Abstract
Aptamers are structured oligonucleotides that recognize molecular targets and can function as direct protein inhibitors. The best-known example is the thrombin-binding aptamer, TBA, a single-stranded 15-mer DNA that inhibits the activity of thrombin, the key enzyme of coagulation cascade. TBA folds as a G-quadruplex structure, as proved by its NMR structure. The X-ray structure of the complex between TBA and human α-thrombin was solved at 2.9-Å resolution, but did not provide details of the aptamer conformation and the interactions with the protein molecule. TBA is rapidly processed by nucleases. To improve the properties of TBA, a number of modified analogs have been produced. In particular, a modified TBA containing a 5′-5′ polarity inversion site, mTBA, has higher stability and higher affinity toward thrombin with respect to TBA, although it has a lower inhibitory activity. We present the crystal structure of the thrombin–mTBA complex at 2.15-Å resolution; the resulting model eventually provides a clear picture of thrombin–aptamers interaction, and also highlights the structural bases of the different properties of TBA and mTBA. Our findings open the way for a rational design of modified aptamers with improved potency as anticoagulant drugs.
INTRODUCTION
Aptamers are single-stranded nucleic acids, both DNA (1) and RNA (2), which bind molecular targets, including proteins, with high affinity and specificity. These peculiar features are related to a tertiary structure, which presents a good shape complementarity with the target molecule (3). Aptamers have been developed for several different fields of applications, in particular, as diagnostic and therapeutic agents (4). The best-known example is that of the thrombin-binding aptamer (TBA), a DNA 15-mer consensus sequence, namely 5′-GGTTGGTGTGGTTGG-3′, discovered in 1992 through the SELEX (Systematic Evolution of Ligands by Exponential Enrichment) methodology (1) when 1013 different DNA molecules were synthesized and screened for thrombin binding.
α-thrombin (thrombin) is a trypsin-like serine protease that plays a pivotal role in haemostasis. Indeed, it is the only enzyme capable of catalyzing the conversion of soluble fibrinogen in insoluble fibrin strands and is the most potent platelet activator. Apart from these procoagulant functions, thrombin plays also an anticoagulant and antifibrinolytic activity in the presence of thrombomodulin (5).
The capability of inhibiting and regulating thrombin activity in vivo by synthetic compounds is an important goal in prevention of thrombosis. The presence on the thrombin surface of two anion-binding subsites or ‘exosites’, distinct from the catalytic center, makes it a more discriminating enzyme as compared to other proteases (6). Exosite I is the recognition site of thrombin physiological substrate fibrinogen and is also involved in the binding of leech anticoagulant hirudin, protease-activated receptor-1, thrombomodulin, factors V and VIII, glycoprotein-1ba and the acid domain of the serpin heparin cofactor II, whereas exosite II, which is located on the opposite side of thrombin, is the binding site of heparin and heparin-dependent serpins.
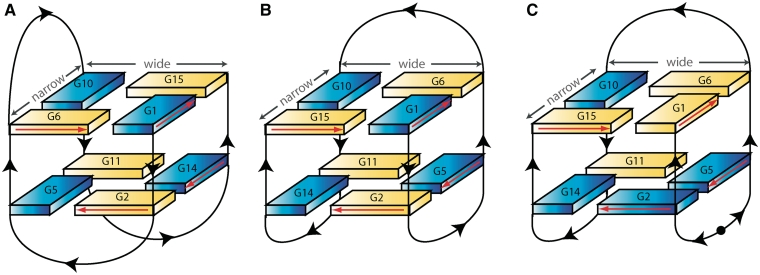
It has been shown that TBA is an ‘exosite inhibitor’ (7–9). It has a strong anticoagulant activity in vitro, it is efficient at nanomolar concentration (1) and acts on the two procoagulant functions of the enzyme: the activation of fibrinogen and the platelet aggregation. Moreover, it is able to bind both free and clot-bound thrombin (10,11), whereas binding to other serum proteins or proteolytic enzymes is essentially undetectable. The solution structure of TBA, determined by NMR (12,13), revealed that it folds in a unimolecular antiparallel quadruplex, with a chair-like conformation. This conformation is stabilized by cyclic hydrogen bonding of four guanines (G-tetrads or G-quartets) and by ions coordination between adjacent stacks of G-tetrads that are surrounded by two TT loops on one side and a TGT loop on the other side (12,13). The crystal structure of the thrombin–TBA complex was obtained at 2.9-Å resolution (Model 1) (14). The aptamer adopts a very different quadruplex topology with respect to the NMR structure. Indeed, the core formed by two G-quartets is the same in the two models, but structural differences exist in the way the central bases are connected. In particular, there is a difference concerning the disposition of the loops with respect to the grooves (Figure 1A and B). An alternative model of the crystal structure of the thrombin–TBA complex (Model 2), built on the basis of the NMR structure of the aptamer, refined to the same R value as the former Model 1, thus leaving the doubt on which structure was the correct one (15). In both models the TBA is sandwiched between two symmetry-related thrombin molecules and interacts with exosite I of a thrombin molecule and exosite II of the second one. In Model 1, the TGT loop is connected to exosite I and the TT loops are connected to exosite II. Model 2 shows an inverted pattern of protein–ligand interactions (15).
Figure 1.
Schematic illustration of (A) X-ray model of TBA (14), (B) NMR model of TBA (12,13), (C) solution (21) and crystallographic (present paper) model of the modified aptamer (mTBA). Black arrows indicate 5′→3′ polarity of the strands. Black circle in mTBA represents the 5′–5′ polarity inversion site. Anti and syn guanines are depicted as yellow and blue solids, respectively. Wide and narrow grooves are explicitly indicated in the three pictures. Red arrows indicate the direction of the proton donors and acceptors in Hoogsteen hydrogen bonds.
The uncertainty between these two models was caused by the absence of electron density in the region of TT and TGT loops connecting the G-tetrads. In a more systematic analysis (16), eight models of the thrombin–aptamer complex, different for the orientation of the NMR model of TBA, were tested on the previously used X-ray diffraction data (14,15). Subtle differences in the crystallographic R-factors and the analysis of the aptamer–protein interactions indicated that Model 2 was most likely the correct one. However, due to the missing density in the loop regions of the aptamer, the details of the ligand–protein interactions could not be properly addressed. Moreover, even recent papers still discuss aptamer-thrombin interactions on the basis of both models (17). In addition, also the stoichiometry of the complex in solution has been recently questioned, as two calorimetric studies suggest either a 2:1 (18) or a 1:1 (19) thrombin to aptamer molar ratio.
In recent years, several modified TBA have been produced and characterized, with the aim to obtain oligonucleotides with improved pharmacological properties, such as higher stability, higher thrombin affinity, longer life times in vivo, etc. (3). In particular, the main problem associated with the use of TBA is a rapid degradation by nucleases in vivo. Since it has been reported that the insertion of a chain polarity inversion site increases resistance to endogenous nucleases (20), a modified aptamer (mTBA) containing a 5′-5′ inversion between Thy3 and Thy4 was produced (21). mTBA presents higher thermal stability and higher thrombin affinity with respect to its unmodified counterpart. The NMR study of mTBA has revealed that it folds in a chair-like quadruplex structure, somehow similar to that of TBA (Figure 1C). However, because of the polarity inversion site, mTBA presents a (3+1) topology (22–24), with three strands parallel to each other and the fourth one oriented in the opposite direction; this topology results in a different syn and anti alternation of the bases within the tetrads and in different groove sizes. The differences between the two molecules do not provide a clear justification of the different properties deriving from the inversion site.
Here, we report the crystallographic analysis of the complex between thrombin and mTBA at 2.15-Å resolution. The higher resolution of the diffraction data, with respect to that of thrombin–TBA complex, has provided a unique, well defined model of the complex, which leaves no doubt on thrombin–aptamer interface. Moreover, the details of the interactions that the protein molecule makes with mTBA in comparison to TBA also allows to rationalize on structural grounds the different behavior of the two aptamers.
MATERIALS AND METHODS
Crystallization, structure determination and refinement
The thrombin–mTBA complex was prepared and crystallized as previously described (25). Briefly, mTBA dissolved in potassium–phosphate buffer 10 mM pH 7.1 was added to human α-thrombin inhibited by d-Phe-Pro-Arg-chloromethylketone (PPACK) in KCl 750 mM. After extensive washing, the resulting complex in potassium phosphate 50 mM pH 7.1 and KCl 100 mM was crystallized at 20°C using polyethylene glycol 20 000 20% w/v, ammonium sulfate 200 mM, n-propanol 3% v/v, sodium acetate 100 mM at pH 5.8. The structure was solved by the molecular replacement method, using the program AMoRe (26) and the PPACK-inhibited thrombin (PDB code 1PPB) (27) as a search model. To avoid bias, inhibitor and water molecules were removed from the model. The correlation for the highest solution was 0.59, with an R-factor of 0.36. The starting model was subjected to few cycles of rigid body refinement with CNS (28) followed by several cycles of coordinate minimization and B-factor refinement with CNS. Each run was alternated with manual model building using the program O (29). Fourier difference maps, calculated with (Fo–Fc) and (2Fo–Fc) coefficients, showed continuous electron density in the active site and in proximity of thrombin exosite I. The analysis of these maps allowed the fitting of PPACK in the active site, the building of the whole aptamer molecule, the identification of an N-acetyl glucosamine residue linked to Asn60G of the heavy chain, which is not present in the search model, and the positioning of several water molecules. At the end of the refinement, two strong electron density peaks were assigned to ions: one, placed near the segment 220–225 of thrombin, was attributed to a sodium ion, according to previous studies (30). The second one, placed between the two G-quartets and almost equidistant from the O6 atoms of the eight core guanines, was attributed to a potassium ion. The final model has R-factor/Rfree values of 0.196/0.252. The protein geometry of the refined structure was monitored using PROCHECK (31) and WHATCHECK (32). Statistics and parameters of the refinement are given in Supplementary Table S1. The drawings were prepared with Pymol (http://pymol.org). The coordinates of the structure have been deposited in the Protein Data Bank (Code 3QLP).
Structural analysis of mTBA and thrombin–mTBA complex
The geometry of the base assembly in mTBA was analyzed using a program that calculates the least squares plane of the two tetrads, the root mean square deviation of guanine atoms from the best planes, the angle that each guanine residue makes with the best plane of the tetrad to which it belongs, and the angle between the two G-quartet planes and their distance along the chain axis.
The network of water molecules bound to mTBA was identified with HBPLUS (33) and by visual inspection of the structure. Unconventional CH•••O hydrogen bonds were identified using HBAT (34).
Features of the protein–aptamer interface, including number and types of residues at the interface, and gap volume index, were calculated using the Protein–Protein Interaction Server (http://www.bioinformatics.sussex.ac.uk/protorp/). For comparison, the same parameters were calculated as average values from a non-redundant dataset of 25 protein–double-stranded DNA complexes solved at a resolution better than 2.4 Å. The shape complementarity score, Sc, as defined by Lawrence and Colman (35), was calculated using the CCP4 package (36), opportunely modified to include nucleic acid parameters. For atomic groups in DNA, radii standard values by Nadassy et al. (37) were used.
RESULTS
Overall structure
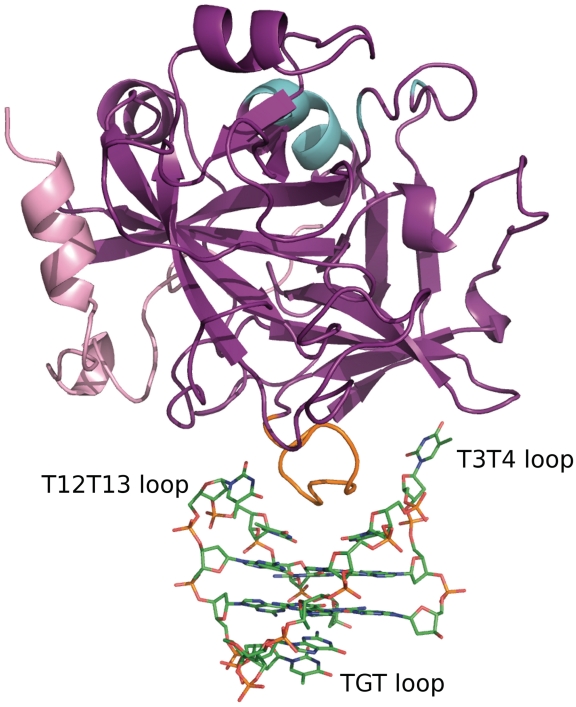
The structure of the complex between thrombin and the modified aptamer (mTBA) has been solved at 2.15-Å resolution (Figure 2). Detailed statistics of the refinement are reported in Supplementary Table S1.
Figure 2.
Overall structure of the thrombin–mTBA complex. Thrombin molecule is represented as cartoon, with heavy chain colored purple, light chain colored light pink, exosite I colored orange and exosite II colored cyan. mTBA molecule is represented as sticks.
The polypeptide chain of thrombin is very well defined for the whole heavy chain (residues 16–247) and for residues 1H–14L of the light chain. All residues in the refined structure lie within ‘allowed’ regions of the Ramachandran plot. As expected, the final model maintains all the structural features of the uncomplexed molecule. Indeed, in comparison to the unbound PPACK-inhibited enzyme (PDB code 1PPB) (27) the RMSD after the superposition of all the Cα protein atoms is only 0.67 Å. The largest deviations are localized in the light chain, at the residues of the flexible N- and C-terminal tails, whose conformation is usually found to be strongly affected by crystal packing. Similar results are also obtained when the present thrombin structure is compared to that of the molecule in complex with TBA (PDB code 1HAO) (15). In this case, the slightly larger RMSD of 0.95 Å is evenly distributed along the molecule and is likely related to the significantly lower resolution and quality of the latter structure. For this reason further comparisons have been done with the uncomplexed molecule (PDB code 1PPB).
mTBA molecule
The X-ray analysis of mTBA in complex with thrombin reveals that the molecule folds as a chair-like quadruplex with two stacked G-quartets, the TGT loop on one side and the two TT loops on the other side. Because of the presence of the chain inversion (Supplementary Figure S1A), mTBA possesses three strands parallel to each other and the fourth one oriented in the opposite sense, as also found in the solution structure of free mTBA (21) (Figure 1C).
Geometrical details of the G-quartet assembly are given in Supplementary Table S2 in comparison to those derived from solution structure. For each tetrad, the four guanine moieties that constitute each quartet are practically coplanar and their relative orientation and separation along the chain axis are in close agreement with the expected values for the π-stacking of aromatic bases. In comparison to the solution structure of free mTBA (21), the analysis reveals that in our crystallographic model the G-tetrads are less distorted from the ideal planar geometry. Interestingly, the separation of the tetrads in the former structure is significantly larger. In this context, it should be pointed out that in the crystallographic structure a potassium ion, which efficiently stabilizes quadruplex structures (38), is sandwiched between the two quartets (Supplementary Figure S1B). The ion has a coordination number of eight and a distorted antiprismatic geometry, at an average distance of 2.8 Å from the eight O6 atoms. In the refinement of the NMR structure, the presence of the metal ion was not contemplated and this may be the reason for the more compact structure of the two tetrads observed in the crystal state.
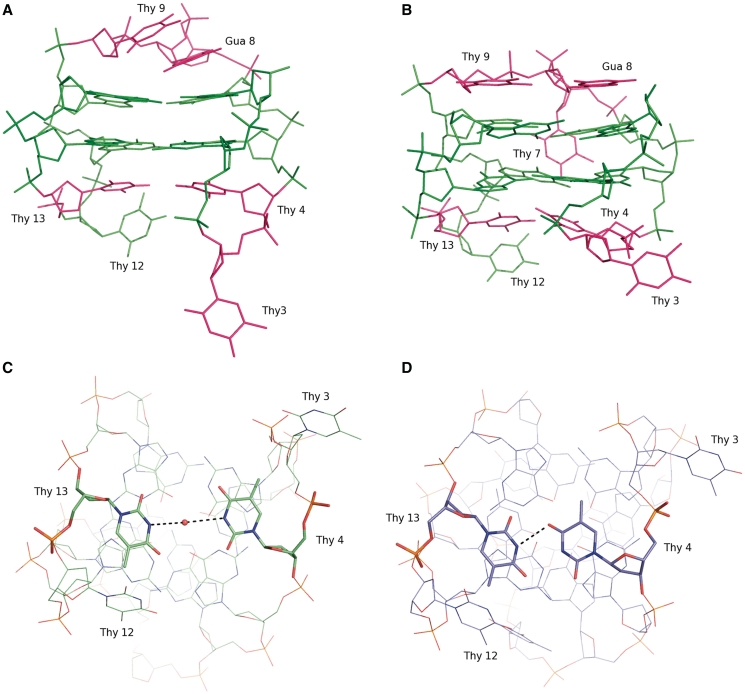
A detailed comparison between the unbound and complexed aptamer is of particular interest as it gives a deeper insight into the dynamics of the aptamer and the way its structure modifies to optimize the interaction with the protein molecule. With respect to free mTBA, the two TT loops, which form the most extended interacting surface with the thrombin molecule, move away from each other (Figure 3A and B) and the hydrogen bond between Thy4 and Thy13 is lost with the insertion of a bridging water molecule (Figure 3C and D). These two bases lie on a plane that stacks on the second G-quartet formed by Gua2, Gua5, Gua11, Gua14. Furthermore, Thy3 adopts a conformation that is completely different in the two models (Figure 3A and B). Interestingly, the more compact structure of the aptamer caused by the presence of the potassium ion in between the two tetrads combined to the binding of TT loops to thrombin, may also explain modifications observed in the TGT loop placed on the other side of the tetrads with respect to the TT loops. The TGT loop adopts in the crystal structure a different conformation, which is unlikely caused by the few and weak packing interactions with a second thrombin molecule (see below). Gua8 and Thy9, which in the solution structure stack on the quartet formed by Gua1, Gua6, Gua10 and Gua15, loose the interaction with the G-tetrad, become more parallel to each other and make direct stabilizing interactions (Figure 3A and B). This conformational change probably determines the expulsion of Thy7 from the wide groove so that the base becomes more exposed to the solvent and disordered.
Figure 3.
Comparison between the crystallographic structure (A and C) and the free structure in solution (21) (B and D) of mTBA. (A and B) Residues that adopt different conformations in the two models are depicted in magenta. (C and D) Thy4-Thy13 pair of mTBA. (C) The bridging water molecule in the crystallographic structure is also shown.
The analysis of mTBA structure highlights some interesting features. Whereas in nucleic acids structures the C8 of purine in anti conformation has been reported to form weak intraresidue C-H•••O interactions with the O5' (39–41), in mTBA inter-residue contacts between C8 and O4′ are observed (Supplementary Figure S2). A brief analysis of other quadruplex structures (PDB codes: 2AVH, 1JPQ, 1I34) shows that this type of interaction is not unusual in antiparallel quadruplex, where guanine residues with syn and anti conformations alternate along the strand. On the contrary it is absent in parallel structures with all anti guanines. This interaction is likely to contribute to some extent to the stability of the G-quadruplex.
Special attention was devoted to investigating the nucleotide-water interactions that are decisive for stability, dynamics, and recognition. The high resolution of the diffraction data gives the opportunity to study in details the organization of the water molecules bound to the aptamer, particularly in the four grooves. In these regions, the water molecules arrange themselves in a continuous network of hydrogen bonds, linking the backbone and the bases of the quadruplex. In G-quadruplex, water molecules are preferentially placed close to N2 and C8 atoms of guanines (42–44). Indeed in the modified aptamer structure, a conserved network of sequential N2–water–water–C8 hydrogen bonds is present in the narrow groove and in one of the two medium grooves; it connects adjacent strands and involves five of the eight core guanines (Gua2, Gua10, Gua11, Gua14, Gua15) (Supplementary Figure S3). New and less regular patterns of hydration, involving N2, N3 or backbone oxygens and a greater number of water molecules between adjacent strands, are observed in the wide groove and in the other medium groove. The mTBA model also reveals new features: the traditional C8–water–phosphate oxygen network is not predominant with respect to interactions where water molecules bridge C8 and oxygen atoms (O4′ or O3′) of the backbone or other water molecules.
Thymine bases, being primarily involved in the binding with thrombin, are less hydrated than guanines. The non-conventional binding of waters to the C6 atom observed, for example, for thymines of the quadruplex of Oxytricha nova telomeric sequence (42) or for all thymines of B-DNA (45), in the case of mTBA involves only Thy4. Finally, a water molecule is hydrogen bonded to the methyl group of Thy12.
This rich pattern of hydration involves both the four grooves and the three loops and it is likely to play a key role in maintaining the loop conformation.
Thrombin–mTBA interactions
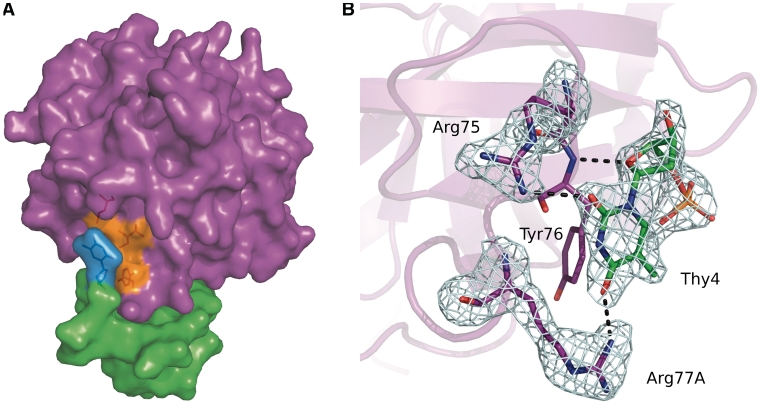
The structure of the complex definitely shows that mTBA interacts with thrombin exosite I by its TT loops with a further contribution of the backbone of Gua2 (Table 1). The recognition involves primarily the protein residues Leu65, Arg75, Tyr76, Glu77, Arg77A, Asn78, Ile79, Ile82. Upon binding, mTBA undergoes to a slight structural rearrangement, particularly in the TT loops (see above), whereas the thrombin molecule strictly retains its original structure (observed in the PPACK-thrombin structure, PDB code 1PPB), except for the flipping of the peptide group between Arg77A and Asn78 (Supplementary Figure S4). In the unbound molecule, these residues are in the second and third position, respectively, of a type I β-turn. In the complex, the turn is converted into a type II β-turn with Asn78 in a Lα conformation. The peptide flipping is favored by the presence of Asn in the third position of the turn, since this residue is known to populate the Lα region of the Ramachandran map (46). This modification of the protein appears to be functional, as it optimizes the contacts with the bound aptamer: the carbonyl oxygen of Arg77A points toward the base of Thy13, thus improving the complementarity between the interacting surfaces; moreover, the guanidium group is salt-bridged to the phosphate of Gua2 and makes a hydrogen bond with Thy4. Interactions between mTBA and thrombin are both hydrophobic and hydrophilic (Table 1). Thy3 occupies a hydrophobic patch lined by Leu65, Ile82 and Tyr76 (Figure 4A) that is in turn in contact with Thy4 (Figure 4B); Thy12 and Thy13 are in contact with Asn78 and Ile79. On the other hand, hydrogen bonds between the guanidium group of Arg75 and Thy4 and Thy13, together with those provided by Arg77A, contribute substantially to the stabilization of this TT pair. Thy12 also interacts with Glu77. Moreover, several water-mediated interactions between mTBA and thrombin residues (Thr74, Gly69, Gly25, His71, Lys81, Ser115, Tyr117) have been also identified.
Table 1.
Interactions between thrombin and mTBA at the most extended interface
| mTBA residue | Hydrogen bonds |
Hydrophobic interaction |
|||
|---|---|---|---|---|---|
| Atom | Thrombin residue | Atom | Distance (Å) | Thrombin residue | |
| Gua2 | O2P | Arg77A | NH1, NH2 | 3.17, 3.27 | |
| Thy3 | – | – | – | – | Tyr76, Leu65, Ile82 |
| Thy4 | O4′ | Tyr76 | N | 2.83 | Tyr76 |
| Thy4 | O2 | Arg75 | NH2 | 3.07 | |
| Thy4 | O4 | Arg77A | NH1 | 2.90 | |
| Thy12 | N3 | Glu77 | OE2 | 2.88 | Ile79 |
| Thy13 | O4 | Arg75 | NH1, NH2 | 3.15, 2.57 | Asn78, Ile79 |
| Thy13 | O3′ | Asn78 | ND2 | 2.81 | |
Figure 4.
(A) Surface representation of thrombin–mTBA complex. Thrombin molecule is colored purple, mTBA green, Thy3 is marked in cyan and the hydrophobic cavity lined by Leu65,Tyr76 and Ile82 side chains is marked in orange, (B) hydrogen bonds involving Thy4. Omit Fo−Fc electron density maps (5.0 σ level) of Thy4, Arg75 and Arg77A are also shown as example.
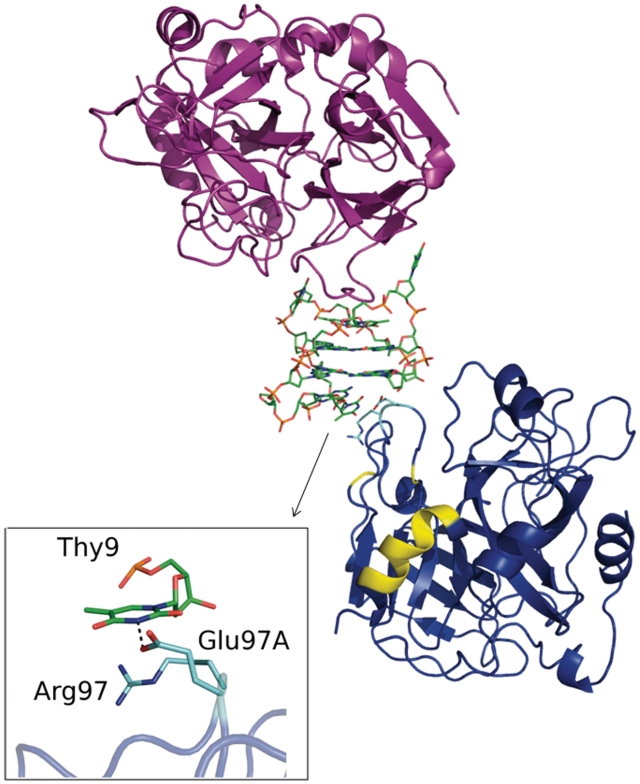
Interestingly, in the crystal structure mTBA interacts with a second thrombin molecule by its TGT loop. This interaction buries a contact area that is less than half of that involving exosite I (248 Å2 versus 570 Å2). In particular contacts between TGT loop and the second thrombin molecule are limited to Thy9 that is involved in cation–π interaction with Arg97 and in a hydrogen bond with Glu97A, which in turn is hydrogen bonded to Gua15. These two thrombin residues are external to exosite II (Figure 5).
Figure 5.
Packing contacts involving mTBA. Thrombin molecules are represented as cartoon, aptamer molecule as sticks. The symmetry related thrombin molecule (the symmetry relationship is −x + 1/2, y − 1/2, −z + 1/2) is colored in blue, with residues interacting with mTBA marked in cyan and residues belonging to exosite II marked in yellow. A zoomed vision of the interactions between Thy9 and the second thrombin molecule is reported in the panel.
Comparison with thrombin–TBA and other protein–DNA complexes
In order to obtain a deeper characterization of the complex, we calculated several interface parameters, such as the buried surface area (ΔASA), the ‘gap volume index’, number and type of protein interacting residues, the contribution of polar, non-polar and neutral atoms. These values are given in Table 2 and compared with those obtained for Model 2 of thrombin–TBA complex (PDB code 1HAO) (15) and with the mean values obtained by a non-redundant dataset of 25 protein–DNA complexes.
Table 2.
Protein–DNA interface features
| Thrombin– mTBA | Thrombin– TBA (Model 2) (15) | Protein– DNA complexesa | |
|---|---|---|---|
| Sequence segmentation | 4 | 5 | 7 (4) |
| Δ ASA (Å2) | 570 | 462 | 1320 (495) |
| Δ %ASA | 4.8 | 4.0 | 13 (7) |
| Atoms in interface | 60 | 40 | 122 (51) |
| Polar atoms contribution to interface (%) | 24.3 | 25.0 | 30 (7) |
| Non-polar atoms contribution to interface (%) | 54.2 | 38.0 | 30 (11) |
| Neutral atoms contribution to interface (%) | 45.5 | 37.7 | 40 (9) |
| Residues in interface (%) | 15 | 12 | 43 (20) |
| Polar residues in interface (%) | 46.7 | 58.3 | 37 (10) |
| Non-polar residues in interface (%) | 33.3 | 25.0 | 28 (9) |
| Charged residues in interface (%) | 20.0 | 16.7 | 35 (9) |
| Hydrogen bonds | 11 | 2 | 21 (9) |
| Hydrogen bonds (/100 Å2 ΔASA) | 1.9 | 0.5 | 1.6 (0.4) |
| Bridging water molecules | 4 | 0 | 17 (10) |
| Bridging water molecules (/100 Å2 ΔASA) | 0.7 | 0 | 1.3 (0.7) |
| Gap volume (Å3) | 2710 | 2997 | 7408 (3050) |
| Gap volume index (Å) | 2.38 | 3.49 | 3.0 (1.0) |
| Sc | 0.73 | 0.56 | 0.65 (0.05) |
aProperties were calculated from a non-redundant data set of 25 protein–DNA complexes belonging to different protein–structure families with resolution better than 2.4 Å.
Interestingly, the thrombin–mTBA interface is much smaller than that generally found in protein–DNA complexes, it contains an unusual large number of non-polar atoms and it is constituted by a number of residues that have a low propensity to bind nucleic acids, such as leucine, isoleucine and glutamic acid (47). A remarkable feature of the interface is its tight packing (Figure 4A), as judged by the surface complementarity index (Sc) that is one of the highest observed for protein–DNA complexes. In particular, with respect to TBA, the chain polarity inversion causes a better fit of the nucleotide in thrombin exosite I (Sc value 0.73 versus 0.56) and allows a larger number of hydrogen bonds to be formed at the interface.
DISCUSSION
Aptamers possess several properties that make them very promising molecules; indeed, thanks to their high affinity and specificity toward the target protein, therapeutic dosing at sub-micromolar levels should be allowed, by reducing potential non-specific effects. Furthermore, they are supposed to be non-immunogenic and antidotes can be designed to control their pharmacologic activity (48), they can be completely engineered and possess desirable storage properties. They are also readily produced and easily modified by chemical synthesis. The structural modifications are generally aimed to increase thermal and nucleases stability, cellular delivery and improve surface attachment to the target.
Since thrombin is the key enzyme of the coagulation cascade, it represents an ideal target for anticoagulant therapeutics and indeed a few years ago the Phase I clinical trial of TBA was started. Despite this encouraging scenario, up to now the rational design of new aptamer-based antithrombotic agents has been hampered by the absence of a clear knowledge of interactions between thrombin and TBA, due to the low quality of the diffraction data used for structure refinement and analysis (16).
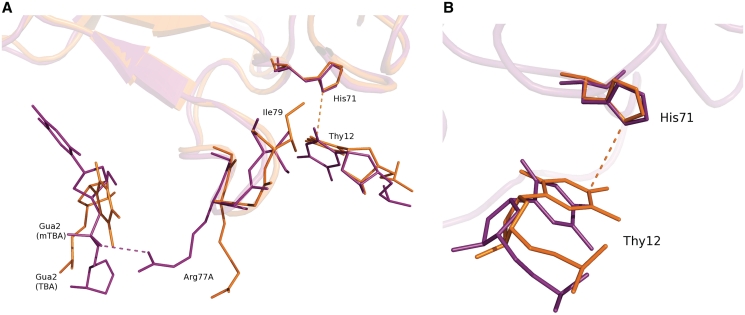
In this context, the higher resolution structure of the complex between thrombin and the modified aptamer mTBA is of particular interest, because it allows identifying the molecular details of the protein–nucleotide recognition. The aptamer tightly binds to thrombin exosite I by its TT loops, through a mix of hydrophobic and polar interactions, consistently with previous biochemical studies (8) on TBA. In agreement with the revised interpretation of the crystallographic data on the thrombin–TBA complex (16), our results on the thrombin–mTBA complex definitely identify in the TT loops a general structural feature for the binding of aptamers to thrombin. In both cases, these loops act as a pincer-like system that embraces the protruding region of exosite I (Figure 2). However, the details of the interactions are different for the two aptamers (Table 1). Indeed the chain inversion in mTBA determines the formation of a greater number of contacts with the enzyme with respect to the unmodified nucleotide and more generally, leads to an increased shape complementarity, as shown by the surface complementarity index Sc (Table 2). These results may well account for the higher affinity toward thrombin of mTBA as compared to TBA (18). On the other hand, the interaction between Thy12 and His71, which is present in the TBA complex (15), is lost in the case of mTBA. His71 plays a key role in the recognition of fibrinogen and its interaction with TBA is considered important for the inhibition of thrombin clotting activity by the aptamer (8). The position of this histidine is the same in the two complexes, but the chain inversion in mTBA causes a subtle reorganization of the 77–79 protein region located at the interface close to Thy12 (Figure 6A). As result a new salt bridge is formed between the side chain of Arg77A and the phosphate oxygen atom of Gua2, which is in close proximity because of the 3'→5' inverted polarity of the strand. In Model 2 TBA backbone is far away from Arg77A, whose side chain is flipped in the opposite direction. In addition, the movement of Ile79, which is in close contact with Thy12, causes in mTBA a displacement of this thymine away from His71 (Figure 6B). Thus, the greater disponibility of His71 to interact with the fibrinogen could explain the lower anticoagulant activity of mTBA (18).
Figure 6.
Superposition of thrombin–mTBA (purple) on the Model 2 of thrombin–TBA (PDB code 1HAO) (15) (orange): (A) interactions between TBA Thy12 and His71 and between mTBA Gua2 and Arg77A, (B) displacement of mTBA Thy12 from His71.
A stabilizing interface of the aptamer with exosite II of a second thrombin molecule was also suggested on the basis of the crystallographic data on the TBA complex (14,15). In the mTBA complex, which assembles in the solid state with a different crystal packing, the surface contact with exosite I is maintained, whereas there are no interactions with exosite II. Contacts with other regions of the protein are in general fewer when compared with those involving exosite I. On the other hand, although the association of mTBA with exosite I of thrombin involves a rather small interface area when compared with those usually found in other DNA–protein complexes, it is characterized by an excellent shape complementarity and a high number of intermolecular interactions, which are typical for aptamers. These findings, together with the fact that in the crystals of both complexes the asymmetric unit contains one protein molecule and one aptamer, strongly suggest that thrombin–aptamers complexes have also in solution a 1:1 stoichiometry, in agreement with the more recent ITC data (19).
In conclusion, the high-resolution structure of thrombin–mTBA complex here reported clarifies several questions regarding thrombin–aptamers interaction. It definitely establishes the way in which the aptamer interacts with thrombin and allows a properly characterization of the contact surface between the two molecules. Moreover, the results highlight the structural bases of the different properties of TBA and mTBA, and in particular their different anticoagulant activities.
The X-ray analysis confirms the general folding (‘3+1’ arrangement) found by NMR studies of free mTBA, with three strands parallel to each other and the forth one oriented in the opposite sense. This finding further enforces the idea that the ‘3+1’ core topology is not to be considered an anomaly, but rather a robust G-quadruplex scaffold, on which new modifications can be inserted (22–24). Indeed, starting from the mTBA structure, it is possible to design modified aptamers that preserve both stability against nucleases and high affinity toward thrombin characteristic of mTBA, and in addition, possess a more effective antithrombotic action. A preliminary modeling suggests that modifications in the Thy12–Thy13 loop could restore the interaction, active in the thrombin–TBA complex, of the aptamer with His71, a key residue for the recognition between the fibrinogen and exosite I. In particular, chemical modification of Thy12 could create new contacts with this protein residue thus improving the anticoagulant action of the aptamer. Experiments are in progress along this line.
More in general, the present investigation provides the scientific community with an enhanced structural and conformational knowledge that will serve as a platform for a rational design of modified aptamers for use in anticoagulant therapies.
ACCESSION NUMBER
3QLP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: financial support from Ministero dell'Istruzione, dell'Università e della Ricerca.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Giosuè Sorrentino and Maurizio Amendola for technical assistance.
REFERENCES
- 1.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 2.Burke JM, Berzal-Herranz A. In vitro selection and evolution of RNA: applications for catalytic RNA, molecular recognition, and drug discovery. FASEB J. 1993;7:106–112. doi: 10.1096/fasebj.7.1.8422956. [DOI] [PubMed] [Google Scholar]
- 3.Lancellotti S, De Cristofaro R. Nucleotide-derived thrombin inhibitors: a new tool for an old issue. Cardiovasc. Hematol. Agents Med. Chem. 2009;7:19–28. doi: 10.2174/187152509787047658. [DOI] [PubMed] [Google Scholar]
- 4.Gold L. Oligonucleotides as research, diagnostic, and therapeutic agents. J. Biol. Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 5.Huntington JA. Molecular recognition mechanisms of thrombin. J. Thromb. Haemost. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 6.Bode W, Turk D, Karshikov A. The refined 1.9-A X-ray crystal structure of D-Phe-Pro-Arg chloromethylketone-inhibited human alpha-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paborsky LR, McCurdy SN, Griffin LC, Toole JJ, Leung LL. The single-stranded DNA aptamer-binding site of human thrombin. J. Biol. Chem. 1993;268:20808–20811. [PubMed] [Google Scholar]
- 8.Tsiang M, Jain AK, Dunn KE, Rojas ME, Leung LL, Gibbs CS. Functional mapping of the surface residues of human thrombin. J. Biol. Chem. 1995;270:16854–16863. doi: 10.1074/jbc.270.28.16854. [DOI] [PubMed] [Google Scholar]
- 9.Wu Q, Tsiang M, Sadler JE. Localization of the single-stranded DNA binding site in the thrombin anion-binding exosite. J. Biol. Chem. 1992;267:24408–24412. [PubMed] [Google Scholar]
- 10.Li WX, Kaplan AV, Grant GW, Toole JJ, Leung LL. A novel nucleotide-based thrombin inhibitor inhibits clot-bound thrombin and reduces arterial platelet thrombus formation. Blood. 1994;83:677–682. [PubMed] [Google Scholar]
- 11.Griffin LC, Tidmarsh GF, Bock LC, Toole JJ, Leung LL. In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood. 1993;81:3271–3276. [PubMed] [Google Scholar]
- 12.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang KY, McCurdy S, Shea RG, Swaminathan S, Bolton PH. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 14.Padmanabhan K, Padmanabhan KP, Ferrara JD, Sadler JE, Tulinsky A. The structure of alpha-thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 15.Padmanabhan K, Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D Biol. Crystallogr. 1996;52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 16.Kelly JA, Feigon J, Yeates TO. Reconciliation of the X-ray and NMR structures of the thrombin-binding aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 17.Pasternak A, Hernandez FJ, Rasmussen LM, Vester B, Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 39:1155–1164. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagano B, Martino L, Randazzo A, Giancola C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008;94:562–569. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nallagatla SR, Heuberger B, Haque A, Switzer C. Combinatorial synthesis of thrombin-binding aptamers containing iso-guanine. J. Comb. Chem. 2009;11:364–369. doi: 10.1021/cc800178m. [DOI] [PubMed] [Google Scholar]
- 20.McGuffie EM, Catapano CV. Design of a novel triple helix-forming oligodeoxyribonucleotide directed to the major promoter of the c-myc gene. Nucleic Acids Res. 2002;30:2701–2709. doi: 10.1093/nar/gkf376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martino L, Virno A, Randazzo A, Virgilio A, Esposito V, Giancola C, Bucci M, Cirino G, Mayol L. A new modified thrombin binding aptamer containing a 5'-5' inversion of polarity site. Nucleic Acids Res. 2006;34:6653–6662. doi: 10.1093/nar/gkl915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang N, Phan AT, Patel DJ. (3 + 1) Assembly of three human telomeric repeats into an asymmetric dimeric G-quadruplex. J. Am. Chem. Soc. 2005;127:17277–17285. doi: 10.1021/ja0543090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo Krauss I, Merlino A, Randazzo A, Mazzarella A, Sica F. Crystallization and preliminary X-ray analysis of the complex of human a-thrombin with a modified thrombin-binding aptamer. Acta Crystallographica Sec. F. 2010;66:961–963. doi: 10.1107/S1744309110024632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navaza J, Saludjian P. AMoRe: an automated molecular replacement program package. Method Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 27.Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J. 1989;8:3467–3475. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 29.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 30.Di Cera E, Guinto ER, Vindigni A, Dang QD, Ayala YM, Wuyi M, Tulinsky A. The Na+ binding site of thrombin. J. Biol. Chem. 1995;270:22089–22092. doi: 10.1074/jbc.270.38.22089. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 32.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 33.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 34.Tiwari A, Panigrahi SK. HBAT: a complete package for analysing strong and weak hydrogen bonds in macromolecular crystal structures. In Silico Biol. 2007;7:651–661. [PubMed] [Google Scholar]
- 35.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project, Number 4, (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Nadassy K, Tomas-Oliveira I, Alberts I, Janin J, Wodak SJ. Standard atomic volumes in double-stranded DNA and packing in protein–DNA interfaces. Nucleic Acids Res. 2001;29:3362–3376. doi: 10.1093/nar/29.16.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hud NV, Smith FW, Anet FA, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 39.Shefter E, Trueblood KN. The crystal and molecular structure of D(+)-barium uridine-5'-phosphate. Acta Crystallogr. 1965;18:1067–1077. doi: 10.1107/s0365110x65002530. [DOI] [PubMed] [Google Scholar]
- 40.Rubin J, Brennan T, Sundaralingam M. Crystal and molecular structure of a naturally occurring dinucleoside monophosphate. Uridylyl-(3'-5')-adenosine hemihydrate. Conformational ‘rigidity’ of the nucleotide unit and models for polynucleotide chain folding. Biochemistry. 1972;11:3112–3128. doi: 10.1021/bi00766a027. [DOI] [PubMed] [Google Scholar]
- 41.Wahl MC, Rao ST, Sundaralingam M. Crystal structure of the B-DNA hexamer d(CTCGAG): model for an A-to-B transition. Biophys. J. 1996;70:2857–2866. doi: 10.1016/S0006-3495(96)79855-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horvath MP, Schultz SC. DNA G-quartets in a 1.86 A resolution structure of an Oxytricha nova telomeric protein-DNA complex. J. Mol. Biol. 2001;310:367–377. doi: 10.1006/jmbi.2001.4766. [DOI] [PubMed] [Google Scholar]
- 43.Phillips K, Dauter Z, Murchie AI, Lilley DM, Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 44.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 45.Kielkopf CL, Ding S, Kuhn P, Rees DC. Conformational flexibility of B-DNA at 0.74 A resolution: d(CCAGTACTGG)(2) J. Mol. Biol. 2000;296:787–801. doi: 10.1006/jmbi.1999.3478. [DOI] [PubMed] [Google Scholar]
- 46.Deane CM, Allen FH, Taylor R, Blundell TL. Carbonyl-carbonyl interactions stabilize the partially allowed Ramachandran conformations of asparagine and aspartic acid. Protein Eng. 1999;12:1025–1028. doi: 10.1093/protein/12.12.1025. [DOI] [PubMed] [Google Scholar]
- 47.Jones S, van Heyningen P, Berman HM, Thornton JM. Protein-DNA interactions: a structural analysis. J. Mol. Biol. 1999;287:877–896. doi: 10.1006/jmbi.1999.2659. [DOI] [PubMed] [Google Scholar]
- 48.Nimjee SM, Rusconi CP, Harrington RA, Sullenger BA. The potential of aptamers as anticoagulants. Trends Cardiovasc. Med. 2005;15:41–45. doi: 10.1016/j.tcm.2005.01.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.