Abstract
Gene targeting can be achieved with lentiviral vectors delivering donor sequences along with a nuclease that creates a locus-specific double-strand break (DSB). Therapeutic applications of this system would require an appropriate control of the amount of endonuclease delivered to the target cells, and potentially toxic sustained expression must be avoided. Here, we show that the nuclease can be transferred into cells as a protein associated with a lentiviral vector particle. I-SceI, a prototypic meganuclease from yeast, was incorporated into the virions as a fusion with Vpr, an HIV accessory protein. Integration-deficient lentiviral vectors containing the donor sequences and the I-SceI fusion protein were tested in reporter cells in which targeting events were scored by the repair of a puromycin resistance gene. Molecular analysis of the targeted locus indicated a 2-fold higher frequency of the expected recombination event when the nuclease was delivered as a protein rather than encoded by a separate vector. In both systems, a proportion of clones displayed multiple integrated copies of the donor sequences, either as tandems at the targeted locus or at unrelated loci. These integration patterns were dependent upon the mode of meganuclease delivery, suggesting distinct recombination processes.
INTRODUCTION
The toxic effects of uncontrolled transgene insertions in the genome have been documented in clinical trials where patients had been treated with retroviral or lentiviral vectors. A transcriptional activation of neighbouring genes by regulatory elements contained in the vector genomes was observed in patients with X-linked severe combined immunodeficiency, chronic granulomatous disease and sickle cell anaemia (1–4). In another situation, a transcriptional shut off of the transgene was induced by chromatin remodelling at the site of insertion, leading to cessation of the therapeutic effect (5). These adverse events may be avoided with a gene transfer technology able to target the chromosomal insertion of therapeutic sequences.
Efforts to target the insertion of retroviral and lentiviral vectors have first focused on modifications of the integrase that result in its catalytic inactivation or on the design of chimeras with a swapped DNA-binding domain. The first approach is used in integration-deficient lentiviral vectors (IDLVs) which can mediate stable gene transfer in a number of cellular targets but are eliminated from actively replicating cells (6–8). A limitation of these vectors is the low levels of transgene expression, compared to the integrative vectors (9). The DNA-binding activity of the integrase can be modified by swapping DNA-binding domains or by using tethering domains linked to LEDGF, a cellular integrase binding protein (10–14). These approaches either result in severely reduced titres or require engineering of the target cell, making them unfit for clinical applications at the present stage.
Viral vectors have been designed to carry DNA recombination substrates in which sequences identical to a targeted locus in the genome allow for a precise genetic modification by homologous recombination. The efficiency of this process is considerably enhanced when a site-specific endonuclease creates a DNA double-strand break (DSB) close to the region of homology. Such targeting endonucleases can now be engineered for virtually any genomic locus, using either the zinc finger technology or by engineering naturally occurring meganucleases (15–17). Several studies have reported high levels of homologous recombination in a variety of cell lines and primary cell cultures, following treatment with IDLV or adeno-associated viral vectors that encode a site-specific endonuclease and a recombination substrate (18–22). The frequencies of gene targeting are usually in the 0.1–10% range depending on the vector architecture, the readout, the targeted locus and the host cell (23).
This could be relevant in certain clinical situations, but the vector system still needs to be improved. First, multiple vectors are required to ferry the different components of the recombination system into the cell. Second, the nuclease coding sequences are expressed for several days, a situation that would not be optimal in a clinical setting due to the background off-target generation of DNA DSBs (24,25). Nuclease toxicity can be reduced by the addition of drug-responsive destabilization domains (25). Here, we have developed a simplified lentiviral system in which a single non-integrating lentiviral vector is used to introduce a repair template and a meganuclease into the cell. The latter is packaged into the lentiviral particle as a protein fused to Vpr, an HIV-1 accessory protein. Following transfer into the cell cytoplasm, the nuclease retains its activity and readily accesses the nucleus where it recognizes and cleaves its target sequence, eventually leading to high rates of homologous recombination at the targeted locus.
MATERIALS AND METHODS
CHOπ10 reporter cell line
The π10 target locus was constructed by: (i) inserting 132 bp downstream of the ATG of the puromycin resistance gene a 55-bp fragment containing the I-SceI recognition sequence and (ii) placing the defective puromycin resistance gene under the control of the promoter region from the human translation elongation factor 1 α subunit (EF1α) gene including exon1, intron 1 and a part of exon 2 (EIE) and c) adding in the 3′ position an IRES-green fluorescent protein (GFP) cassette and a neomycin resistance cassette. The construct was transfected into CHO-K1 cells. Single copy integrants were characterized by Southern blot analysis of G418-resistant clones. CHOπ10 cells (GFP+/Puro−) were maintained in F12-K medium supplemented with G418 as described (26).
Vector design
The repair matrix (RMA) contains the EIE sequences from the human EF1-α gene followed by a functional puromycin resistance gene and 153 bp from the EMCV IRES (Figure 1A), corresponding to 1.15 kb of 5′ and 0.6 kb of 3′ homology around the I-SceI site at the π10 locus. The pHAGE.cppt.RMA.wpre plasmid was generated by introducing the RMA into a promoter-less pHAGE.cppt.wpre lentiviral construct (27), in reverse orientation in order to avoid splicing out of the EF1α intron from the lentiviral genomic RNA. The pHAGE.cppt.CMV.I-SceI.wpre plasmid was generated by introducing the 714 bp I-SceI coding sequence from pCLS0197 (28) into the pHAGE.cppt.CMV.wpre lentiviral construct.
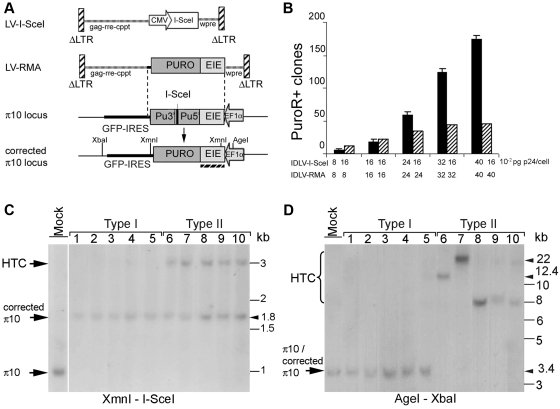
Figure 1.
Targeting the π10 locus with lentiviral vectors. (A) (Top) Lentiviral vectors encoding the I-SceI meganuclease (LV-I-SceI) or a π10 recombination template (LV-RMA). The RMA contains donor sequences homologous to exon1–intron1–exon2 (EIE) sequences of the Elongation factor 1-α (EF1α) gene, the puromycin resistance gene (Puro) and part of the internal ribosomal entry site (IRES) from EMCV. LTR, long terminal repeat; wpre, woodchuck hepatitis virus post-transcription regulatory element; rre, rev responsive element; cppt, central polypurine tract. (Middle) Organization of the π10 locus, including the promoter and EIE sequences from the EF1α gene driving a defective Puromycin resistance gene followed by an IRES–GFP cassette. The Puro marker is interrupted by 55 bp containing the I-SceI recognition sequence. (Bottom) Structure of the π10 locus after homologous recombination with the RMA. (B) 105 CHOπ10 cells were treated with the indicated doses of IDLVs (0.08–0.4 pg HIV-1 p24 Gag/cell). Seventy-two hours after transduction, cells were treated with puromycin during 15 days. The histogram shows the number of PuroR+ clones obtained. The data are representative for three independent experiments for black bars, and two independent experiments for striped bars. (C) Genomic DNAs from the randomly isolated and amplified PuroR+ clones were digested with XmnI and I-SceI and analysed by Southern blot with a 32P-labelled EIE probe [underlined by hatched line in (A)]. Fragment sizes are: Mock, 1 kb; type I, targeted clones, 1.8 kb; type II, targeted clones with head to tail concatemers (HTC) of the LV-RMA, 3 kb. (D) Genomic DNAs from the same PuroR+ clones were digested with AgeI and XbaI and analysed by Southern blot with the same 32P-labelled EIE probe. Fragment sizes for: mock, 3.4 kb; type I, targeted clones, 3.4 kb; type II, targeted clones with head to tail concatemers (HTC) of the LV-RMA, 8–22 kb depending on the size of the tandem repeat. The calculated size for the concatemers are 7.979 kb (n = 2, lanes 8–10); 12.486 kb (n = 3, lane 6); 22.302 kb (n = 5, lanes 7, 9 and 10). Variations in band sizes (i.e. between lanes 8 and 9) probably reflect the presence of 1 or 2 LTR (calculated difference of 401 bp). The π10 locus, type I and II structures are further detailed in Figure 6A–C.
All I-SceI-Vpr expression constructs were obtained by replacing I-SceI in pCLS0197 by fusion-polymerase chain reaction (PCR) assembled fragments containing an HA tagged I-SceI fused in frame to an HIV-1 gag-derived protease cleavage site (Pr7.1 or Pr24.2) followed by Vpr. Fusion-PCR amplification products for ISVP7.1 or ISVP24.2 (pCMV-I-SceI-Ha-Pr7.1-Vpr or pCMV-I-SceI-Ha-Pr24.2-Vpr, respectively) were obtained in three steps: (i) PCR amplification of I-SceI from pCLS0197 [Fw-5′AAAGAACGTGTTAACCACCT, Rev-5′CCGAAACTTTCCTGAAATACCCATACGACGTCCCA]; (ii) PCR amplification of Ha-Pr7.1(or Pr24.2)-Vpr fragment using pCMV-Ha-Pr7.1-Vpr or pCMV-Ha-Pr24.2-Vpr (29) as templates [Fw-5′TACCCATACGACGTCCCAGA, Rev- 5′ATTACTCGAGCTAGGATCTACTGGCTCCATTTC]; and (iii) fusion PCR using the PCR products obtained in steps i and ii [Fw-5′AAAGAACGTGTTAACCACCT, Rev- 5′ATTACTCGAGCTAGGATCTACTGGCTCCATTTC].
For the ISΔVP7.1 fusion (pCMV-I-SceI-Ha-Pr7.1-ΔVpr14-88), the I-SceI fragment was amplified from pCLS0197 [Fw-5′TACCCATACGACGTCCCAGA, Rev-5′ TCCATTCATTGTGTGGCTTGCCCAGGAAGTTGG] and the truncated Vpr (ΔVpr14-88) was amplified with [Fw-5′CCACACAATGAATGGACACTA, Rev-5′ATTACTCGAGTTATCTCCTCTGTTGAGTAACGCCTA] using ISVP7.1 as a template. The IS expression construct was obtained by replacing the I-SceI-Ha-Pr7.1-Vpr fragment from ISVP7.1 by a PCR-amplified fragment containing HA tagged I-SceI. VP construct is described in ref. 29.
Lentiviral vector production and analysis
HEK293T cells were plated at 2.5 × 106 cells/15-cm Petri dishes in Dulbecco's Modified Eagle's Medium with Glutamax and 4.5 g/l glucose (Gibco-Invitrogen, Cergy Pontoise, France) supplemented with 10% fetal bovine serum (FBS) and antibiotics. Cells were transfected with Calcium phosphate precipitates after 72 h. Packaging plasmids, pHDMg-D64L, p-Rev and p-Tat and the transfer vector plasmid were co-transfected with a molar ratio of 1.9/1/4.6/25. For the production of lentiviral particles containing Vpr fusions, the corresponding construct was added at a molar ratio of 17.5. Culture media was harvested after 48 and 72 h, passed through a 0.45-µm filter (Millipore), and ultracentrifuged at 100 000g for 2 h. Pellets were re-suspended in phosphate-buffered saline–bovine serum albumin (PBS–BSA) 1% and stored at −80°C. The amount of p24Gag was measured using the QuickTiter™ lentivirus Titer Kit (Euromedex, Souffelweyersheim, France). Lentiviral particle (LP) titre was calculated following the manufacturer recommendation of 1 ng p24 = 1.25 × 107 LPs. Yield ranged from 1 to 25 µg/ml p24 corresponding to 1.25 × 1010 to 0.3 × 1012 LPs/ml. For western blot analysis lentiviral particles were lysed in 2× Laemmli buffer (Sigma-Aldrich, Saint Quentin Fallavier, France), denatured at 95°C for 5 min, separated on 10–12% sodium dodecyl sulphate– polyacrylamide gel electrophoresis (SDS–PAGE) gels, transferred to Hybond-CExtra membrane (GE Healthcare Europe GmBH, Saclay, France) and probed with antibodies against the Ha tag (Roche Diagnostics), p24 (Euromedex, Souffelweyersheim, France) and VSV-G (Sigma-Aldrich, Saint Quentin Fallavier, France).
Homologous recombination assay
CHOπ10 cells were plated at 105 cells/well in six-well plates. The following day, cells were co-infected with the indicated doses of vectors, all added together, in the presence of 10 µg/ml polybrene (Euromedex, Souffelweyersheim, France). After 72 h cells were plated at 30% confluence and 12 h later the medium was supplemented with 10 µg/ml puromycin (Invitrogen, Cergy Pontoise, France). During 15 days, puromycin-containing media was removed and replaced every other day. PuroR+ clones were counted, randomly isolated and grown for further analysis. The gene targeting frequency was calculated as the ratio of PuroR+ clones after the 15-day selection over the number of cells at the time of selection (Supplementary Table S1).
DNA analysis in targeted clones
Genomic DNA was isolated with Genomic DNA Purification Kit (Fermentas, St Remy les Chevreuses, France) and 200 ng were used to detect homologous recombination by PCR (2× PCR MasterMix, Promega, Charbonnieres Les Bains, France) with the following primers: Fw1-5′CCGCCACCATGACCGAGTACAA, Fw2-5′ACGAAGTTATGGTCACCGAG and Rev-5′CTCGTAGAAGGGGAGGTTGCG. The following conditions for amplification were used: 94°C for 5 min, then 25 cycles at 94°C for 1 min, 62°C for 1 min and 72°C for 1 min, followed by extension at 72°C for 5 min. Clones were further analysed by Southern blot. Fifteen micrograms of genomic DNA were digested with the indicated enzymes overnight. Digested DNA was separated on 1% agarose gel, blotted to Hybond-N+ nylon membrane (GE Healthcare Europe GmBH, Saclay, France) and hybridized with a 32P-radiolabelled probe prepared from a 883-bp XmnI–EcoRI fragment from the EIE sequence, using Random Primers DNA Labelling System (Invitrogen, Cergy Pontoise, France). After washing (2× SSC/0.1% SDS, room temperature; 0.2× SSC/0.1% SDS, room temperature; 0.1× SSC/0.1% SDS, 42°C and 65°C, 2× SSC, room temperature) membranes were placed 3 days at −80°C for autoradiography.
RESULTS
I-SceI- mediated chromosomal targeting by IDLV delivery
The efficiency of I-SceI-mediated homologous recombination by lentiviral delivery was evaluated in a reporter cell line in which a single chromosomal copy of an inactivated puromycin resistance gene has been introduced. The puromycin resistance gene is interrupted by a 55-bp insert containing a recognition site for I-SceI (see ‘Material and Methods’ section). Transfection of plasmids encoding I-SceI and a repair matrix (RMA) containing a functional but promoter-less puromycin resistance gene results in targeting events in about 0.1% of cells (26). Here CHOπ10 cells were treated with a combination of IDLVs encoding I-SceI or the RMA (Figure 1A). The cells were counted after 72 h and placed under puromycin selection. Cell counting revealed a dose-dependent growth inhibition suggesting toxicity of lentiviral I-SceI expression (Figure 3B and Supplementary Table S1). The number of puromycin-resistant colonies increased in an IDVL dose-dependent manner (Figure 1B, black bars). A gene targeting frequency of 1 × 10−3 was estimated by counting the number of puromycin-resistant (PuroR+) clones. Only four PuroR+ clones were obtained in the absence of I-SceI at the maximal dose of IDLV-RMA (0.4 pg HIV-1 Gag p24/cell). This suggests that the frequency of PuroR+ clones that could have arisen from non-targeted events (or from targeted events in the absence of meganuclease) was 1.6 × 10−5 in accordance with previous reports (30). No PuroR+ clones were obtained in the presence of IDLV-I-SceI alone (0.4 pg HIV-1 Gag p24/cell), excluding the possibility of gene repair by I-SceI-induced non-homologous end joining (NHEJ). When the dose of IDLV–I-SceI was kept constant (0.16 pg HIV-1 p24/cell), while the dose of RMA-encoding lentivirus was increased, a lower number of PuroR+ clones was obtained for all experimental points, indicating that the amount of I-SceI was limiting (Figure 1B, stripped bars). I-SceI-mediated CHOπ10 locus targeting by IDLVs was confirmed by PCR (as described in Supplementary Figure S2) and Southern blot analysis. Genomic DNAs prepared from 36 randomly picked and amplified PuroR+ clones were digested by two combinations of enzymes in order to assess the structure of the π10 locus (Figure 1C and D). A probe with the exon1–intron1–exon2 (EIE) sequence of the EF1-α gene, present on both the RMA and the targeted locus, was used. Digestion with XmnI and I-SceI revealed a band at 1.0 kb in the control reporter cell line. This band is expected to disappear once HR converts the I-SceI-interrupted Puro sequences at the locus into functional ones, and be replaced by a band of 1.8 kb (Figure 6A and B). Cutting outside of the targeted sequences with AgeI and XbaI yields a 3.4-kb band, which is 55 bp shorter after HR (not resolved in Figure 1D). As indicated in Figure 5 (left column), 22% of the clones analysed presented this profile indicative of a simple targeting event (type I clones). Most other clones, designated as type II, presented an additional 3.0-kb band in the XmnI–I-SceI digest and an AgeI–XbaI band of more than 8 kb. These profiles correspond to a repaired π10 locus also containing tandem copies of the proviral genome (Figure 6C), as previously observed by others (19).
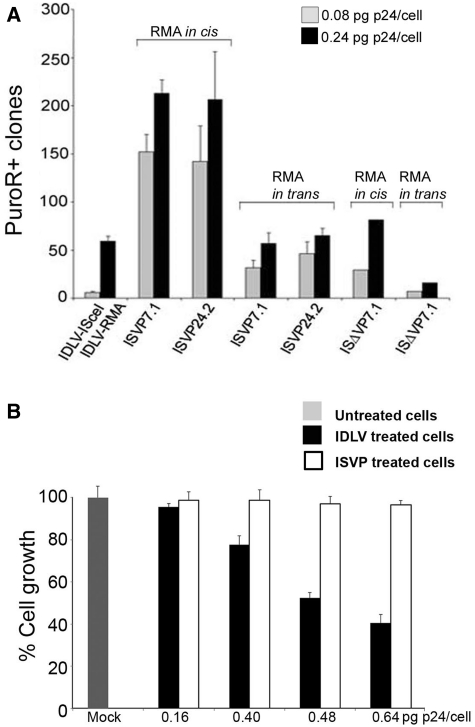
Figure 3.
π10 locus targeting with I-SceI-containing lentiviral particles. (A) 105 reporter cells were treated with IDLVs prepared in the presence of ISVP7.1 or ISVP24.2 protein expression constructs at doses of 0.08 pg (grey bars) or 0.24 pg (black bars) HIV-1 p24 Gag/cell. The RMA was brought either in cis or in trans. The same doses of IDLV-I-SceI that encodes I-SceI (Figure 1A) were used in controls. Seventy-two hours after transduction, cells were treated with puromycin during 15 days and PuroR+ clones were counted. (B) Cell growth following I-SceI delivery. CHOπ10 cells were plated at 105 cell/well in six-well plates. The following day, cells were treated with the indicated doses of IDLVs (transduction) or ISVP transducing particles (protein delivery). After 72 h, cells were counted (Supplementary Table S1) and plated at 30% confluence for puromycin selection. Mock (black bar), untreated cells.
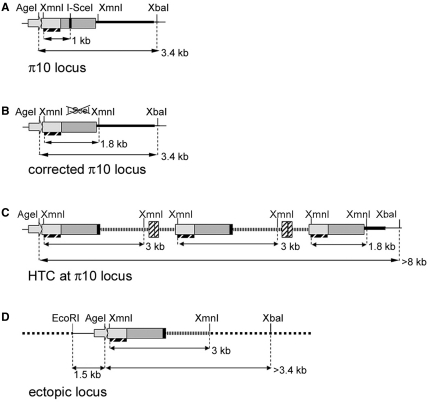
Figure 6.
Structure of the π10 locus after I-SceI-mediated targeting. (A) Genetic structure of the π10 locus including the I-SceI recognition site. The position of restriction enzymes used for Southern blot analysis is indicated and the probe used is shown as a hatched bar. (B) Corrected π10 locus in type I clones in which the I-SceI recognition site was eliminated. (C) Genetic structure of head-to-tail concatemers (HTC) of the RMA vector genome at the π10 locus in type II clones. The I-SceI recognition is eliminated but n ≥ 2 copies of the RMA vector genome [hatched boxes (LTR) and lines] are integrated downstream of the repair sequences. Digestion by AgeI and XbaI yield fragments of over 8, 12 and 22 kb depending on the number of vector genomes in tandem. (D) Genetic structure of an ectopic locus where a recombined RMA is integrated with 5′ sequences from the π10 locus containing the EF1α promoter. Dotted lines represent sequences at the ectopic locus. Type III clones contain both the intact π10 locus [see (A), 3.4 kb AgeI–XbaI fragment] and an ectopic recombined structure (AgeI–XbaI fragment >3.4 kb).
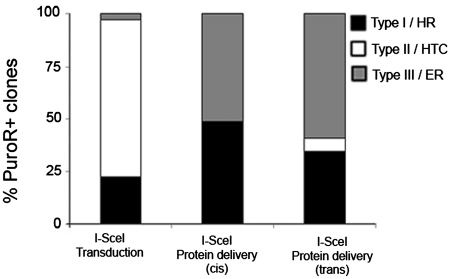
Figure 5.
I-SceI-mediated gene targeting events in PuroR+ clones. The distributions of type I–III clones among PuroR+ clones obtained after conventional I-SceI transduction (left, n = 36 clones analysed), virion-associated protein delivery in cis (middle, n = 35 clones analysed) or virion-associated protein delivery in trans (right, n = 32 clones analysed) are shown.
I-SceI protein incorporation into viral particles
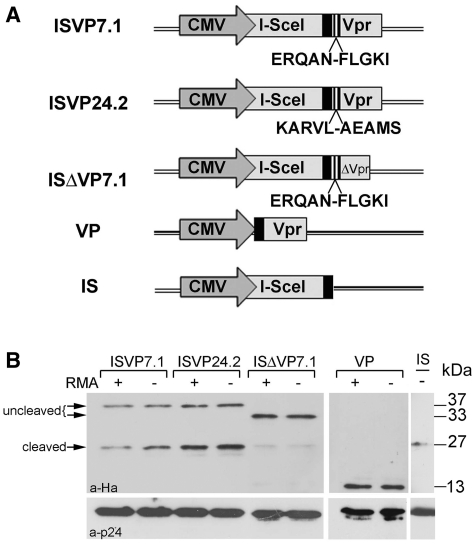
The system described above requires the use of multiple vectors, expressed for several days, which may cause toxicity (Figure 3B, Supplementary Table S1) possibly due to meganuclease off-target activities (24). For this reason, we have combined lentivirus-based nucleic acid delivery with protein transducing technology to avoid prolonged chromosome exposure to DSB. In this system, a single IDLV is used to introduce the recombination template together with the I-SceI protein. We generated different constructs where I-SceI carried a C-terminal HA tag and was fused in-frame to the N-terminus of the viral accessory protein Vpr or of ΔVpr, a Vpr fragment comprising aminoacids 14–88, minimally required for virus incorporation (31). Two different cleavage sites for the HIV protease (p7/1 or p24/2) were introduced upstream of Vpr (Figure 2A), in order to generate a Vpr-free meganuclease after processing inside the virion (32). To characterize these fusion constructs named ISVP7.1 (I-SceI-Ha-p7/1-Vpr), ISVP24.2 (I-SceI-Ha-p24/2-Vpr) and ISΔVP7.1 (I-SceI-Ha-p7/1-ΔVpr), the corresponding plasmids were transfected in HEK293-T cells. Cell lysates were prepared after 32 h and analysed by western blot with an antibody directed against HA. The expected bands of 37 kDa or 33 kDa were obtained corresponding to ISVP7.1, ISVP24.2 or ISΔVP7.1, respectively (Supplementary Figure S1A). I-SceI-containing particles were generated in the presence or the absence of a transfer vector containing the RMA or a GFP expression cassette (data not shown), purified and analysed by western blot (see ‘Materials and Methods’ section). High concentrations of p24 were obtained with each construct indicating that Vpr-based incorporation of the meganuclease does not affect production and release of Gag proteins. The western blot analysis showed that I-SceI fusion proteins were correctly packaged into VSV-G pseudotyped virions. Over 50% of fusion proteins were processed by the viral protease (Figure 2B). In a control experiment, VP or IS particles containing respectively Vpr or I-SceI without Vpr were also produced, purified and analysed by western blot. As expected, Vpr was readily incorporated into the virion. A low level of virion-associated I-SceI was found in the absence of Vpr.
Figure 2.
Packaging I-SceI into lentiviral particles. (A) I-SceI was fused to the N terminus of full-length (ISVP7.1, ISVP24.2) or truncated (ISΔVP7.1) Vpr. I-SceI was HA tagged (black box) and viral HIV-1 protease cleavage sites in Gag (p7/1 or p24/2) were introduced upstream of Vpr (stripped box). These fused proteins were used for I-SceI packaging with or without an RMA vector genome in the same particle. VP and IS constructs expressing Ha tagged Vpr or I-SceI respectively, were used as control. (B) Western blot analysis of I-SceI-containing particles. Viral particles (45–50 ng HIV-1 p24 Gag) were run on a 10% SDS–PAGE gel and the blot was probed with an antibody against the HA tag or against HIV-1 Gag p24 (lower panel). Two bands corresponding to ∼60% cleaved (27 kDa) and ∼40% uncleaved (37 or 33 kDa) proteins were obtained for ISVP7.1 and ISVP24.2. Note the lesser incorporation and cleavage efficiency (∼10%) for the ISΔVP7.1 fusion. The expected 13-kDa band was obtained using the VP control. The IS control lane contains a 27-kDa band indicating a background level (<10%) of I-SceI association with the viral particles in the absence of Vpr.
To ensure that the meganuclease was localized in the particle and not trapped on the viral membrane, ISVP24.2-containing particles were treated with increasing doses of proteinase K (0.02–0.1 mg/ml) and analysed by western blot using antibodies directed against HA or VSV-G. The VSV-G protein present at the virion surface was readily digested while most of the I-SceI-containing fusions were resistant to digestion (Supplementary Figure S1B).
Targeting the π10 locus with I-SceI-containing lentiviral particles
To evaluate the activity of the packaged meganuclease, CHOπ10 cells were treated with I-SceI-containing viral particles carrying a puromycin repair template (IDLV-RMA cis, 0.08 or 0.24 pg HIV-1 p24/cell). Alternatively, the RMA was brought by a separate lentivirus (IDLV-RMA trans) (Figure 3A). Cell counting before puromycin selection did not show the dose-dependent growth inhibition observed previously (Figure 3B; Supplementary Table S1). This was consistent with the minute amounts of I-SceI that could be detected in the cells after lentiviral-mediated protein transfer (Supplementary Figure S1C). Compared to the previous series of experiments in which I-SceI was encoded by one of the vectors, about four times more PuroR+ clones (210 clones versus 59 clones) were obtained following treatment with the vector in cis configuration. The efficiency was decreased when the repair matrix was brought in trans, suggesting that the presence of both the RMA and the meganuclease in the same particle was facilitating recombination. A lower number of clones was obtained with ISΔVP7.1-containing particles, possibly because of the less efficient processing of this fusion by the viral protease.
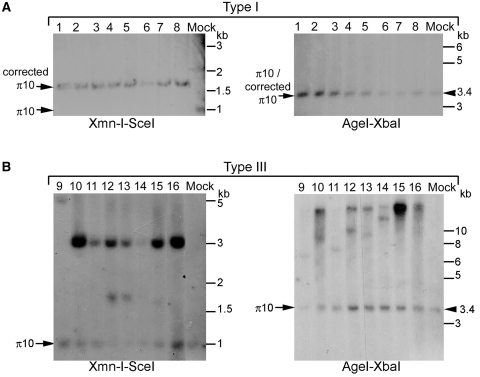
Targeting of the π10 locus was confirmed on 67 of the PuroR+ clones obtained with the cis or trans IDLV-RMA vectors which were grown and analysed by PCR (Supplementary Figure S2) and Southern blot (Figure 4). Three different profiles were obtained on Southern blots, including the type I (Figure 4A, clones 1–8) and type II (data not shown) along with a third profile (type III, Figure 4B, clones 9–16). The proportions of perfectly targeted clones (type I) were 48% and 33% for cis and trans delivery, respectively, a higher level than the 22% obtained after IDLV–I-SceI transduction (Figure 5).
Figure 4.
Analysis of PuroR+ clones obtained by I-SceI-containing lentiviral particles. (A) Southern blot analysis of type I clones (from 1 to 8) obtained after lentiviral particle-associated I-SceI delivery. Clones from both cis and trans experiments are shown. (Left) Genomic DNA samples from PuroR+ clones were digested with XmnI and I-SceI and analysed with a 32P-labelled EIE probe. Fragment sizes for: mock, 1 kb; type I clones, 1.8 kb. (Right) The same genomic DNAs were digested with AgeI and XbaI and hybridized with the 32P-labelled EIE probe. Fragment sizes for: mock and type I clones, 3.4 kb. (B) Southern blot analysis of type III clones (from 9 to 16) obtained under the same conditions. (Left) Genomic DNAs from PuroR+ clones were digested with XmnI and I-SceI and analysed with a 32P-labelled EIE probe. Fragment sizes for: mock, 1 kb; type III clones, 1 kb (unmodified π10 locus) + 3 kb (RMA vector sequences). (Right) The same genomic DNAs were digested with AgeI and XbaI and hybridized with the 32P-labelled internal EIE probe. Fragment sizes for: mock, 3.4 kb; type III clones, 3.4 kb (unmodified π10 locus) and additional species at 7–25 kb, corresponding to RMA sequences at an ectopic locus.
Upon XmnI–I-SceI digest, all type III clones displayed a copy of the original π10 locus (1.0 kb band, Figure 4B, clones 9–16). While most of them only had one additional 3.0-kb band that could originate from the RMA vector genome (Figure 6D), a minority had different profiles (e.g. clone 9) suggestive of a heterogeneous recombination process (Figure 4B, clone 9 and Supplementary Figure S4, clones 22 and 23). Cutting with AgeI and XbaI resulted in the π10 specific 3.4-kb band as well as in several high-molecular-weight bands whose sizes are not consistently multiple of the RMA provirus size, as in type II clones. We conclude that in type III clones, the active puromycin resistance gene was not inserted at the π10 locus. There are two possible ways of explaining their drug resistance phenotype. First, they could arise from integration of the provirus without recombination, for instance, at randomly occurring DSB, followed by activation of the promoter-less resistance gene by nearby cellular transcriptional signals. We consider this scenario unlikely because of the high number of type III clones obtained and of the very low background of Puro+ clones in the absence of I-SceI (see above). A second possibility is ectopic recombination, whereby recombination at the π10 locus creates a puromycin resistance gene linked to the EF1-α promoter, which subsequently integrates at another locus (33–35). This hypothesis was supported by further Southern blot analysis of type III clones using probes outside of the homology region contained in the RMA construct. This analysis suggests that the 3′ end of the π10 locus which includes IRES–GFP sequences remains intact, while the EF1-α promoter and at least 1 kb of upstream sequences were amplified and presumably moved to an ectopic locus (Figure 6D and Supplementary Figure S3).
In a control experiment the I-SceI protein, without Vpr, was expressed during vector production and viral particles were analysed by western blot. A low level of I-SceI was found to be associated to the particles in the absence of Vpr. These low amounts of virion-associated meganuclease, in the absence of fusion to Vpr, resulted in only seven type III PuroR+ clones (Supplementary Figure S4, clones 17–23), suggesting that Vpr is not a major determinant of these unconventional recombination events.
DISCUSSION
This work demonstrates the possibility of delivering meganucleases into cells in a transient and dose-controlled manner for the purpose of gene targeting. Gene targeting was obtained using IDLVs encoding a repair matrix and containing a meganuclease as a virion-associated protein. The frequency of targeting events was comparable to the one obtained using conventional means of introduction of the nuclease and repair matrix by transfection (26) or lentiviral transduction (Figure 1). Lentiviral virions can be used to ferry heterologous proteins as fusions with viral proteins such as Vpr, Nef, IN or Gag (36–39). The delivery of active proteins at the time of virion entry and trafficking into the cell has been reported (40). The technology was recently used for the administration the Cre recombinase, showing that an active DNA-modifying enzyme acting in the nucleus could be delivered in that way (41). Here, we have delivered a meganuclease that creates site-specific DNA DSBs at nanomolar concentrations (42). For a DSB to occur on the chromosome, we estimate that a minimum of 1000 molecules of the protein per cell are required (43). Since up to 700 Vpr molecules are present in each virion (44), a transient peak of nanomolar concentration of meganuclease can be achieved using standard multiplicities of infection.
When the meganuclease was packaged into virions separate from those encoding the recombination template (trans configuration), the number of recombination events scored was comparable to the one obtained with an I-SceI-encoding vector. In contrast, it was higher when the nuclease was in the same virion as the repair sequences. This suggests that colocalization of the two elements of the recombination system into the pre-integration complex is important. It could be simply due to a local increase in the nuclease concentration and a higher rate of DSB. It also physically brings the repair template close to the DSB, thereby potentially helping recombination.
Our Southern blot analysis of the targeted π10 locus in PuroR+ clones defines three types of recombination events. Type I represents perfect gene targeting events where a single copy of the DNA template carried by the lentiviral genome is used for DSB repair, resulting in the conversion of the puromycin resistance gene. Their proportion is twice higher when the nuclease is packaged into the virions. The Southern blot signature of type II clones includes additional bands consistent with the presence of concatemers of the vector genome (n = 2, 3, 5 on Figure 1D). These structures are a common feature of studies using retroviral or integration deficient lentiviral vectors for DSB-induced gene targeting (19,30). In the absence of active integrase, circle and/or concatemer formation is a default pathway for eliminating the free DNA ends of the linear proviral genome (45). Type II structures may arise from copying the repair template on the concatemer, beyond the first proviral copy. Alternatively, head-to-tail concatemers could be produced by iterative copying of a circular, monomeric vector genome containing the repair template (46). The presence of concatemers represents a problem for gene targeting because they may interrupt an otherwise repaired gene and because they bring lentiviral sequences with potential for transcriptional interference in the proximity of the targeted locus.
Type III clones appear almost exclusively when the meganuclease is virion-associated. I-SceI-loaded particles bring the nuclease and the recombination template in the nucleus at the same time, and in the same pre-integration complex when in the cis configuration. Under these conditions, DSB occurs early after transduction, when most of the proviral genomes are still linear DNA products of reverse transcription. In contrast, when I-SceI is encoded by the vector, its biosynthesis requires a lag period during which the repair template provirus dissociates from the pre-integration complex and becomes circularized or concatemerized. The type III profiles are reminiscent of ectopic recombination in which a free 3′ end from the repair template is extended by copying the homologous chromosomal sequences (33,35). This process would generate hybrid molecules linking the EF1-α promoter and upstream genomic sequences to the puromycin resistance gene from the vector DNA. The recombinant structures then become integrated, often in the vicinity of the targeted locus. Early experiments using non-integrative retroviral vectors for gene targeting had already revealed similar ectopic recombination events (30). However, it remains to be explained why such events would be more frequent with the protein delivery system. One can hypothesize that recombination events are initiated at an earlier step when the viral DNA is still linear. However, strand invasion of the extrachromosomal template by DNA ends generated by I-SceI is not supposed to initiate DNA synthesis from 3′ extremities on the template. Thus, one has to envision that invading 3′ extremities can be more efficiently formed on the linear provirus engaged in a recombination process.
In conclusion, both modes of meganuclease delivery, either as a protein or encoded by a vector, lead to gene targeting events, a proportion of which involves non-homologous recombination. The frequency of bona fide recombination event is about 2-fold higher when the nuclease is delivered as a protein. We suggest that the approach described here is potentially safer because a single burst of enzyme is delivered to the cell. It may also be applied to a large number of single chain meganucleases such as those engineered from I-CreI (47).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
This work was supported by grants from the Association Française contre les Myopathies (AFM) and Institut National de la Santé et de la Recherche Médicale (INSERM). AI held a fellowship from AFM. Funding for open access charges: AFM.
Conflict of interest statement. Preliminary disclosure documents have been filed in pursuit of a patent application for this system.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arnold Munnich (Inserm U781) for support.
REFERENCES
- 1.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, Glimm H, Kuhlcke K, Schilz A, Kunkel H, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 2.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 4.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein S, Ott MG, Schultze-Strasser S, Jauch A, Burwinkel B, Kinner A, Schmidt M, Kramer A, Schwable J, Glimm H, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 6.Apolonia L, Waddington SN, Fernandes C, Ward NJ, Bouma G, Blundell MP, Thrasher AJ, Collins MK, Philpott NJ. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol. Ther. 2007;15:1947–1954. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- 7.Yanez-Munoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, Buch P, MacLaren RE, Anderson PN, Barker SE, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat. Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- 8.Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu XJ, Yang C, Bahner I, Kohn DB. Transient gene expression by nonintegrating lentiviral vectors. Mol. Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Bayer M, Kantor B, Cockrell A, Ma H, Zeithaml B, Li X, McCown T, Kafri T. A large U3 deletion causes increased in vivo expression from a nonintegrating lentiviral vector. Mol. Ther. 2008;16:1968–1976. doi: 10.1038/mt.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan W, Dong Z, Wilkinson TA, Barbas CF, III, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J. Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferris AL, Wu X, Hughes CM, Stewart C, Smith SJ, Milne TA, Wang GG, Shun MC, Allis CD, Engelman A, et al. Lens epithelium-derived growth factor fusion proteins redirect HIV-1 DNA integration. Proc. Natl Acad. Sci. USA. 2010;107:3135–3140. doi: 10.1073/pnas.0914142107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 2006;360:760–773. doi: 10.1016/j.jmb.2006.04.073. [DOI] [PubMed] [Google Scholar]
- 13.Silvers RM, Smith JA, Schowalter M, Litwin S, Liang Z, Geary K, Daniel R. Modification of integration site preferences of an HIV-1-based vector by expression of a novel synthetic protein. Hum. Gene Ther. 2010;21:337–349. doi: 10.1089/hum.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gijsbers R, Ronen K, Vets S, Malani N, De Rijck J, McNeely M, Bushman FD, Debyser Z. LEDGF hybrids efficiently retarget lentiviral integration into heterochromatin. Mol. Ther. 2010;18:552–560. doi: 10.1038/mt.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 16.Galetto R, Duchateau P, Paques F. Targeted approaches for gene therapy and the emergence of engineered meganucleases. Expert Opin. Biol. Ther. 2009;9:1289–1303. doi: 10.1517/14712590903213669. [DOI] [PubMed] [Google Scholar]
- 17.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 18.Cornu TI, Cathomen T. Targeted genome modifications using integrase-deficient lentiviral vectors. Mol. Ther. 2007;15:2107–2113. doi: 10.1038/sj.mt.6300345. [DOI] [PubMed] [Google Scholar]
- 19.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 20.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 21.Porteus MH, Cathomen T, Weitzman MD, Baltimore D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell. Biol. 2003;23:3558–3565. doi: 10.1128/MCB.23.10.3558-3565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch ML, Green L, Porteus MH, Samulski RJ. Self-complementary AAV mediates gene targeting and enhances endonuclease delivery for double-strand break repair. Gene Ther. 2010;17:1175–1180. doi: 10.1038/gt.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gellhaus K, Cornu TI, Heilbronn R, Cathomen T. Fate of recombinant adeno-associated viral vector genomes during DNA double-strand break-induced gene targeting in human cells. Hum. Gene Ther. 2010;21:543–553. doi: 10.1089/hum.2009.167. [DOI] [PubMed] [Google Scholar]
- 24.Petek LM, Russell DW, Miller DG. Frequent endonuclease cleavage at off-target locations in vivo. Mol. Ther. 2010;18:983–986. doi: 10.1038/mt.2010.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruett-Miller SM, Reading DW, Porter SN, Porteus MH. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLoS Genet. 2009;5:e1000376. doi: 10.1371/journal.pgen.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabaniols JP, Paques F. Robust cell line development using meganucleases. Methods Mol. Biol. 2008;435:31–45. doi: 10.1007/978-1-59745-232-8_3. [DOI] [PubMed] [Google Scholar]
- 27.Mostoslavsky G, Fabian AJ, Rooney S, Alt FW, Mulligan RC. Complete correction of murine Artemis immunodeficiency by lentiviral vector-mediated gene transfer. Proc. Natl Acad. Sci. USA. 2006;103:16406–16411. doi: 10.1073/pnas.0608130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selig L, Pages JC, Tanchou V, Preveral S, Berlioz-Torrent C, Liu LX, Erdtmann L, Darlix J, Benarous R, Benichou S. Interaction with the p6 domain of the gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J. Virol. 1999;73:592–600. doi: 10.1128/jvi.73.1.592-600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis J, Bernstein A. Gene targeting with retroviral vectors: recombination by gene conversion into regions of nonhomology. Mol. Cell. Biol. 1989;9:1621–1627. doi: 10.1128/mcb.9.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao XJ, Kobinger G, Dandache S, Rougeau N, Cohen E. HIV-1 Vpr-chloramphenicol acetyltransferase fusion proteins: sequence requirement for virion incorporation and analysis of antiviral effect. Gene Ther. 1999;6:1590–1599. doi: 10.1038/sj.gt.3300988. [DOI] [PubMed] [Google Scholar]
- 32.Serio D, Rizvi TA, Cartas M, Kalyanaraman VS, Weber IT, Koprowski H, Srinivasan A. Development of a novel anti-HIV-1 agent from within: effect of chimeric Vpr-containing protease cleavage site residues on virus replication. Proc. Natl Acad. Sci. USA. 1997;94:3346–3351. doi: 10.1073/pnas.94.7.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adair GM, Nairn RS, Wilson JH, Seidman MM, Brotherman KA, MacKinnon C, Scheerer JB. Targeted homologous recombination at the endogenous adenine phosphoribosyltransferase locus in Chinese hamster cells. Proc. Natl Acad. Sci. USA. 1989;86:4574–4578. doi: 10.1073/pnas.86.12.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCulloch RD, Read LR, Baker MD. Strand invasion and DNA synthesis from the two 3' ends of a double-strand break in mammalian cells. Genetics. 2003;163:1439–1447. doi: 10.1093/genetics/163.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mangerich A, Scherthan H, Diefenbach J, Kloz U, van der Hoeven F, Beneke S, Burkle A. A caveat in mouse genetic engineering: ectopic gene targeting in ES cells by bidirectional extension of the homology arms of a gene replacement vector carrying human PARP-1. Transgenic Res. 2009;18:261–279. doi: 10.1007/s11248-008-9228-x. [DOI] [PubMed] [Google Scholar]
- 36.Singh SP, Lai D, Cartas M, Serio D, Murali R, Kalyanaraman VS, Srinivasan A. Epitope-tagging approach to determine the stoichiometry of the structural and nonstructural proteins in the virus particles: amount of Vpr in relation to Gag in HIV-1. Virology. 2000;268:364–371. doi: 10.1006/viro.2000.0191. [DOI] [PubMed] [Google Scholar]
- 37.Muratori C, D'Aloja P, Superti F, Tinari A, Sol-Foulon N, Sparacio S, Bosch V, Schwartz O, Federico M. Generation and characterization of a stable cell population releasing fluorescent HIV-1-based virus like particles in an inducible way. BMC Biotechnol. 2006;6:52. doi: 10.1186/1472-6750-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schenkwein D, Turkki V, Karkkainen HR, Airenne K, Yla-Herttuala S. Production of HIV-1 integrase fusion protein-carrying lentiviral vectors for gene therapy and protein transduction. Hum. Gene Ther. 2010;21:589–602. doi: 10.1089/hum.2009.051. [DOI] [PubMed] [Google Scholar]
- 39.Voelkel C, Galla M, Maetzig T, Warlich E, Kuehle J, Zychlinski D, Bode J, Cantz T, Schambach A, Baum C. Protein transduction from retroviral Gag precursors. Proc. Natl Acad. Sci. USA. 2010;107:7805–7810. doi: 10.1073/pnas.0914517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Link N, Aubel C, Kelm JM, Marty RR, Greber D, Djonov V, Bourhis J, Weber W, Fussenegger M. Therapeutic protein transduction of mammalian cells and mice by nucleic acid-free lentiviral nanoparticles. Nucleic Acids Res. 2006;34:e16. doi: 10.1093/nar/gnj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michel G, Yu Y, Chang T, Yee JK. Site-specific gene insertion mediated by a Cre-loxP-carrying lentiviral vector. Mol. Ther. 2010;18:1814–1821. doi: 10.1038/mt.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redondo P, Prieto J, Munoz IG, Alibes A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 43.Milo R, Jorgensen P, Moran U, Weber G, Springer M. BioNumbers–the database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010;38:D750–D753. doi: 10.1093/nar/gkp889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson CM, Malim MH. SnapShot: HIV-1 proteins. Cell. 2008;133:742, 742.e1. doi: 10.1016/j.cell.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 47.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.