mammalian mitochondria (25) has been recently rediscussed (26–28). Coexistence of recombination-dependent replication and recombination-independent rolling-circle replication has been proposed in plant mitochondria (29), but these processes remain little documented. Besides the main chromosomal genome, mitochondria from plants and fungi contain autonomous circular or linear plasmids (30,31). Plant circular plasmids are interesting models for mitochondrial replication (32), but the substrate for mitochondrial import is linear DNA (13) and the information on the replication of linear plasmids is limited (33). Although relevant activities were detected, recombination in mammalian mitochondria in turn remains a contentious subject and in any case is rare (34). On the contrary, homologous recombination based on sequence repeats is considered to shape the mtDNA in plant organelles, generating a dynamic set of subgenomic molecules (35,36). These complex processes lead to a stoichiometrically variable multipartite genome organization (37). However, recombination in plant mitochondria involves specific large size repeats and is under the surveillance of nuclear-encoded protein factors (38,39).
In the present work, we made use of DNA constructs consisting of a portion of the GFP (green fluorescent protein) reporter gene flanked by mtDNA sequences. We show that such exogenous DNA fragments imported into plant mitochondria can get through the different controls and limitations mentioned above, join the mtDNA and integrate into the organellar genome through homologous recombination. Analysis of the 5′ and 3′ junctions showed accurate integration of the reporter gene into the mtDNA, without sequence duplication or deletion.
MATERIALS AND METHODS
Gene constructs and plasmids
The first of the two constructs used for recombination studies was based on the 5.27-kb repeat present in the maize (Zea mays) mitochondrial DNA (40). Part of this repeat (i.e. nucleotides 3881–6927 in the complete maize mitochondrial genome, accession number AY506529) was amplified by PCR from maize mtDNA with the primers DR-5 and DR-3. The PCR product was cloned into the SacI and XhoI sites of the pBluescript KS vector (Stratagene). Its sequence fitted the database genomic sequence (accession number AY506529), apart from a few single nucleotide divergences. A 438-nt fragment of the gfp (green fluorescent protein) gene (nucleotides 98–535 of the coding sequence), amplified from plasmid pCK-GFP3 (41) with primers gfp_int_Hind_5 and gfp_int_Hind_3, was subsequently inserted into the central HindIII site (nucleotides 5419–5424 in the AY506529 sequence) of the cloned maize sequence. The insertion was in antisense orientation versus the maize sequence. The resulting plasmid was called pBDR-Zm/gfp.
For the second construct, a section (i.e. nucleotides 8166–10 261 in the sequence with the accession number X93575) of the potato (Solanum tuberosum) mitochondrial gene nad2, encoding subunit 2 of the NADH dehydrogenase, was amplified by PCR from potato mtDNA with primers nad2-5 and nad2-3. The PCR product was cloned into the HindIII and XbaI sites of the pBluescript KS vector. Sequencing revealed significant divergence versus the database information (accession number X93575), essentially residing in nad2 intron 4 which was longer by 131 nt (see Supplementary Data and Supplementary Figure S1, for the full sequence of the obtained product). The central SphI fragment (i.e. nucleotides 780–1181) of the cloned nad2 sequence was excised and replaced by the same partial gfp sequence as above. In this case, the 438 nt fragment of the gfp gene was amplified from plasmid pCK-GFP3 with primers gfp_int_Sph_5 and gfp_int_Sph_3. The insertion was also in antisense orientation versus the nad2 sequences. The resulting plasmid was called pBnad2-St/gfp.
The primers used to build these constructs had the following sequences:
DR-5 (SacI site underlined)
5′-AGCAAGAGCTCATACCGCTCCGTTAGGTACTA-3′
DR-3 (XhoI site underlined)
5′-GTACACTCGAGTCTTTGTTGAGTGTACCCCGA-3′
gfp_int_Hind_5 (HindIII site underlined)
5′-AGCAAAAGCTTGTGAAGGTGATGCAACATACG-3′
gfp_int_Hind_3 (HindIII site underlined)
5′-GTACAAAGCTTCTAGTTGAACGCTTCCATCTTC-3′
nad2-5 (HindIII site underlined)
5′-GTACAAAGCTTAGCGGCGAATTTCAAACTTGTG-3′
nad2-3 (XbaI site underlined)
5′-AGCAATCTAGAAACGACTTGTCACGATCCATTG-3′
gfp_int_Sph_5 (SphI site underlined)
5′-AGCAAGCATGCGTGAAGGTGATGCAACATACG-3′
gfp_int_Sph_3 (SphI site underlined)
5′-GTACAGCATGCCTAGTTGAACGCTTCCATCTTC-3′
DNA substrate labeling
To generate radioactive DNA substrates for mitochondrial import assays, the sequences of interest were amplified by PCR from plasmids pBDR-Zm/gfp and pBnad2-St/gfp with the couples of primers DR-5/DR-3 and nad2-5/nad2-3 (see above), respectively. Fifty nanograms of PCR product were subsequently submitted to a single additional cycle of PCR with a 20-min elongation step in the absence of unlabeled dCTP and in the presence of 100 µCi of [α-32P]dCTP (3000 Ci/mmol). During the last 5 min of elongation, 0.2 mM of unlabeled dCTP was added. After the reaction, the unincorporated radioactivity was eliminated by gel-filtration through a Sephadex G-50 spin-column.
Plant material and isolation of mitochondria
In the case of maize, mitochondria were isolated from etiolated 4-day-old seedlings grown in complete darkness. Freshly cut epicotyls (200–300 g) were homogenized in a Waring blender with three volumes of isolation buffer (300 mM sucrose, 20 mM KH2PO4, 10 mM KCl, 5 mM EDTA, 1 mM MgCl2, 0.1% w/v BSA, 6 mM ß-mercaptoethanol, pH 8.2). Following filtration through a nylon net (100 µm mesh), the homogenate was centrifuged at 1500g for 15 min to remove cell debris and nuclei. Crude mitochondria were subsequently spun down at 16 000g for 15 min. The pellet was resuspended in washing buffer (300 mM sucrose, 20 mM KH2PO4, 10 mM KCl, 5 mM EDTA, 1 mM MgCl2, 0.1% w/v BSA, pH 7.2), loaded onto a sucrose cushion (1 M sucrose, 20 mM KH2PO4, 5 mM EDTA, 1 mM MgCl2, pH 7.2) and centrifuged at 40 000g for 15 min. The upper layer containing the mitochondria was transferred to a new tube, diluted with washing buffer and centrifuged at 16 000g for 15 min. The final pellet was resuspended in a small volume of washing buffer.
For potato, mitochondria were isolated from tubers according to Neuburger et al. (42) and Koulintchenko et al. (13). Tobacco mitochondria were isolated from wild type BY-2 Nicotiana tabacum cell suspensions. A 4-day-old BY-2 cell suspension was filtered through a 100-µm mesh nylon net. To prepare protoplasts, the recovered cells were resuspended in 50 ml of enzymatic solution (0.1% w/v pectolyase Y23, 1% w/v cellulase RS (Onozuka), 0.45 M mannitol, 3.5 mM MES, pH 5.5). The suspension was transferred to a Petri dish and the cell wall was digested for 2 h at 30°C in the dark. Protoplasts were centrifuged at 800g for 5 min and washed with 40 ml of 0.45 M mannitol, 3.5 mM MES, pH 5.5. After a 10 min centrifugation at 800g, the pellet was resuspended in 40 ml of extraction buffer (0.3 M sucrose, 30 mM sodium diphosphate, 2 mM EDTA, 0.3% w/v BSA, 0.8% w/v polyvinyl pyrrolidone 25 K, 0.5% w/v cysteine, 5 mM glycine, 2 mM ß-mercaptoethanol). The protoplasts were disrupted by passing three times through a 30-µm mesh nylon net. Mitochondria were further isolated according to Delage et al. (43). After centrifugation of the filtrate at 2000g for 15 min, the supernatant was recovered and centrifuged at 11 000g. The pellet was resuspended in 2–3 ml of washing buffer (300 mM sucrose, 10 mM K2HPO4, 1 mM EDTA, 5 mM glycine, 0.1% w/v BSA, pH 7.5) and mitochondria were purified by centrifugation through discontinuous Percoll (Sigma) gradients (successive layers of 45, 21 and 13.5% v/v Percoll in 250 mM sucrose, 50 mM Tris–HCl, 3 mM EDTA, pH 7.5) for 1 h at 40 000g. Intact mitochondria collected from the 13.5/21% interphase were diluted 10 times with washing buffer and pelleted by centrifugation at 12 500g for 15 min. This washing step was repeated twice.
Mitochondrial import and in organello recombination assays
Mitochondrial import of DNA constructs was carried out in 40 mM potassium phosphate, 0.4 M sucrose, pH 7.0 (13) in the case of potato and tobacco mitochondria and in 40 mM potassium phosphate, 0.3 M sucrose, pH 7.0 in the case of maize seedling mitochondria. The samples (1 ml) containing up to 50 ng of [32P]-labeled DNA and an amount of purified mitochondria corresponding to 1 mg of proteins were incubated at 25°C for 40 min under mild shaking. Following addition of 200 µg of DNase I and 10 mM MgCl2, the incubation was continued for 20 min in the same conditions. Mitochondria were subsequently washed two times by resuspension in 1 ml of 10 mM potassium phosphate, 300 mM sucrose, 10 mM EDTA, 10 mM EGTA, 0.1% w/v BSA, 5 mM glycine, pH 7.5 and centrifugation for 5 min at 10 000g. The final pellet was resuspended in DNA synthesis buffer. In the case of potato mitochondria, the DNA synthesis buffer contained 330 mM sucrose, 90 mM KCl, 10 mM MgCl2, 12 mM tricine, 5 mM KH2PO4, 1.2 mM EGTA, 1 mM GTP, 2 mM DTT, 2 mM ADP, 10 mM sodium succinate, 50 µM of each dNTP (dATP, dGTP, dTTP, dCTP), pH 7.2 (13). For the experiments with maize and tobacco mitochondria, the DNA synthesis buffer was 300 mM sucrose, 10 mM KCl, 5 mM MgCl2, 12 mM tricine, 5 mM KH2PO4, 1.2 mM EGTA, 1 mM GTP, 2 mM DTT, 2 mM ADP, 10 mM sodium succinate, 0.15 mM of each dNTP (dATP, dGTP, dTTP, dCTP), pH 7.2. Upon further incubation for 1–2 h at 25°C under mild shaking, mitochondria were pelleted and extracted with one volume of 10 mM Tris–HCl, 1 mM EDTA, 1% w/v SDS, pH 7.5 and one volume of phenol. The nucleic acids recovered in the aqueous phase were ethanol precipitated.
Electrophoretic analysis of import and in organello recombination samples
Different gel electrophoresis methods were used to analyze the nucleic acid samples from mitochondrial import and in organello recombination assays. Native electrophoresis was on 1% w/v agarose gels in TAE buffer (40 mM Tris–acetate, 1 mM EDTA, pH 8.0) and containing 0.5 µg/ml ethidium bromide. Before loading on the gel, samples were completed with 0.2 volume of glycerol 50% v/v, SDS 1% w/v, EDTA 1 mM, bromphenol blue 0.1% w/v, xylene cyanol 0.1% w/v. Alkaline electrophoresis was on 1% w/v agarose gels in alkaline buffer (50 mM NaOH, 10 mM EDTA). Before loading on the gel, samples were completed with 0.2 volume of 50 mM NaOH, 1 mM EDTA, 3% w/v Ficoll (Type 400), 0.025% w/v bromcresol green, 0.05% w/v xylene cyanol. Migration was carried out at 3.5 V/cm. Gels were neutralized by soaking in 1 M Tris–HCl pH 7.6, 1.5 M NaCl for 40 min and subsequently stained with 0.5 µg/ml ethidium bromide in TAE buffer for 30 min. As an alternative method for denaturing analysis, samples were heated for 5 min at 100°C in 20 mM EDTA, 0.05% w/v bromphenol blue, 0.05% w/v xylene cyanol, 95% v/v formamide, chilled on ice for at least 2 min and loaded on a native 1% w/v agarose gel. All samples were finally transferred from the agarose gels to nylon membranes and revealed by autoradiography.
Analysis of import and in organello recombination samples following restriction digestion
For such assays, nucleic acids extracted from the mitochondria after import and post-incubation in DNA synthesis conditions were separated on a 0.8% w/v native agarose gel. The area of the gel containing the high molecular weight DNA was cut out and the DNA was eluted by centrifugation through glass-wool. The recovered material was phenol-extracted and ethanol-precipitated. After quantification, 5–10 µg of DNA were digested for 18 h with BamHI and ScaI restriction enzymes at 37°C in buffer appropriate for double digestion. To inactivate the enzymes, the temperature was raised to 70°C for 15 min. The final samples were run on a 0.8% w/v agarose gel in TAE buffer, transferred to a nylon membrane and submitted to autoradiography.
Southern hybridization
To confirm the migration of the mtDNA, aliquots of nucleic acid samples from mitochondrial import and in organello recombination assays were run on the agarose gels and transferred onto nylon membranes in alkaline conditions. Membranes were soaked for 10 min in 5× SSPE (750 mM NaCl, 200 mM NaH2PO4, 200 mM EDTA, pH 7.4) containing 1% w/v SDS. Pre-hybridization was run for 4 h at 42°C in hybridization buffer (5× SSPE, 50% v/v formamide, 5× Denhardt’s solution, 1% w/v SDS, 20 µg/ml denatured herring sperm DNA). Hybridization with a [32P]-labeled probe for the cob gene was run for 40 h at 42°C. Finally, the membranes were sequentially washed in 5× SSPE with 0.1% w/v SDS, 1× SSPE with 0.1% w/v SDS and 0.2× SSPE with 0.2% w/v SDS, before detection by autoradiography.
The cob probe (343 bp) was amplified from tobacco mtDNA with the following primers:
-
cob5
5′-CCAACCCCGAGCAATCTTAG-3′
-
cob3
5′-AAGCTCATCTGACCCCAAGG-3′
PCR analyses
PCR reactions were run with a high fidelity polymerase (Expand High Fidelity PCR System, Roche). For inverse PCR analyses, 1 µg of total DNA extracted from the mitochondria after import and in organello recombination assays was digested with the MseI restriction enzyme. After phenol extraction and ethanol precipitation, 100 ng of the DNA were used for ligation. To favor intramolecular ligation events, the reaction was carried out overnight at 11°C with a concentration of 5′ ends as low as 5 pM and 2.5 U of T4 DNA ligase. Following phenol extraction and ethanol precipitation, the ligated DNA served as a template for PCR reactions with the primers gfp_int3 and nad2St_inv (for the assays run with potato mitochondria) or gfp_int3 and nad2Nt_inv (for the assays run with tobacco mitochondria). For direct PCR reactions, we used as a template 100 pg of plasmid DNA or 100 ng of mitochondrial DNA extracted from the mitochondria after import and in organello recombination assays. To analyze in detail the recombination junctions, the appropriate regions were amplified with different combinations of primers. For DNA samples from the assays run with potato mitochondria, each junction was amplified independently. The 5′-junction region was amplified with primers ex2St and gfp_int5, whereas the 3′-junction region was obtained with primers gfp_int3 and ex3St. For DNA samples from the assays run with tobacco mitochondria, we first amplified, with primers ex1Nt and ex4Nt, a 2.5-kb fragment spanning the whole recombined region. This fragment was subsequently used as a template for nested PCR with primers ex2Nt and gfp_int5 (to amplify the 5′ junction) or gfp_int3 and ex3Nt (to amplify the 3′ junction).
The primers used for PCR strategies had the following sequences:
-
gfp_int3
5′-CTAGTTGAACGCTTCCATCTTC-3′
-
gfp_int5
5′-GTGAAGGTGATGCAACATACG-3′
-
nad2St_inv
5′-AGCAAGCTTGCCTAGCAGAGACGTGG-3′
-
nad2Nt_inv
5′-AATTTTTTATTATAAAGGGCAGGC-3′
-
ex2St
5′-GATAACCACGTCTCTGCTAGGCA-3′
-
ex3St
5′-AAAGAATGAAGTAATGAAAGAGG-3′
-
ex1Nt
5′-TTTTTGGATATATAATCCAAGTCG-3′
-
ex2Nt
5′-CTTTTTTCGTTGAGAATTCCTCG-3′
-
ex3Nt
5′-AAGTAATGAAAGAGGAAGTC-3′
-
ex4Nt
5′-TTTAAAGATATGAACTGAGTGCC-3′
RESULTS
DNA substrates for mitochondrial import and in organello homologous recombination
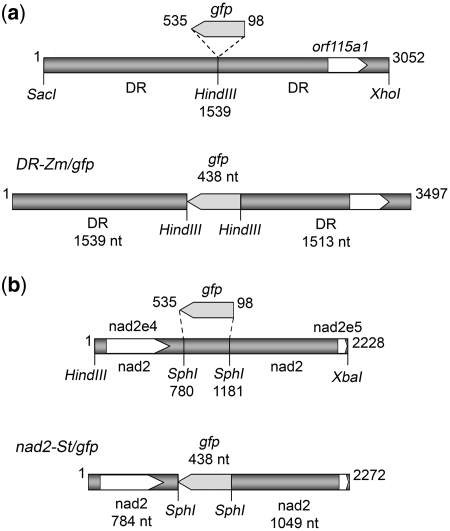
To evaluate the possibility to integrate exogenous imported DNA into the plant mitochondrial genome, two constructs were designed (schemes in Figure 1; full sequences in Supplementary Data, Supplementary Figures S2 and S3). Both of them consisted of a part of the gfp gene flanked by fragments of mitochondrial DNA. In the first construct, DR-Zm/gfp (Figure 1a), the partial gfp sequence was flanked by fragments of a 5.27-kb repeated DNA element present in the mitochondrial genome of maize fertile cytoplasm and not found in other plant species. This element is repeated twice in direct orientation in the maize mtDNA and is considered as a major site of recombination within the mitochondrial genome. Sequences related to the Rl (Sl) and R2 (S2) linear episomes also reside immediately adjacent to this repeat, with a short overlap at one of the two boundaries (44). In the second construct, nad2-St/gfp (Figure 1b), the partial gfp sequence was flanked by fragments of the nad2 gene from the potato mitochondrial genome: the end of intron 3, exon 4 and a piece of intron 4 on one side, the 3′ half of intron 4 and part of exon 5 on the other side. The choice of potato sequences, despite limited available genomic information, was justified by the fact that tubers from this species are a specially valuable source of functional mitochondria. On the other hand, the organization of the nad2 gene is similar in the species for which the sequence is known. The exon sequences of this gene are highly conserved, whereas the intron sequences present in our construct have inessential differences between the species. Thus, the nad2-St/gfp construct could be used to transfect potato, maize and tobacco isolated mitochondria. For import experiments, the DR-Zm/gfp and nad2-St/gfp constructs were synthesized as [32P]-radiolabeled linear DNA amplified by PCR as described previously (13) from plasmids pBDR-Zm/gfp and pBnad2-St/gfp, using the corresponding direct and reverse primers (see ‘Materials and Methods’ section).
Figure 1.
Organization of the gene constructs used as substrates for mitochondrial import and in organello recombination assays. Details of cloning and assembly are given in ‘Materials and Methods’ section. (a) Construct DR-Zm/gfp was composed of nucleotides 3881–6927 of the maize mtDNA (accession AY506529) with nucleotides 98–535 of the gfp coding sequence inserted in antisense orientation into the central HindIII site. (b) Construct nad2-St/gfp was composed of the end of intron 3, exon 4, intron 4 and part of exon 5 of the potato mitochondrial nad2 gene with nucleotides 98–535 of the gfp coding sequence replacing in antisense orientation the central SphI fragment. The number of nucleotides in the different components is indicated.
The imported DNA specifically associates with its counterpart sequences in the mitochondrial genome
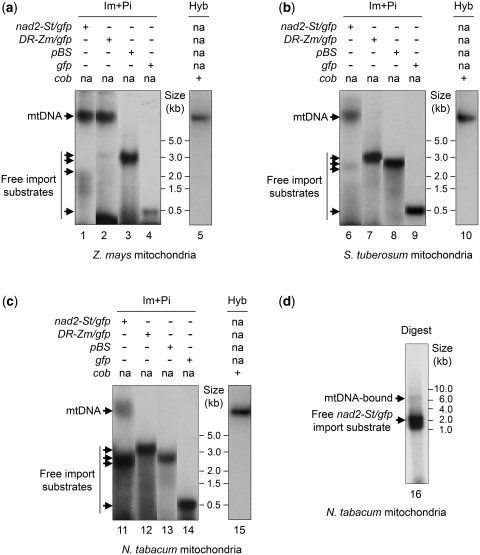
PCR-amplified radioactively labeled constructs nad2-St/gfp and DR-Zm/gfp were used as substrates for import into isolated maize or potato mitochondria, based on mitochondrial natural competence (13). Linear pBluescript plasmid (pBS, Stratagene), as well as the gfp fragment alone were used as control DNAs without homology to mitochondrial genome sequences. DNA import was followed by incubation of the organelles in DNA synthesis conditions. The latter were optimized in preliminary experiments according to the source of the mitochondria (see ‘Materials and Methods’ section). The result of the assays was assessed by analyzing the distribution of the radiolabeled substrate in the final mitochondrial fraction recovered from the organelles after extensive DNase treatment. Following import and post-incubation, DNA corresponding to constructs with homology to mtDNA sequences was associated with the high molecular weight mtDNA. The extent of association varied with the efficiency of individual experiments but could reach complete shift. When using maize mitochondria, the nad2-St/gfp DNA and the DR-Zm/gfp DNA were both co-migrating on gel with the mtDNA (Figure 2a, lanes 1 and 2). On the other hand, no shift towards the high molecular weight fraction was detected for linearized pBluescript and for the gfp fragment alone (Figure 2a, lanes 3 and 4). In the case of potato mitochondria, efficient association with the mtDNA took place with the nad2-St/gfp construct, but not with DR-Zm/gfp (Figure 2b, lanes 6 and 7). This was consistent with the fact that the maize DR region has no equivalent in the potato mitochondrial genome. Again, neither linearized pBluescript, nor the gfp fragment alone, co-migrated with the mtDNA (Figure 2b, lanes 8 and 9). As a whole, band shifting of the imported DNA appeared to be specific for the sequence content and could result from hybridization or recombination of the homologous sequences with their counterparts in the mitochondrial genome. That no label appeared at the level of the mtDNA in control assays also meant that no artifact occurred which would have been due to the release of radioactive nucleotides from the imported DNA and their re-incorporation into the organelle DNA. The nad2-St/gfp and DR-Zm/gfp constructs associated with the maize mtDNA to a similar extent (Figure 2a, lanes 1 and 2). This suggested that the process did not depend on the nature of the mitochondrial sequences present in the construct but only on the fact that the constructs contained sequences homologous to the mtDNA.
Figure 2.
Specific association of the imported DNA with the main mitochondrial DNA. (a–c) Different [32P]-labeled DNA substrates (nad2-St/gfp, DR-Zm/gfp, pBluescript and gfp) were imported into isolated maize (a), potato (b) or tobacco (c) mitochondria. The organelles were subsequently incubated in DNA synthesis buffer for 2 h. Nucleic acids were extracted and directly separated on native agarose gels that were transferred onto nylon membranes for autoradiography (Im + Pi). Migration of the mtDNA was confirmed through Southern blot hybridization (Hyb) with a probe for the cob gene (lanes 5, 10 and 15). ‘na’ stands for ‘not applicable’. (d) Following import of radiolabeled nad2-St/gfp construct into isolated tobacco mitochondria and incubation in DNA synthesis buffer, nucleic acids were extracted from the organelle sample and fractionated on native agarose gel. The high molecular weight mtDNA was recovered from the gel and digested with BamHI and ScaI. The digested DNA was in turn fractionated on native agarose gel and transferred onto a nylon membrane for autoradiography (Digest). Migration of import substrates and of reference fragments is indicated in the different panels.
Unfortunately, more detailed manipulations failed with maize mitochondria, due to low stability of the isolated organelles. Further experiments were run with mitochondria from potato tubers and tobacco BY-2 cell cultures. Tobacco was advantageous because its mitochondrial genome has been entirely sequenced (36). Alignment of the potato nad2 sequences used in construct nad2-St/gfp with the corresponding tobacco mitochondrial sequences revealed that the organization of this gene is similar in the two species. At the same time, the alignment revealed local sequence differences which could in turn serve as markers to localize the borders of the homologous recombination events. In line with the results obtained for potato organelles, import/post-incubation assays with tobacco mitochondria revealed mtDNA association of the labeled nad2-St/gfp construct (Figure 2c, lane 11), whereas no migration shift was detected in the case of DR-Zm/gfp, pBluescript or gfp alone (Figure 2c, lanes 12–14). In these assays, we observed a lower proportion of nad2-St/gfp co-migrating with the mtDNA, as compared to maize and potato organelles (Figure 2a–c). To confirm that the imported nad2-St/gfp DNA only associated with its homologous mtDNA region, standard uptake of the radiolabeled construct into tobacco mitochondria was as previously followed by incubation in DNA synthesis conditions. The nucleic acids extracted from the organelles were fractionated on native agarose gel. The high molecular weight DNA was recovered from the gel, to eliminate the excess of free imported DNA, and digested with two restriction enzymes, BamHI and ScaI, for which no recognition sites were present in the nad2-St/gfp sequence. The size of the predicted BamHI–ScaI target sequence of the construct was estimated as 6.8 kb by computing from the known tobacco genomic sequence. The autoradiogram of the experiment (Figure 2d) revealed some remaining free imported DNA and a radioactive product of ∼7 kb. This suggested that the imported nad2-St/gfp DNA indeed specifically associated with a single specific site in the mitochondrial genome through its target-homologous sequences.
The imported DNA recombines with the main mitochondrial genome
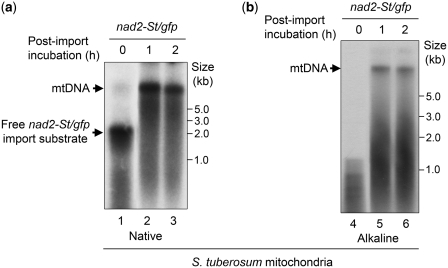
The above digestion assay showed that the nad2-St/gfp DNA specifically interacted with the main mtDNA. To establish to which extent the association was stable, DNA samples were further analyzed in denaturing conditions. Aliquots of mtDNA obtained in standard import/post-incubation experiments with radiolabeled nad2-St/gfp and isolated potato mitochondria were fractionated in parallel on native (Figure 3a) and on alkaline (Figure 3b) agarose gels. Even in denaturing conditions, part of the radioactivity remained associated with the high molecular weight mtDNA, supporting recombination (Figure 3b, lanes 5 and 6).
Figure 3.
Stable association of the imported DNA with the main mitochondrial DNA. Following import of [32P]-labeled nad2-St/gfp into isolated potato mitochondria and incubation in DNA synthesis buffer for 1 or 2 h, nucleic acids were extracted from the organelle samples and directly fractionated on native (a) or denaturing (b) agarose gel. Radioactivity was detected by autoradiography after transfer onto nylon membranes. Migration of the mtDNA, of import substrates and of reference fragments is indicated.
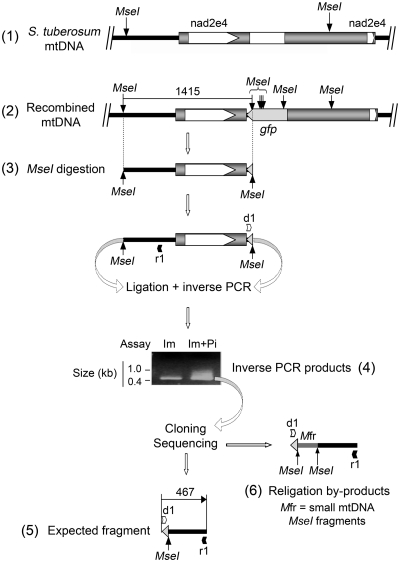
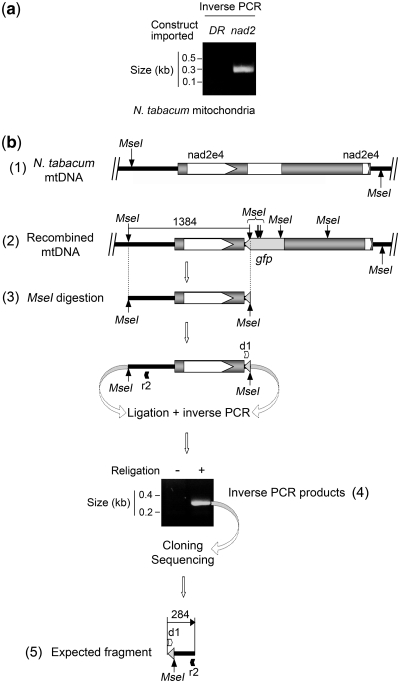
To prove the integration of the gfp reporter sequence into the potato mtDNA, an inverse PCR approach was designed (Figure 4). The DNA samples from import/post-incubation assays with nad2-St/gfp and potato mitochondria were digested with the MseI restriction enzyme and religated. Diverging primers were designed to amplify by inverse PCR a chimeric fragment which could only be obtained if the nad2-St/gfp was integrated into the mtDNA, as religation between an MseI site present in the mtDNA and a site brought in by the gfp sequence was needed (Figure 4). One primer was annealing in opposite orientation upstream of the region of homology between the construct and the mtDNA. The other was specific for the gfp sequence. In the most efficient experiments, specific inverse PCR amplification was obtained both from organelle samples collected already after the import step and from samples collected after post-incubation in DNA synthesis buffer, implying that some recombination could start at the import stage (Figure 4). Higher signals were nevertheless generated from post-incubated samples. The obtained inverse PCR products (Figure 4) were cloned and 89 clones were sequenced. From these, in 49 cases the PCR products had the predicted structure and contained the expected fragments from the gfp gene and the mtDNA region [Figure 4, scheme (5); full sequence in Supplementary Data, Supplementary Figure S4a]. The inserts in the remaining 40 clones were also representative for the recombination event but contained additional sequences of 50–150 bp in length corresponding to mtDNA-derived small MseI fragments inserted, during the religation reaction, between the mtDNA MseI site and the gfp MseI site considered for the inverse PCR strategy [Figure 4, scheme (6)]. Thus, inverse PCR provided direct evidence for the in organello integration of the foreign sequences into the potato mitochondrial genome.
Figure 4.
Integration of the imported nad2-St/gfp construct into the potato mitochondrial genome. Radiolabeled nad2-St/gfp construct was imported into isolated potato mitochondria. Half of the suspension was kept at that stage, whereas the other half was incubated in DNA synthesis buffer for 1 h. Nucleic acids were extracted from both samples, digested with MseI and religated. The religation mixes were used for inverse PCR. Both samples yielded inverse PCR products (4), implying that recombination can start during the import step. The PCR products generated from the import/post-incubation sample (Im + Pi) were cloned and sequenced. Sequencing revealed the fragment expected from integration by homologous recombination [(5); full sequence in Supplementary Data and Supplementary Figure S4a]. Religation by-products were also detected in which mtDNA-derived small MseI fragments were inserted between the MseI sites considered for the inverse PCR strategy (6). (1) Organization of the target region in the potato mtDNA with the original MseI sites; (2) expected organization and MseI site distribution following recombination of the imported nad2-St/gfp construct with the target region in the mtDNA and integration of the gfp sequence; (3) MseI fragment expected from the recombined target region (1415 nt); (4) agarose gel analysis of the inverse PCR products obtained with nucleic acids from organelle samples collected after the import step (Im) or after import and incubation in DNA synthesis buffer (Im + Pi), migration of reference fragments is indicated; (5) content of the inverse PCR products (467 nt) representative for homologous recombination; and (6) content of religation by-products; d1, primer gfp_int3; r1, primer nad2St_inv.
Similar experiments were developed with isolated tobacco mitochondria (Figure 5). Import/post-incubation assays were carried out as previously with both the nad2-St/gfp construct and the DR-Zm/gfp construct. The DNA samples obtained after phenol extraction were fractionated on agarose gel after denaturation and submitted as above to inverse PCR after MseI digestion and religation. Confirming the specificity of our observations, no amplification was obtained from assays involving the DR-Zm/gfp construct (Figure 5a). On the contrary, assays with the nad2-St/gfp construct yielded an inverse PCR product with the expected size [Figure 5a and b(4)]. Cloning and sequencing of the amplified fragment [Figure 5, scheme (5); full sequence in Supplementary Data, Supplementary Figure S4b] showed that the nad2-St/gfp construct based on the potato nad2 sequences could also recombine in organello with the tobacco mitochondrial genome.
Figure 5.
Integration of the imported nad2-St/gfp construct into the tobacco mitochondrial genome. (a) Following import of radiolabeled DR-Zm/gfp construct or nad2-St/gfp construct into isolated tobacco mitochondria and incubation in DNA synthesis buffer for 2 h, nucleic acids were extracted, digested with MseI and religated. The religation mixes were used for inverse PCR and the reaction products were analyzed on agarose gel; (DR) assay run with the DR-Zm/gfp construct, (nad2) assay run with the nad2-St/gfp construct; migration of reference fragments is indicated. (b) A similar assay as in (a) was run with the nad2-St/gfp construct and the inverse PCR products were cloned and sequenced. Sequencing revealed the fragment expected from integration by homologous recombination [(5); Supplementary Data and Supplementary Figure S4b]. (1) Organization of the target region in the tobacco mtDNA with the original MseI site; (2) expected organization and MseI site distribution following recombination of the imported nad2-St/gfp construct with the target region in the mtDNA and integration of the gfp sequence; (3) MseI fragment expected from the recombined target region (1384 nt); (4) agarose gel analysis of the inverse PCR products obtained after religation of the MseI digest ( + ); as a control, no inverse PCR products were obtained when omitting religation (–); migration of reference fragments is indicated; and (5) content of the inverse PCR products (284 nt) representative for homologous recombination; d1, primer gfp_int3; r2, primer nad2Nt_inv.
Analysis of the 5′ and 3′ junctions of the recombined region
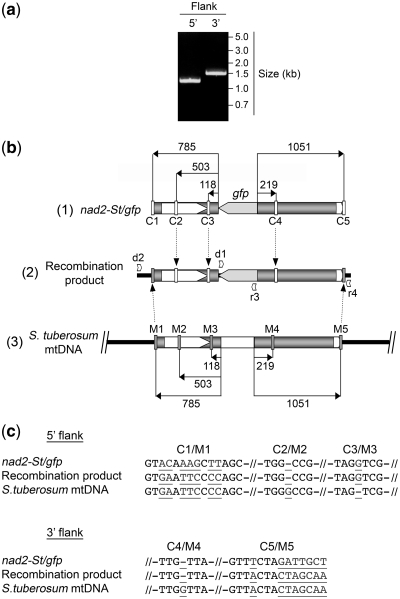
To analyze the sequences involved in the recombination process between the nad2-St/gfp construct and the potato mtDNA region, as well as the accuracy of the integration, the 5′ and 3′ flanking sequences of the integrated partial gfp gene were amplified independently by PCR (Figure 6a). For these reactions, one primer was specific for an mtDNA sequence upstream or downstream of the region of homology with the construct and the second was derived from the partial gfp gene [Figure 6b, scheme (2)]. The amplified products (Figure 6a) thus contained a fragment of the gfp sequence, the 5′ or 3′ mitochondrial sequence of the construct and a piece of mtDNA upstream or downstream of the sequences present in the construct. Scheme (2) in Figure 6b combines the two separate PCR products and the gfp sequence. The 5′ and 3′ mitochondrial sequences in the nad2-St/gfp construct had a few nucleotide differences versus the corresponding mtDNA sequences. These differences are indicated in Figure 6b and could serve as markers to distinguish the origin of the fragments. The corresponding sequence motifs are detailed in Figure 6c.
Figure 6.
Sequence analysis of the recombined region resulting from the integration of the imported nad2-St/gfp construct into the potato mitochondrial genome. Following import of radiolabeled nad2-St/gfp construct into isolated mitochondria and incubation in DNA synthesis buffer for 2 h, nucleic acids were extracted and used for direct PCR amplification of the 5′ and 3′ recombined regions flanking the integrated gfp sequence. The amplified products were cloned and sequenced. (a) analysis of the direct PCR products on agarose gel; position of the primers is indicated in (b), scheme (2); migration of reference fragments is indicated. (b) Integration of the nad2-St/gfp construct into the potato mtDNA; (1) organization of the nad2-St/gfp construct with the distribution of the sequence markers versus the potato mtDNA (C1–C5); (2) organization of the recombined region in the potato mtDNA showing the origin of the sequence markers, as determined experimentally; the 5′ part of the recombined region was amplified with primers d2 (ex2St) and r3 (gfp_int5); the 3′ part of the recombined region was amplified with primers d1 (gfp_int3) and r4 (ex3St); the two products were sequenced (full sequences in Supplementary Data and Supplementary Figure S5) and the sequences were assembled to yield the recombination product; (3) organization of the target region in the potato mtDNA with the distribution of the sequence markers versus the nad2-St/gfp construct (M1–M5). Distances from the borders of the gfp sequence or of the central mtDNA SphI fragment are indicated in number of nucleotides. (c) Details of the sequence marker motifs as positioned in (b); nucleotides differing between the nad2-St/gfp construct and the potato mtDNA are in underlined grey.
The obtained PCR products (Figure 6a) were cloned and sequenced (Supplementary Data and Supplementary Figure S5). In the cloned inserts, the sequences corresponding to the very ends of the regions of homology between the nad2-St/gfp construct and the potato mitochondrial genome came from the mtDNA itself (Figure 6b and c). On the 5′ side, the first sequences deriving from the construct, according to the markers, appeared at ∼500-bp upstream of the 5′-end of the partial gfp gene. On the 3′ side, we had only one internal marker in this system, at ∼200-bp downstream of the 3′-end of the gfp gene, and this marker corresponded to the construct sequence (Figure 6b and c). From these first data, it appeared that the exchange of homologous sequences extended up to at least 0.5 kb during the recombination process.
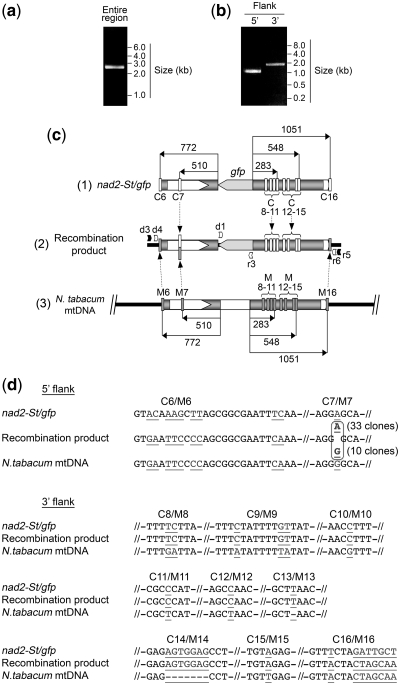
The investigations were further developed with tobacco mitochondria, taking advantage of the short local sequence differences between the nad2-St/gfp construct and the corresponding N. tabacum genomic region (Figure 7c and d). Although a single PCR step also proved sufficient to amplify the recombined regions, in this case we opted for a nested PCR strategy for detailed analyses, so as to favor maximum specificity. Thus, following the import and in organello recombination assay, a first PCR reaction was performed with primers specific for mtDNA sequences upstream and downstream of the region of homology between the construct and the mtDNA (Figure 7a and c). Nested PCR was subsequently run with the first PCR product to amplify independently the 5′ and 3′ flanking sequences of the integrated partial gfp gene (Figure 7b and c). Scheme (2) in Figure 7c combines the PCR products and the gfp sequence and indicates the position of the local differences between the 5′ and 3′ potato mitochondrial sequences in the nad2-St/gfp construct and the corresponding tobacco mtDNA regions. The sequence motifs are detailed in Figure 7d. As previously, the obtained PCR products were cloned and sequenced (Supplementary Data and Supplementary Figure S6). The sequences corresponding to the very ends of the regions of homology between the nad2-St/gfp construct and the tobacco mitochondrial genome came from the mtDNA in the recombined products (Figure 7c and d). On the 5′ side, the single specific internal marker was at 510-bp upstream of the 5′-end of the gfp sequence. In ∼80% of the clones sequenced (33 out of 43), this marker came from the nad2-St/gfp construct. However, 20% of the clones (10 out of 43) contained the mtDNA-derived sequence at this position (Figure 7c and d), implying that the cutting edge of the homologous sequence exchange was indeed ∼0.5 kb. For the 3′ junction, all markers within the 548-bp region downstream of the 3′-end of the gfp gene had the construct-specific sequence (Figure 7c and d).
Figure 7.
Sequence analysis of the recombined region resulting from the integration of the imported nad2-St/gfp construct into the tobacco mitochondrial genome. Following import of radiolabeled nad2-St/gfp construct into isolated mitochondria and incubation in DNA synthesis buffer for 2 h, nucleic acids were extracted and used for direct PCR amplification of the entire recombined region. The obtained product served as a template to amplify independently by nested PCR the 5′ and 3′ recombined regions flanking the integrated gfp sequence. Nested PCR products were cloned and sequenced. (a) and (b) analysis of the direct PCR products on agarose gel; original amplification of the entire recombined region (a) and nested PCR for the 5′ and 3′ flanks (b); position of the primers is indicated in (c), scheme (2); migration of reference fragments is indicated. (c) Integration of the nad2-St/gfp construct into the tobacco mtDNA; (1) organization of the nad2-St/gfp construct with the distribution of the sequence markers versus the tobacco mtDNA (C6–C16); (2) organization of the recombined region in the tobacco mtDNA showing the origin of the sequence markers, as determined experimentally; marker 7 was of mixed origin; a fragment spanning the recombined region was first amplified with primers d3 (ex1Nt) and r5 (ex4Nt); the PCR product was subsequently used as a template for nested PCR, so as to amplify the 5′ part of the recombined region with primers d4 (ex2Nt) and r3 (gfp_int5) and the 3′ part of the recombined region with primers d1 (gfp_int3) and r6 (ex3Nt); the two nested PCR products were sequenced (full sequences in Supplementary Data and Supplementary Figure S6) and the sequences were assembled to yield the recombination product; (3) organization of the target region in the tobacco mtDNA with the distribution of the sequence markers versus the nad2-St/gfp construct (M6–M16). Distances from the borders of the gfp sequence or of the central mtDNA SphI fragment are indicated in number of nucleotides. (d) Details of the sequence marker motifs as positioned in (c); nucleotides differing between the nad2-St/gfp construct and the tobacco mtDNA are in underlined grey.
Altogether, sequence analyses of the recombined products from the assays with potato and tobacco mitochondria showed that: (i) the gfp reporter gene was integrated into the mtDNA in an accurate way without duplications or deletions; (ii) the recombination events occurred in the flanking sequences upstream and downstream of the reporter gene; (iii) the exchange of homologous sequences likely involved ∼0.5–0.6 kb on each side of the reporter gene, i.e. less than the flanking sequences available in the construct; and (iv) the process tolerated short local sequence differences between the imported DNA and the corresponding regions in the mtDNA.
DISCUSSION
The present work establishes that exogenous DNA can enter plant mitochondria, recombine with the mtDNA on the basis of homologous regions and transfer a sequence of interest into the resident genome. This makes it unlikely that the so far reported failure to obtain mitochondrial transformation in plant cells be due to a resistance of the mitochondrial genetic processes towards integration of the exogenous DNA. It rather suggests that unsuccessful attempts resulted from an inappropriate transfection approach or from the lack of a proper selection marker. Our results thus shed a new light on mitochondrial transformation in vivo.
We also report here the first direct investigation of an homologous recombination process in plant mitochondria. Endogenous recombination in plant mtDNA is based on specific repeats up to several kilobases in size and one could wonder whether these have special properties. In this context, it is interesting to point out that in our experiments recombination did not require specific features in the involved mitochondrial sequences, but only needed the presence of sequences homologous to the mtDNA in the imported construct. This suggests that most regions in the mitochondrial genome can be targeted for functional studies or insertion and expression of new genes, paving the way for a general use of such a strategy.
Deeper analysis based on sequence tags enabled further characterization of the mitochondrial recombination mechanism which appeared to be extremely precise. Sequencing results implied the accurate insertion of a continuous exogenous DNA fragment composed of the gfp sequence and at least 0.5 kb of intact construct sequence on each side. These observations somehow contrast with the data available for the recombination of synthetic DNA with the chloroplast genome, although such information remains scarce despite increasing development of plastid biotechnology [e.g. (45–47)]. Organelle DNA sequences present in transformation vectors also serve as targeting regions to direct integration into the plastid genome. Similar to our results, plastid targeting sequences have no special properties and may derive from most parts of the plastid genome. Foreign genes have been inserted in at least 14 intergenic regions (48). Based on polymorphic markers leading to antibiotic resistance, it has originally been considered that integration of the exogenous DNA resulted in complete or nearly complete replacement of the resident chloroplast DNA sequence by the corresponding sequence in the donor plasmid. In tobacco, the entire length of the homologous region seemed to be incorporated and retained in the transformed plastid genome (49). Recent analyses with new synthetic vectors carrying a number of sequence tags in the plastid targeting regions revealed a more contrasted situation. It was shown that recombination with the organelle DNA actually occurred at either end of the homologous sections, yielding recombination products with entirely synthetic (construct sequence inserted) or entirely wild-type (resident sequence kept) plastid DNA in the targeted region (46). The left targeting sequence of the construct was reported to be incorporated into the plastid genome in only half of the cases and partial replacement was also observed. Recombination could take place in the range of ∼120–250 nt upstream of the non-plastid sequence of the construct (46), whereas in our assays at least ∼0.5 kb of exogenous mitochondrial sequence was incorporated together with the gfp marker. Recombination close to the ends of the homologous regions was proposed to proceed through heteroduplex formation between the entire synthetic sequence and the plastid genome, followed by repair using either the organelle strand or the exogenous strand as a template (46). According to our data, a similar hypothesis for mitochondria would imply a full bias for the exogenous strand in the repair reaction, which in first instance seems unlikely. These data altogether suggest that recombination between the organelle genome and an exogenous DNA proceeds in a different way in plastids and in plant mitochondria. Also noticeable, diverging sequence stretches present in the transformation vector regions homologous to the organelle DNA appear to be deleted during recombination in chloroplasts, presumably by looping out, whereas diverging sequences specific for the targeted plastid genome region can be maintained (45).
A further observation unfavorable for heteroduplex repair in plant mitochondria is the 20% occurrence of recombination events clearly located inside the left targeting sequence, as revealed by the appearance of an mtDNA marker in an intermediate position (Figure 7c and d). Interestingly, as mentioned, this position where the sequence was distributed between the construct marker and the mtDNA marker was representative for the exchange of ∼500 bp of homologous sequence, a result which can be related to earlier data on recombination control within the resident mtDNA. Three nuclear-encoded factors have been shown to be involved in higher plant mitochondrial recombination surveillance and control of mtDNA rearrangements, MSH1 (37), RECA3 (39) and OSB1 (38). They appear to take part in the repression of homologous recombination between small or intermediate size repeats, so as to avoid extensive rearrangements of the mitochondrial genome. Similar mechanisms based in particular on the RECA1 protein have been described in the moss Physcomitrella patens (50). Whereas in wild-type Arabidopsis thaliana mitochondrial repeats up to 556 bp do not recombine (51), they become recombinationally active in msh1 mutants (52). Whether repression of recombination extends to intermediate repeats >556 bp could not be determined because there are no repeats in the size range between 556 bp and 4.3 kb in the A. thaliana mitochondrial genome and a similar situation occurs in other species [e.g. (12) and references therein]. Our observation of recombination events involving the exchange of only ∼500 bp of DNA from an otherwise longer homologous sequence suggests that the range of 500–600 bp is indeed the upper limit of recombination containment in plant mitochondria. Conversely, our results together with the literature data imply that recombination of an imported exogenous construct with the resident DNA will only be promoted by mtDNA-homologous sequences of at least ∼500 bp. This opens the interesting possibility to design transfection constructs carrying multiple short sequence stretches homologous to the mitochondrial genome (i.e. organelle promoter and regulation motifs associated with the exogenous gene of interest) and still direct the insertion to a single, presumably neutral locus of the mtDNA through longer repeats of >500 bp.
Remarkably, recombination control in C. reinhardtii, one of the two unicellular organisms amenable to mitochondrial transformation, appeared to be much looser. In that case, complementation of a point mutation located at 509 bp inside the region of homology between the mtDNA and a relevant transformation construct could not be obtained, suggesting that the sequence exchange was systematically <0.5 kb (53). Moreover, a construct carrying as little as 0.1 kb of sequence homologous to the mtDNA was still able to recombine with the linear C. reinhardtii mitochondrial genome and to complement a dum mutant with a large deletion in the left terminus (53). The efficiency of C. reinhardtii mitochondrial transformation nevertheless remained proportional to the length of the mtDNA-homologous region present in the transfected construct (53).
Beyond the transfection method, genetic transformation of organelle genomes presents specific requirements with respect to selectable markers. Multiple genome copies must be converted to the transformed genotype and the selection applied during this process should not be lethal, or transformants will be killed before they can be identified. S. cerevisiae and C. reinhardtii, for which the organelle function is expendable, allow for selection based on rescue of a mitochondrial mutation with wild-type gene copies [e.g. (54,55)]. In S. cerevisiae, co-transformation with two plasmids allows for primary selection based on a nuclear marker, which is then followed by selection for restored respiration. Developing a suitable selection strategy for the recovery of mitochondrial genome transformants in obligate aerobes such as land plants still remains an open challenge.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
French Centre National de la Recherche Scientifique (CNRS, UPR2357) through institutional funding and an international cooperation program (PICS); Université de Strasbourg (UdS); Agence Nationale de la Recherche (ANR-06-MRAR-037-02); Russian Academy of Sciences (RAS); Russian Foundation for Basic Research (RFBR). Funding for open access charge: IBMP, CNRS UPR2357.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank our colleagues in both laboratories, as well as R.N. Lightowlers, for help, discussions, suggestions or materials provided during the course of these studies.
REFERENCES
- 1.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta. 2010;1797:113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 3.Bonen L. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion. 2008;8:26–34. doi: 10.1016/j.mito.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Holec S, Lange H, Canaday J, Gagliardi D. Coping with cryptic and defective transcripts in plant mitochondria. Biochim. Biophys. Acta. 2008;1779:566–573. doi: 10.1016/j.bbagrm.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Pelletier G, Budar F. The molecular biology of cytoplasmically inherited male sterility and prospects for its engineering. Curr. Opin. Biotechnol. 2007;18:121–125. doi: 10.1016/j.copbio.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 8.McCauley DE, Olson MS. Do recent findings in plant mitochondrial molecular and population genetics have implications for the study of gynodioecy and cytonuclear conflict? Evolution. 2008;62:1013–1025. doi: 10.1111/j.1558-5646.2008.00363.x. [DOI] [PubMed] [Google Scholar]
- 9.Bonnefoy N, Remacle C, Fox TD. Genetic transformation of Saccharomyces cerevisiae and Chlamydomonas reinhardtii mitochondria. Methods Cell Biol. 2007;80:525–548. doi: 10.1016/S0091-679X(06)80026-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Liu L, Chen J. Mitochondrial DNA heteroplasmy in Candida glabrata after mitochondrial transformation. Eukaryot. Cell. 2010;9:806–814. doi: 10.1128/EC.00349-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyriakouli DS, Boesch P, Taylor RW, Lightowlers RN. Progress and prospects: gene therapy for mitochondrial DNA disease. Gene Ther. 2008;15:1017–1023. doi: 10.1038/gt.2008.91. [DOI] [PubMed] [Google Scholar]
- 12.Kubo T, Newton KJ. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8:5–14. doi: 10.1016/j.mito.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum. Mol. Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 15.Weber-Lotfi F, Ibrahim N, Boesch P, Cosset A, Konstantinov Y, Lightowlers RN, Dietrich A. Developing a genetic approach to investigate the mechanism of mitochondrial competence for DNA import. Biochim. Biophys. Acta. 2009;1787:320–327. doi: 10.1016/j.bbabio.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D'Souza GG, Boddapati SV, Weissig V. Mitochondrial leader sequence–plasmid DNA conjugates delivered into mammalian cells by DQAsomes co-localize with mitochondria. Mitochondrion. 2005;5:352–358. doi: 10.1016/j.mito.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Weissig V, Boddapati SV, Cheng SM, D’Souza GG. Liposomes and liposome-like vesicles for drug and DNA delivery to mitochondria. J. Liposome Res. 2006;16:249–264. doi: 10.1080/08982100600851169. [DOI] [PubMed] [Google Scholar]
- 18.Placido A, Gagliardi D, Gallerani R, Grienenberger JM, Maréchal-Drouard L. Fate of a larch unedited tRNA precursor expressed in potato mitochondria. J. Biol. Chem. 2005;280:33573–33579. doi: 10.1074/jbc.M505269200. [DOI] [PubMed] [Google Scholar]
- 19.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai H, Lo YS, Litvinchuk A, Wang YT, Jane WN, Hsiao LJ, Chiang KS. Structural and functional characterizations of mung bean mitochondrial nucleoids. Nucleic Acids Res. 2005;33:4725–4739. doi: 10.1093/nar/gki783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malka F, Lombes A, Rojo M. Organization, dynamics and transmission of mitochondrial DNA: focus on vertebrate nucleoids. Biochim. Biophys. Acta. 2006;1763:463–472. doi: 10.1016/j.bbamcr.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Park S, Hanekamp T, Thorsness MK, Thorsness PE. Yme2p is a mediator of nucleoid structure and number in mitochondria of the yeast Saccharomyces cerevisiae. Curr. Genet. 2006;50:173–182. doi: 10.1007/s00294-006-0087-9. [DOI] [PubMed] [Google Scholar]
- 23.Kucej M, Butow RA. Evolutionary tinkering with mitochondrial nucleoids. Trends Cell Biol. 2007;17:586–592. doi: 10.1016/j.tcb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 24.Gilkerson RW, Schon EA, Hernandez E, Davidson MM. Mitochondrial nucleoids maintain genetic autonomy but allow for functional complementation. J. Cell. Biol. 2008;181:1117–1128. doi: 10.1083/jcb.200712101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tapper DP, Clayton DA. Precise nucleotide location of the 5′ ends of RNA-primed nascent light strands of mouse mitochondrial DNA. J. Mol. Biol. 1982;162:1–16. doi: 10.1016/0022-2836(82)90159-0. [DOI] [PubMed] [Google Scholar]
- 26.Yasukawa T, Reyes A, Cluett TJ, Yang MY, Bowmaker M, Jacobs HT, Holt IJ. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J. 2006;25:5358–5371. doi: 10.1038/sj.emboj.7601392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008;283:36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohjoismaki JL, Holmes JB, Wood SR, Yang MY, Yasukawa T, Reyes A, Bailey LJ, Cluett TJ, Goffart S, Willcox S, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J. Mol. Biol. 2010;397:1144–1155. doi: 10.1016/j.jmb.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Backert S, Borner T. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.) Curr. Genet. 2000;37:304–314. doi: 10.1007/s002940050532. [DOI] [PubMed] [Google Scholar]
- 30.Brown GG, Zhang M. In: The Molecular Biology of Plant Mitochondria. Levings CS III, Vasil IK, editors. Dordrecht, the Netherlands: Kluwer Academic Publishers; 1995. pp. 61–91. [Google Scholar]
- 31.Griffiths AJ. Natural plasmids of filamentous fungi. Microbiol. Rev. 1995;59:673–685. doi: 10.1128/mr.59.4.673-685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Backert S. R-loop-dependent rolling-circle replication and a new model for DNA concatemer resolution by mitochondrial plasmid mp1. EMBO J. 2002;21:3128–3136. doi: 10.1093/emboj/cdf311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handa H. Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions? Mitochondrion. 2008;8:15–25. doi: 10.1016/j.mito.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Elson JL, Lightowlers RN. Mitochondrial DNA clonality in the dock: can surveillance swing the case? Trends Genet. 2006;22:603–607. doi: 10.1016/j.tig.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, Miyashita N, Nasuda S, Nakamura C, Mori N, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33:6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genomics. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 37.Abdelnoor RV, Yule R, Elo A, Christensen AC, Meyer-Gauen G, Mackenzie SA. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl Acad. Sci. USA. 2003;100:5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaegel V, Guermann B, Le Ret M, Andres C, Meyer D, Erhardt M, Canaday J, Gualberto JM, Imbault P. The plant-specific ssDNA binding protein OSB1 is involved in the stoichiometric transmission of mitochondrial DNA in Arabidopsis. Plant Cell. 2006;18:3548–3563. doi: 10.1105/tpc.106.042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clifton SW, Minx P, Fauron CM, Gibson M, Allen JO, Sun H, Thompson M, Barbazuk WB, Kanuganti S, Tayloe C, et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136:3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menand B, Maréchal-Drouard L, Sakamoto W, Dietrich A, Wintz H. A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 1998;95:11014–11019. doi: 10.1073/pnas.95.18.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neuburger M, Journet EP, Bligny R, Carde JP, Douce R. Purification of plant mitochondria by isopycnic centrifugation in density gradients of Percoll. Arch. Biochem. Biophys. 1982;217:312–323. doi: 10.1016/0003-9861(82)90507-0. [DOI] [PubMed] [Google Scholar]
- 43.Delage L, Duchene AM, Zaepfel M, Maréchal-Drouard L. The anticodon and the D-domain sequences are essential determinants for plant cytosolic tRNA(Val) import into mitochondria. Plant J. 2003;34:623–633. doi: 10.1046/j.1365-313x.2003.01752.x. [DOI] [PubMed] [Google Scholar]
- 44.Houchins JP, Ginsburg H, Rohrbaugh M, Dale RM, Schardl CL, Hodge TP, Lonsdale DM. DNA sequence analysis of a 5.27-kb direct repeat occurring adjacent to the regions of S-episome homology in maize mitochondria. EMBO J. 1986;5:2781–2788. doi: 10.1002/j.1460-2075.1986.tb04568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruhlman T, Verma D, Samson N, Daniell H. The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 2010;152:2088–2104. doi: 10.1104/pp.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinagawa-Garcia SR, Tungsuchat-Huang T, Paredes-Lopez O, Maliga P. Next generation synthetic vectors for transformation of the plastid genome of higher plants. Plant Mol. Biol. 2009;70:487–498. doi: 10.1007/s11103-009-9486-x. [DOI] [PubMed] [Google Scholar]
- 47.Wang HH, Yin WB, Hu ZM. Advances in chloroplast engineering. J. Genet. Genomics. 2009;36:387–398. doi: 10.1016/S1673-8527(08)60128-9. [DOI] [PubMed] [Google Scholar]
- 48.Maliga P. Plastid transformation in higher plants. Annu. Rev. Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- 49.Staub JM, Maliga P. Long regions of homologous DNA are incorporated into the tobacco plastid genome by transformation. Plant Cell. 1992;4:39–45. doi: 10.1105/tpc.4.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Odahara M, Kuroiwa H, Kuroiwa T, Sekine Y. Suppression of repeat-mediated gross mitochondrial genome rearrangements by RecA in the moss Physcomitrella patens. Plant Cell. 2009;21:1182–1194. doi: 10.1105/tpc.108.064709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 52.Arrieta-Montiel MP, Shedge V, Davila J, Christensen AC, Mackenzie SA. Diversity of the arabidopsis mitochondrial genome occurs via nuclear-controlled recombination activity. Genetics. 2009;183:1261–1268. doi: 10.1534/genetics.109.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamasaki T, Kurokawa S, Watanabe KI, Ikuta K, Ohama T. Shared molecular characteristics of successfully transformed mitochondrial genomes in Chlamydomonas reinhardtii. Plant Mol. Biol. 2005;58:515–527. doi: 10.1007/s11103-005-7081-3. [DOI] [PubMed] [Google Scholar]
- 54.Johnston SA, Anziano PQ, Shark K, Sanford JC, Butow RA. Mitochondrial transformation in yeast by bombardment with microprojectiles. Science. 1988;240:1538–1541. doi: 10.1126/science.2836954. [DOI] [PubMed] [Google Scholar]
- 55.Remacle C, Cardol P, Coosemans N, Gaisne M, Bonnefoy N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl Acad. Sci. USA. 2006;103:4771–4776. doi: 10.1073/pnas.0509501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.