Abstract
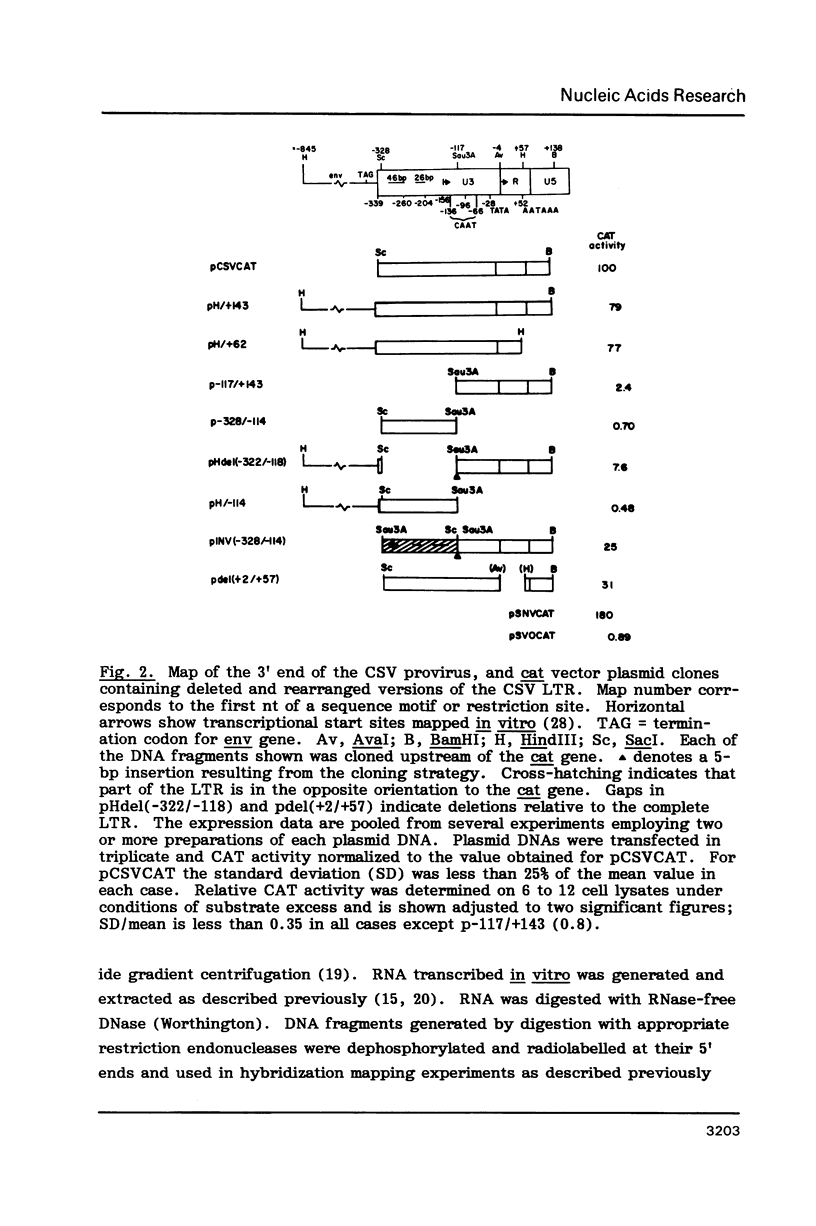
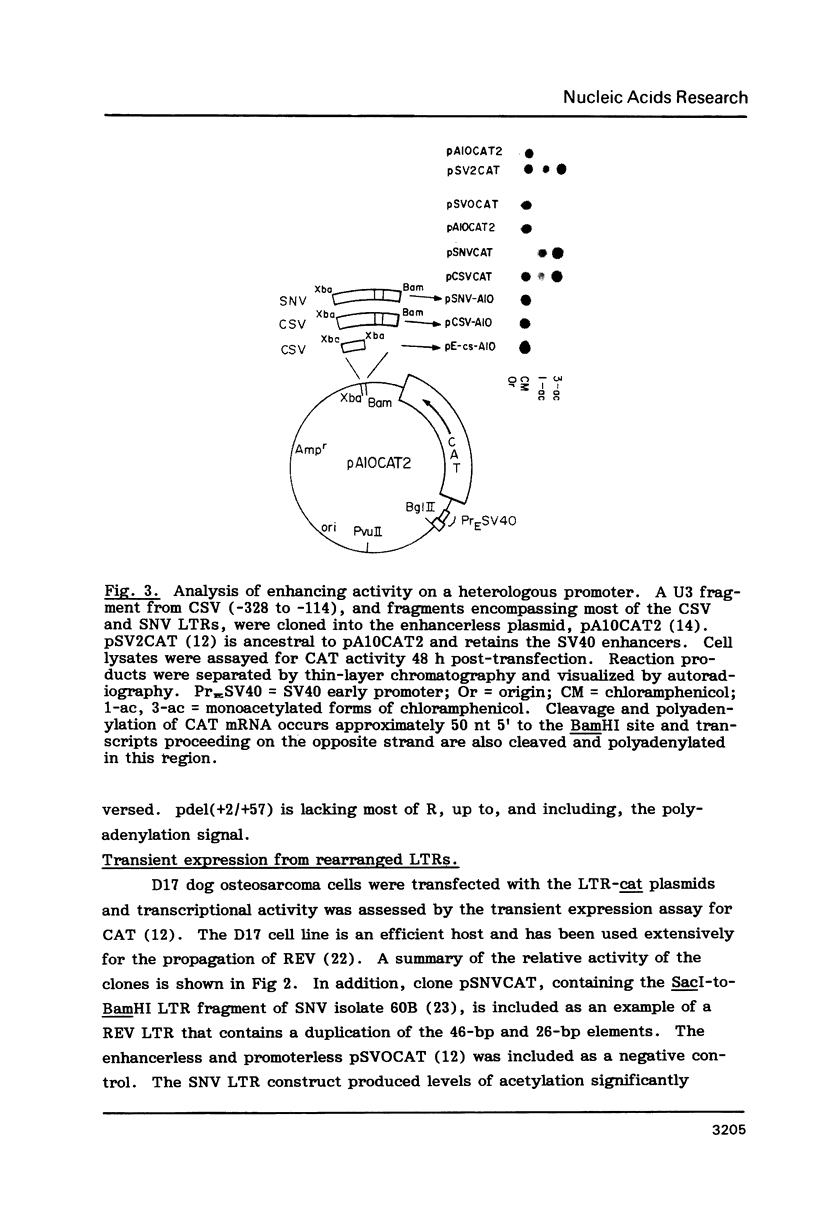
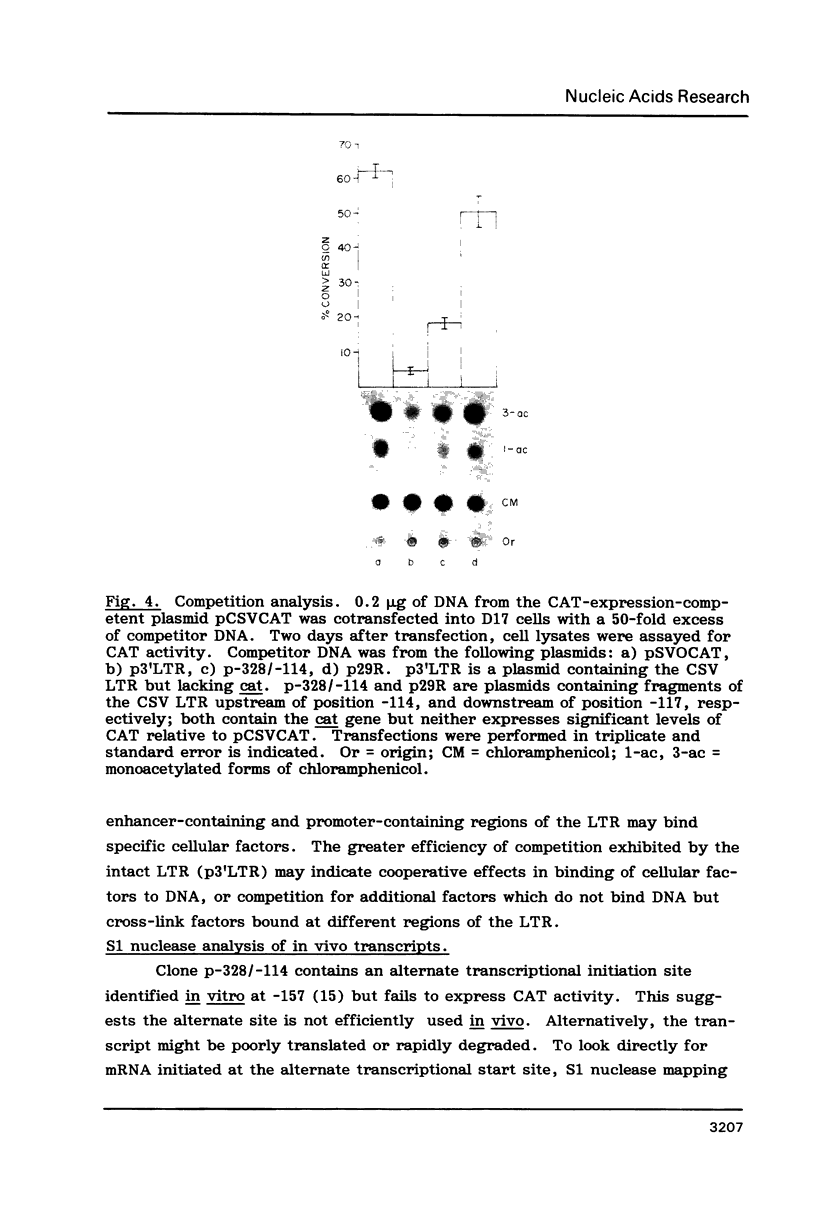
A region of the Reticuloendotheliosis virus (REV) long terminal repeat (LTR) harbouring single or duplicated copies of 46-bp and 26-bp sequence elements is implicated in enhancer activity. Sequences residing upstream from the proviral 3' LTR did not contribute to activity of the intact LTR. Gene expression regulated by a combination of REV enhancer and SV40 early region promoter was 50-fold less than from the analogous construct containing the chicken syncytial virus promoter. Deletion of LTR sequences immediately downstream of the CAP site, which include a region capable of forming a stable hairpin in the mRNA, decreased expression by 70%. Expression assays and S1 nuclease mapping showed that a second transcriptional start site, identified by transcription in vitro using HeLa cell lysates and purified DNA templates, was not used in vivo in the cell lines examined.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bandyopadhyay P. K., Temin H. M. Expression from an internal AUG codon of herpes simplex thymidine kinase gene inserted in a retrovirus vector. Mol Cell Biol. 1984 Apr;4(4):743–748. doi: 10.1128/mcb.4.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz E. W., Jr, Wydro R. M., Nadal-Ginard B., Dina D. Moloney murine sarcoma proviral DNA is a transcriptional unit. Nature. 1980 Dec 25;288(5792):665–669. doi: 10.1038/288665a0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cook M. K. Cultivation of a filterable agent associated with Marek's disease. J Natl Cancer Inst. 1969 Jul;43(1):203–212. [PubMed] [Google Scholar]
- Cullen B. R., Raymond K., Ju G. Functional analysis of the transcription control region located within the avian retroviral long terminal repeat. Mol Cell Biol. 1985 Mar;5(3):438–447. doi: 10.1128/mcb.5.3.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Casey J. W. Two elements in the bovine leukemia virus long terminal repeat that regulate gene expression. Science. 1986 Mar 21;231(4744):1437–1440. doi: 10.1126/science.3006241. [DOI] [PubMed] [Google Scholar]
- Dougherty J. P., Temin H. M. A promoterless retroviral vector indicates that there are sequences in U3 required for 3' RNA processing. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1197–1201. doi: 10.1073/pnas.84.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embretson J. E., Temin H. M. Transcription from a spleen necrosis virus 5' long terminal repeat is suppressed in mouse cells. J Virol. 1987 Nov;61(11):3454–3462. doi: 10.1128/jvi.61.11.3454-3462.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Mittal S., Chute H., Chao E., Pattengale P. K. Rearrangements and insertions in the Moloney murine leukemia virus long terminal repeat alter biological properties in vivo and in vitro. J Virol. 1986 Oct;60(1):204–214. doi: 10.1128/jvi.60.1.204-214.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. V., Bich-Thuy L. T., Stafford J., Queen C. Synergism between immunoglobulin enhancers and promoters. Nature. 1986 Jul 24;322(6077):383–385. doi: 10.1038/322383a0. [DOI] [PubMed] [Google Scholar]
- Gilmartin G. M., Parsons J. T. Identification of transcriptional elements within the long terminal repeat of Rous sarcoma virus. Mol Cell Biol. 1983 Oct;3(10):1834–1845. doi: 10.1128/mcb.3.10.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin G. H. Identification of three sequence-specific DNA-binding proteins which interact with the Rous sarcoma virus enhancer and upstream promoter elements. J Virol. 1988 Jun;62(6):2186–2190. doi: 10.1128/jvi.62.6.2186-2190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves B. J., Eisenman R. N., McKnight S. L. Delineation of transcriptional control signals within the Moloney murine sarcoma virus long terminal repeat. Mol Cell Biol. 1985 Aug;5(8):1948–1958. doi: 10.1128/mcb.5.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanecak R., Mittal S., Davis B. R., Fan H. Generation of infectious Moloney murine leukemia viruses with deletions in the U3 portion of the long terminal repeat. Mol Cell Biol. 1986 Dec;6(12):4634–4640. doi: 10.1128/mcb.6.12.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isfort R., Witter R. L., Kung H. J. C-myc activation in an unusual retrovirus-induced avian T-lymphoma resembling Marek's disease: proviral insertion 5' of exon one enhances the expression of an intron promoter. Oncogene Res. 1987;2(1):81–94. [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Tsichlis P., Khoury G. Multiple enhancer domains in the 3' terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Res. 1984 Aug 24;12(16):6427–6442. doi: 10.1093/nar/12.16.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciw P. A., Bishop J. M., Varmus H. E., Capecchi M. R. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983 Jul;33(3):705–716. doi: 10.1016/0092-8674(83)90013-2. [DOI] [PubMed] [Google Scholar]
- Manley J. L. Analysis of the expression of genes encoding animal mRNA by in vitro techniques. Prog Nucleic Acid Res Mol Biol. 1983;30:195–244. doi: 10.1016/s0079-6603(08)60687-x. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Noori-Daloii M. R., Swift R. A., Kung H. J., Crittenden L. B., Witter R. L. Specific integration of REV proviruses in avian bursal lymphomas. Nature. 1981 Dec 10;294(5841):574–576. doi: 10.1038/294574a0. [DOI] [PubMed] [Google Scholar]
- Norton P. A., Coffin J. M. Characterization of Rous sarcoma virus sequences essential for viral gene expression. J Virol. 1987 Apr;61(4):1171–1179. doi: 10.1128/jvi.61.4.1171-1179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K., Nakamura M., Saito S., Noda T., Ito Y., Sugamura K., Hinuma Y. Identification of two distinct elements in the long terminal repeat of HTLV-I responsible for maximum gene expression. EMBO J. 1987 Feb;6(2):389–395. doi: 10.1002/j.1460-2075.1987.tb04767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. P., Holbrook N., Levens D. Binding of a cellular protein to the gibbon ape leukemia virus enhancer. Mol Cell Biol. 1987 Aug;7(8):2735–2744. doi: 10.1128/mcb.7.8.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway A. A., Swift R. A., Kung H. J., Fujita D. J. In vitro transcription analysis of the viral promoter involved in c-myc activation in chicken B lymphomas: detection and mapping of two RNA initiation sites within the reticuloendotheliosis virus long terminal repeat. J Virol. 1985 Apr;54(1):161–170. doi: 10.1128/jvi.54.1.161-170.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler H. R., Gruss P. Specific interaction between enhancer-containing molecules and cellular components. Cell. 1984 Feb;36(2):403–411. doi: 10.1016/0092-8674(84)90233-2. [DOI] [PubMed] [Google Scholar]
- Sealey L., Chalkley R. At least two nuclear proteins bind specifically to the Rous sarcoma virus long terminal repeat enhancer. Mol Cell Biol. 1987 Feb;7(2):787–798. doi: 10.1128/mcb.7.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Temin H. M. Spontaneous variation and synthesis in the U3 region of the long terminal repeat of an avian retrovirus. J Virol. 1982 Jan;41(1):163–171. doi: 10.1128/jvi.41.1.163-171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A., Reddy E. P., Dunn C. Y., Aaronson S. A. Molecular dissection of transcriptional control elements within the long terminal repeat of the retrovirus. Science. 1984 Jan 20;223(4633):286–289. doi: 10.1126/science.6322296. [DOI] [PubMed] [Google Scholar]
- Swift R. A., Boerkoel C., Ridgway A., Fujita D. J., Dodgson J. B., Kung H. J. B-lymphoma induction by reticuloendotheliosis virus: characterization of a mutated chicken syncytial virus provirus involved in c-myc activation. J Virol. 1987 Jul;61(7):2084–2090. doi: 10.1128/jvi.61.7.2084-2090.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift R. A., Shaller E., Witter R. L., Kung H. J. Insertional activation of c-myc by reticuloendotheliosis virus in chicken B lymphoma: nonrandom distribution and orientation of the proviruses. J Virol. 1985 Jun;54(3):869–872. doi: 10.1128/jvi.54.3.869-872.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornell A., Hallberg B., Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988 Apr;8(4):1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]