Plant protoplasts are often used as experimental material without paying attention to the tissue they are isolated from (a protoplast is a protoplast), whereas in other cases, they are considered not sufficiently able to reproduce the in planta situation.
Here, we show that protoplasts are a very reliable experimental system, as long as we carefully chose their source.
Transient transformation of protoplasts isolated from an adult tissue has been successfully used for developmental studies (Sheen, 2001), biochemical analysis (Sheahan et al., 2007), to examine the influence of hormones and stress factors (Meyer et al., 1984; Pasternak et al., 2002), to investigate cell wall regeneration (Leucci et al., 2007), to determine the subcellular localization of tagged proteins (Goodman et al., 2004), and to analyze protein interactions (Walter et al., 2004). Protoplasts represent a very convenient system as they can efficiently be transformed with several DNA constructs at the same time to study the colocalization and/or interactions of differently labeled proteins (Walter et al., 2004; Chen et al., 2006), and because they allow better imaging (higher resolution) compared to cells in an intact tissue.
The most used sources of protoplasts are leaf mesophyll (Sheen, 2001) as universal system for transient expression of plant genes (Yoo et al., 2007). They are generally believed to provide information about the cellular function of proteins normally expressed in other cell types. Several collections of markers for endocellular compartments and structures are available (e.g. Geldner et al., 2009) as characterized by their expression in leaf cells (intact tissue or protoplasts). It is assumed that the observations done in protoplasts from this tissue can be extended to (any) intact tissue, but no reports have been published in which this aspect has been studied in detail.
In contrast to animal cells, plant cells can easily change their identity when taken out of their environment, and when cell lineages are disrupted, and the position of cells is altered, they rapidly change identity according to their new position (van den Berg et al., 1995). Protoplasts cultured over a period of weeks can regenerate entire plants, indicating that they undergo dedifferentiation. Therefore, protoplasts are generally considered to lose their identity and to be comparable with cells from suspension cultures, which would make them unsuitable to investigate cell-type or tissue-specific processes. This is possibly a consequence of the little attention paid so far to the biological state of freshly made protoplasts or the kinetics by which cell identity changes. No specific studies define whether tissue specificity is retained within the time frame required for isolation, transformation, and transient expression analysis.
The opinion of most researchers about protoplasts falls in two opposite categories: “any type of protoplasts are fine for me as long as they work (and leaf protoplasts are the easiest)” or “protoplasts are just not reliable, so you should not use them.” Here, we try to convince researchers that both views are wrong and that protoplasts can give highly reliable results, if used in an appropriate way based on a proper understanding of their features.
We recently discovered a new pathway involved in the acidification of the vacuole in epidermal cells and identified via mutants several key components such as the tonoplast H+ P-ATPase PH5 (Verweij et al., 2008b) and a novel tonoplast pump encoded by PH1 (F. Quattrocchio, A. Hoshino, K. Spelt, M. Faraco, W. Verweij, G. Di Sansebastiano, and R. Koes, unpublished data). As we noted that these and other tonoplast proteins move in distinct cell types via distinct pathways to the tonoplast, we developed a protocol to efficiently produce and transiently transform protoplasts from petunia (Petunia hybrida) petals and compared their features with those of the widely used leaf mesophyll protoplasts and of cells in intact tissue.
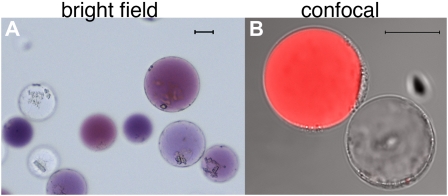
The limb of petunia petals is rather thin and consists of an upper (adaxial) and lower (abaxial) epidermis (colored by anthocyanin pigments) and several layers of mesophyll cells that lack anthocyanins (Koes et al., 1990). The anthocyanins provide a convenient marker to distinguish protoplasts originating from the petal epidermis from those derived from the mesophyll (Fig. 1).
Figure 1.
Protoplasts freshly isolated from petals of the petunia hybrid M1XV30. A, Bright-field image. The two cell types are recognizable by the presence of anthocyanins in the central vacuole of epidermal cells and their absence in the mesophyll cells. B, Confocal image of the same protoplast preparation as in A. The red fluorescence, due to anthocyanin autofluorescence, allows the recognition of the two cell types (both visible in transmitted light) during confocal analysis. The size bar equals 20 μm. Protoplast isolation: Petals or leaves from greenhouse-grown plants were sterilized in 5% hypochlorite solution (for 30 s then rinsed in sterile water) and perforated using a needle bed (a “kenzan” for Japanese ikebana), prior overnight digestion in TEX buffer (B5 salts, 500 mg/L MES, 750 mg/L CaCl2 [2*H2O] 250 mg/L NH4NO3, and 0.4 m Suc [13.7%], pH 5.7), plus 0.2% Macerozyme R10 and 0.4% Cellulase R10 (Yakult). The digested material was filtered through a 150-SIGMA mesh filter (or similar filter) and protoplast suspension was then centrifuged for 10 min at 75g at room temperature in a swing-out rotor to concentrate the protoplasts in a band floating above the medium. After 2× washing with 10 mL of TEX buffer (centrifugation at 75g for 5 min between washing steps) protoplasts were then resuspended in an appropriate volume of MMM solution (0.5 m mannitol, 15 mm MgCl2, 0.1% MES). A total of 300 μL of protoplasts was used for each transformation: 30 μg of (supercoiled) plasmid DNA was added followed by 300 μL of polyethylene glycol solution [0.4 m mannitol, 0.1 m Ca(NO3)2*H2O, 40% polyethylene glycol brought to pH 8.0 with KOH] and 2 mL of TEX. Incubation at 25°C for 2 h was followed by washing with TEX buffer, as described before, and resuspended, after centrifugation, in 2 mL of TEX buffer. We have applied this protocol to flowers of different ages (from nearly open buds to fully expanded petals) and genotypes (different genetic backgrounds and/or mutations in genes affecting pigment deposition and/or vacuolar acidification). Transformation efficiency was in all cases above 60%. Plants were grown in a greenhouse with temperature never below 19°C and never exceeding 30°C, with a cycle of a minimum of 16 h of light in all seasons (supplied with artificial light in the winter). Suboptimal or unstable plant growth conditions can make efficiency of protoplast isolation and transformation drop dramatically.
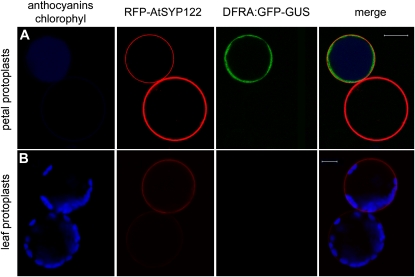
We selected the promoter of the DFRa gene from petunia to determine whether protoplasts isolated from petals retain their tissue-specific promoter activity (within the time course of a transient expression experiment). DFRa encodes dihydroflavonol 4-reductase (DFR), which catalyzes the first anthocyanin-specific reaction in the flavonoid pathway (Koes et al., 2005). DFRa is transcribed in petals, anthers, and in the seed coat but not in unpigmented tissues such as leaves (Huits et al., 1994). In situ hybridization of thin-petal sections has shown that DFRa mRNA is expressed in the epidermis only and is absent from the mesophyll (Quattrocchio et al., 2006). We fused a 2-kb promoter fragment of DFRA to a GFP-GUS reporter. This promoter fragment has been shown to contain all the cis-regulatory elements necessary to drive expression in all pigmented tissues, to rescue a dfra mutant anthocyanin6, and to respond to the transcription regulators that control anthocyanin biosynthesis (Huits et al., 1994; Quattrocchio et al., 1998; Spelt et al., 2000). The DFRa:GFP-GUS construct was transformed in protoplasts together with the 35S:RFP-AtSYP122 construct where the constitutive 35S promoter drives the expression of a translational fusion of RFP and the SNARE (soluble N-ethylmaleimide-sensitive-factor attachment protein receptor) protein SYP122, previously used to label the plasma membrane (Rehman et al., 2008).
By confocal microscopy, we observed that DFRa:GFP-GUS (green fluorescence) was expressed only in the (purple) protoplasts derived from the epidermis (800 cells observed), whereas 35S:RFP-SYP122 (red fluorescence) was expressed in both cell types (Fig. 2A; Supplemental Fig. S1). All colored cells expressing DFRa:GFP-GUS also expressed 35S:RFP-AtSYP122F, showing that these protoplasts were cotransformed at very high efficiency by two distinct constructs. In contrast, leaf mesophyll derived protoplasts transformed with the same constructs did not show any DFRa:GFP expression, even though they efficiently expressed 35S:RFP-AtSYP122 (Fig. 2B; Supplemental Fig. S1). This specific experiment was repeated three times with identical results in independently isolated protoplast populations from petals of the wild-type hybrid line M1XV30. In all these experiments transformation efficiency was above 60%. These results show that protoplasts retain their gene expression program and that within the time frame of the experiment (48 h), no signs of dedifferentiation or loss of cell identity are detectable.
Figure 2.
Transient expression of DFRA-GFP and 35S-RFP-AtSYP122 in petunia petal and leaf protoplasts. A, Petal protoplasts express the 35S:RFP-AtSYP122 marker of the plasma membrane (driven by the ectopic CaMV35S promoter) in both cells accumulating anthocyanins and unpigmented cells (anthocyanins in blue color to distinguish them from the RFP signal). The GFP signal (driven by the DFRa promoter) is only visible in cells accumulating anthocyanins. B, Petunia leaf protoplasts (the blue color now evidences chloroplasts) express the CaMV35S promoter driven RFP-AtSYP122, but the DFRA:GFP construct is not expressed in these cells. Images were acquired with a Zeiss confocal laser microscope (LSM Pascal). Fluorescence was detected using a 488-/543-nm dichroic beam splitter, a 505- to 530-nm band pass filter for GFP, and a 560- to 615-nm band pass filter for RFP; chlorophyll and anthocyanins epifluorescence was detected with the filter set for trimethylrhodamine isothiocyanate (>650 nm). The size bar equals 20 μm.
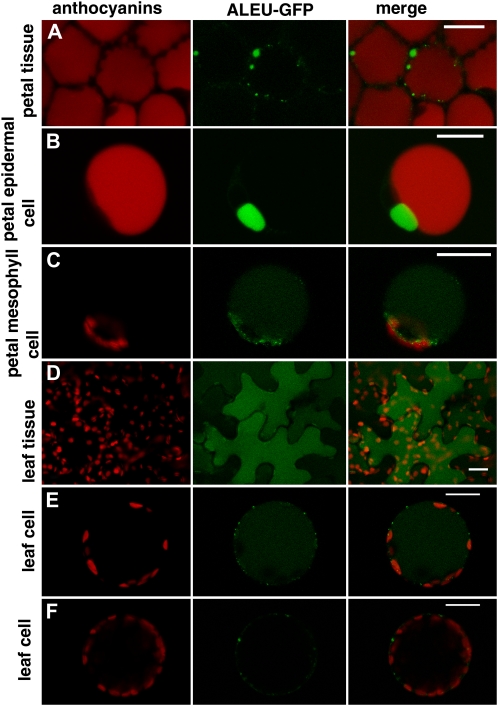
To test whether the use of protoplasts from different tissues could result in different protein localization, we have chosen to transiently express a chimeric protein consisting of GFP and the N-terminal sorting sequence of aleurain (ALEU-GFP; Di Sansebastiano et al., 2001). This marker was shown to be sorted to the central vacuole of root and leaf cells (Fluckiger et al., 2003). We examined the localization of ALEU-GFP in leaf and petal tissue after agroinfiltration (for experimental procedure see, Verweij et al., 2008a) and compared that to the localization in protoplasts derived from the same tissues after transient transformation.
Agroinfection of intact petals results in ALEU-GFP accumulation in epidermal cells in the lumen of small vacuole-like structures that are distinct from the large central vacuole containing the anthocyanins (Fig. 3A; Verweij et al., 2008b). In transiently transformed protoplasts that originate from the petal epidermis ALEU-GFP accumulated in similar vacuolar structures (Fig. 3B), whereas in protoplasts derived from the mesophyll it was targeted to the lumen of the large central vacuole and to small dots in the cytoplasm (probably prevacuolar compartments). We observed the small vacuole-like ALEU-GFP-labeled compartments in 100% of the transformed protoplasts originating from petal epidermis of wild-type lines in three different genetic backgrounds (R27, M1XV30, V23XV30) in 32 independent transformation experiments. These compartments, characteristic of the endomembrane organization in petal epidermal cells (F. Quattrocchio, K. Spelt, M. Faraco, G. Di Sansebastiano, and R. Koes, unpublished data), are not induced by the marker expression, as freshly prepared protoplasts contain transparent bodies clearly separated from the anthocyanin-rich central vacuole (data not shown).
Figure 3.
Transient expression of 35S:ALEU-GFP in petal and leaf protoplasts and in intact tissues. A, Accumulation of ALEU:GFP in petals of petunia flowers 24 h after agroinfiltration. Red autofluorescence of anthocyanins is visible in all cells and the GFP signal accumulates in small compartments independent from the central vacuole. B, Petal epidermal protoplast (recognizable from the presence of red fluorescent anthocyanins in the central vacuole) that accumulate ALEU-GFP in a small compartment independent from the central vacuole. C, Petal mesophyll protoplast (no anthocyanins in the central vacuole) accumulating ALEU-GFP in the central vacuole and in (prevacuolar) small compartments. D, Accumulation of ALEU-GFP in leaf epidermal cells 24 h after infiltration. The GFP signal is present in the central vacuole. E, Leaf protoplast (probably originating from the leaf epidermis) accumulating ALEU-GFP in the central vacuole. F, A leaf protoplast (probably originating from mesophyll) showing a different pattern of accumulation of ALEU-GFP. The size bar equals 20 μm.
Agroinfiltrated leaf epidermal cells accumulate ALEU-GFP in the central vacuole (Fig. 3D), whereas in protoplasts from leaves, we observed two different patterns of accumulation of ALEU-GFP. Some protoplasts show fluorescence in the central vacuole (Fig. 3E), like agroinfiltrated epidermal leaf cells, whereas other protoplasts in the same population show ALEU-GFP accumulation in small prevacuolar compartments. Possibly these cells originate from a different layer in the leaf—probably subepidermal as they have a higher concentration of chloroplasts—and represent a different cell type.
These results show that the intracellular localization of specific proteins is tissue- and cell type dependent.
DISCUSSION
It is widely believed that protoplasts are dedifferentiated cells and therefore the tissue from which they are isolated is of minor importance (as the protoplasts will lose their original identity anyhow). As leaves are an abundantly available tissue from which protoplasts are easily isolated, mesophyll protoplasts from Arabidopsis (Arabidopsis thaliana) or tobacco (Nicotiana tabacum) have been extensively used to determine the subcellular localization of proteins and the activity of genes normally expressed in other tissues (Sheen, 2001; Yoo et al., 2007).
Here, we show that protoplasts isolated from different tissues do display major differences with regard to promoter activity and protein sorting. In particular, the DFRa promoter is only active in transiently transformed protoplasts originating from the petal epidermis, but not in protoplasts originating from the petal mesophyll or the various cell types in leaves. This mirrors the expression pattern of DFRa in the intact plant (Huits et al., 1994; Quattrocchio et al., 2006). Furthermore, ALEU-GFP is targeted to different intracellular domains in protoplasts of different origin, which accurately reflect the targeting of this protein in intact tissues. While studying proteins involved in the acidification of the vacuole in epidermal petal cells, such as the tonoplast pumps PH5 (Verweij et al., 2008b) and PH1 (F. Quattrocchio, A. Hoshino, K. Spelt, M. Faraco, W. Verweij, G. Di Sansebastiano, and R. Koes, unpublished data) and vacuolar SNAREs, we again observed that their trafficking is highly cell specific, both in transiently transformed protoplasts and in intact tissues. These data all indicate that protoplasts derived from distinct tissues do retain their tissue- and cell-specific features within the time frame of a transient expression assay.
In addition, we show that a simple organ like a petal yields a heterogeneous population of protoplasts with different gene expression and protein-sorting features. The specific gene expression and protein-sorting features in petal epidermal protoplasts mirror those of epidermal cells in the intact flower, but are entirely different from those observed in petal mesophyll protoplasts. Therefore, the advantages of protoplasts, such a high-resolution imaging and the ease of manipulation by exogenous application or injection of chemicals, can be exploited to study highly tissue- or cell-type-specific processes, like promoter activation, protein sorting, or vesicle trafficking.
Given that leaves are even more complex than petals and consist of several different cell types, we assume that leaf protoplasts are at least as heterogeneous, which may account for the finding that ALEU-GFP accumulates in at least two distinct patterns in different protoplasts. It was indeed previously shown that different cell types can be recognized in protoplasts preparations from leaves as they display distinct distributions of membrane markers (Di Sansebastiano et al., 2001). This implies that one should interpret results obtained with such heterogeneous cell populations with caution. In this light it is important to realize that the collections of fluorescent markers for different subcellular compartments that recently became available (Nelson et al., 2007; Geldner et al., 2009) have been characterized in mesophyll cells and that (some of) these markers may label different compartments in distinct cell types.
Petal protoplasts represent a fortunate case as cells from the epidermis and the mesophyll can be easily distinguished by their color. In other tissues where such natural markers for distinct cells are not available, they can be easily introduced, for example by using transgenes that are expressed in specific cell types. Protoplasts can be prepared from tissues of a transgenic plant expressing an appropriate marker gene, for example a fluorescent protein expressed from a cell-type-specific promoter. Given the very high frequency of cotransformation, an equally reliable and even easier and more versatile approach, which is compatible with high-throughput screening programs, is to isolate protoplasts from wild-type plants and to cointroduce such a cell-specific marker gene together with the gene constructs that are being studied. For example, a cointroduced DFRa:GFP gene is an equally good marker as the anthocyanins to identify (transformed) protoplasts derived from the epidermis.
The protocol for the isolation and transformation of petal protoplasts presented here (see Fig. 1 legend) is derived from existing protocols used for leaf mesophyll protoplasts with minor modifications. Thus, it is likely that the same procedure may be used to isolate and transform protoplasts from a range of other tissues to provide a convenient, fast, and reliable tool for the analysis of a variety of (cell-specific) biological processes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Activity of the DFRa promoter and of the 35S promoter in different cell types within one petal protoplast population.
Supplementary Material
References
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Di Sansebastiano GP, Paris N, Marc-Martin S, Neuhaus JM. (2001) Regeneration of a lytic central vacuole and of neutral peripheral vacuoles can be visualized by green fluorescent proteins targeted to either type of vacuoles. Plant Physiol 126: 78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckiger R, De Caroli M, Piro G, Dalessandro G, Neuhaus JM, Di Sansebastiano GP. (2003) Vacuolar system distribution in Arabidopsis tissues, visualized using GFP fusion proteins. J Exp Bot 54: 1577–1584 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof Y-D, Chory J. (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, Casati P, Walbot V. (2004) A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16: 1812–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huits HSM, Gerats AGM, Kreike MM, Mol JNM, Koes RE. (1994) Genetic control of dihydroflavonol 4-reductase gene expression in Petunia hybrida. Plant J 6: 295–310 [DOI] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Koes RE, Van Blokland R, Quattrocchio F, Van Tunen AJ, Mol JNM. (1990) Chalcone synthase promoters in petunia are active in pigmented and unpigmented cell types. Plant Cell 2: 379–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci MR, Di Sansebastiano GP, Gigante M, Dalessandro G, Piro G. (2007) Secretion marker proteins and cell-wall polysaccharides move through different secretory pathways. Planta 225: 1001–1017 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Aspart L, Chartier Y. (1984) Auxin-induced regulation of protein synthesis in tobacco mesophyll protoplasts cultivated in vitro: II. Time course and level of auxin control. Plant Physiol 75: 1034–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Pasternak TP, Prinsen E, Ayaydin F, Miskolczi P, Potters G, Asard H, Van Onckelen HA, Dudits D, Fehér A. (2002) The role of auxin, pH, and stress in the activation of embryogenic cell division in leaf protoplast-derived cells of alfalfa. Plant Physiol 129: 1807–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Verweij W, Kroon A, Spelt C, Mol J, Koes R. (2006) PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18: 1274–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing JF, van der Woude K, Mol JNM, Koes R. (1998) Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13: 475–488 [DOI] [PubMed] [Google Scholar]
- Rehman RU, Stigliano E, Lycett GW, Sticher L, Sbano F, Faraco M, Dalessandro G, Di Sansebastiano GP. (2008) Tomato Rab11a characterization evidenced a difference between SYP121-dependent and SYP122-dependent exocytosis. Plant Cell Physiol 49: 751–766 [DOI] [PubMed] [Google Scholar]
- Sheahan MB, Rose RJ, McCurdy DW. (2007) Actin-filament-dependent remodeling of the vacuole in cultured mesophyll protoplasts. Protoplasma 230: 141–152 [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol JN, Koes R. (2000) anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell 12: 1619–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. (1995) Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65 [DOI] [PubMed] [Google Scholar]
- Verweij W, Di Sansebastiano G, Quattrocchio F, Dalessandro G. (2008a) Advanced microscopy techniques as instruments for cell and tissue analysis in plants: Agrobacterium mediated transient expression of vacuolar GFPs in petunia leaves and petals. Plant Biosyst 142: 1–5 [Google Scholar]
- Verweij W, Spelt C, Di Sansebastiano GP, Vermeer J, Reale L, Ferranti F, Koes R, Quattrocchio F. (2008b) A novel type of tonoplast localized H+-ATPase is required for vacuolar acidification and coloration of flowers and seeds. Nat Cell Biol 10: 1456–1462 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.